Effect of Pholiota nameko Polysaccharides Inhibiting Methylglyoxal-Induced Glycation Damage In Vitro
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Sample Preparation
2.3. MG–BSA Glycation Model
2.3.1. Inhibition Effects on the Formation of Amadori Products
2.3.2. MG-Trapping
2.3.3. Inhibition Effects on the Formation of Carbonyl Groups
2.3.4. Inhibition Effects on the Decrease in ε-NH2 Group
2.3.5. Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis
2.3.6. Fluorescent AGE Analysis
2.4. Cell Culture
2.5. Cell Viability
2.6. MG-Induced Cell Oxidative Damage
2.6.1. Determination of Protective Ability of PNPs against MG-Induced Cell Damage
2.6.2. MG-Induced Cell Growth Rate Assay
2.6.3. ROS Generation
2.7. Statistical Analysis
3. Results
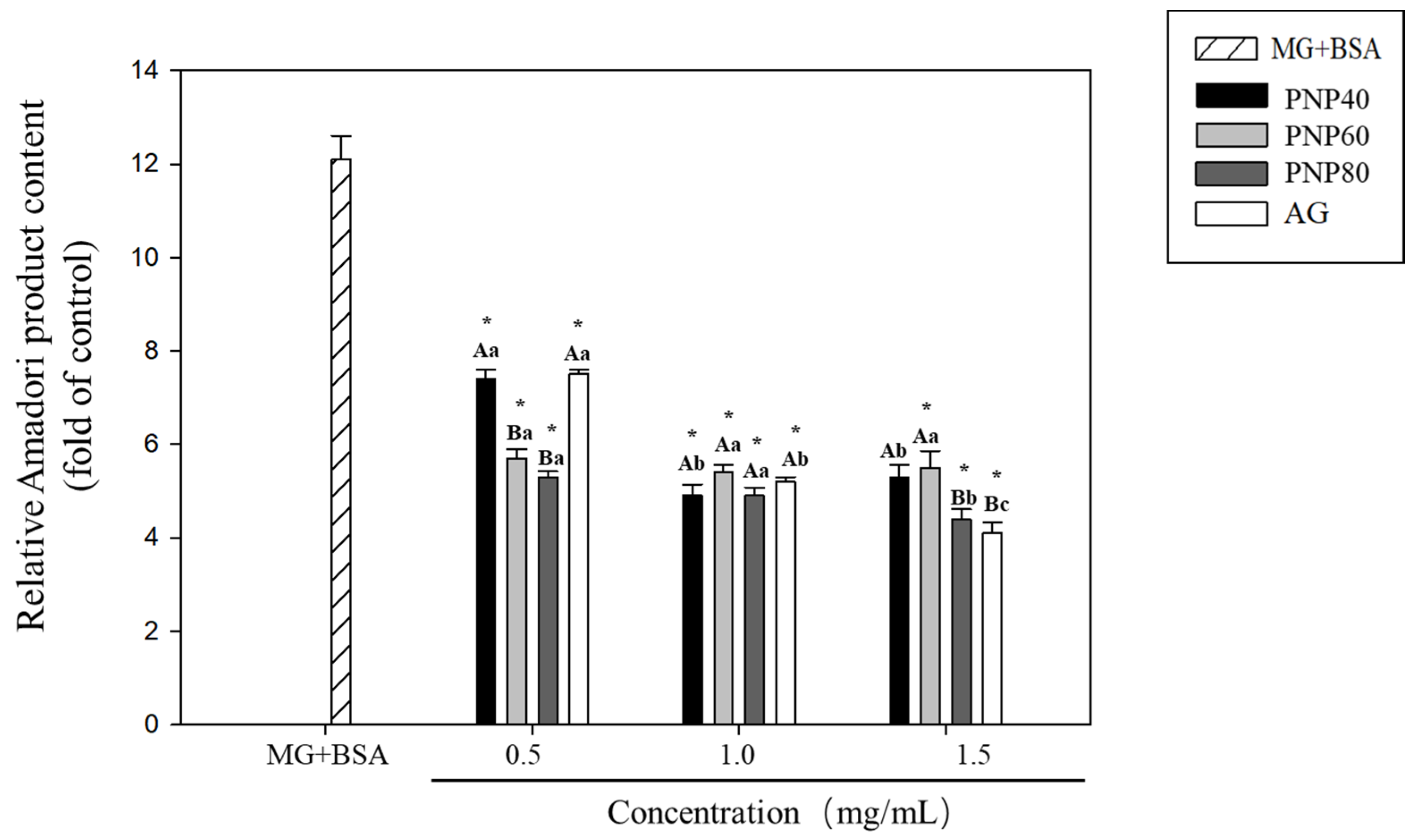
3.1. Inhibition Effects of PNPs on the Formation of Amadori Products
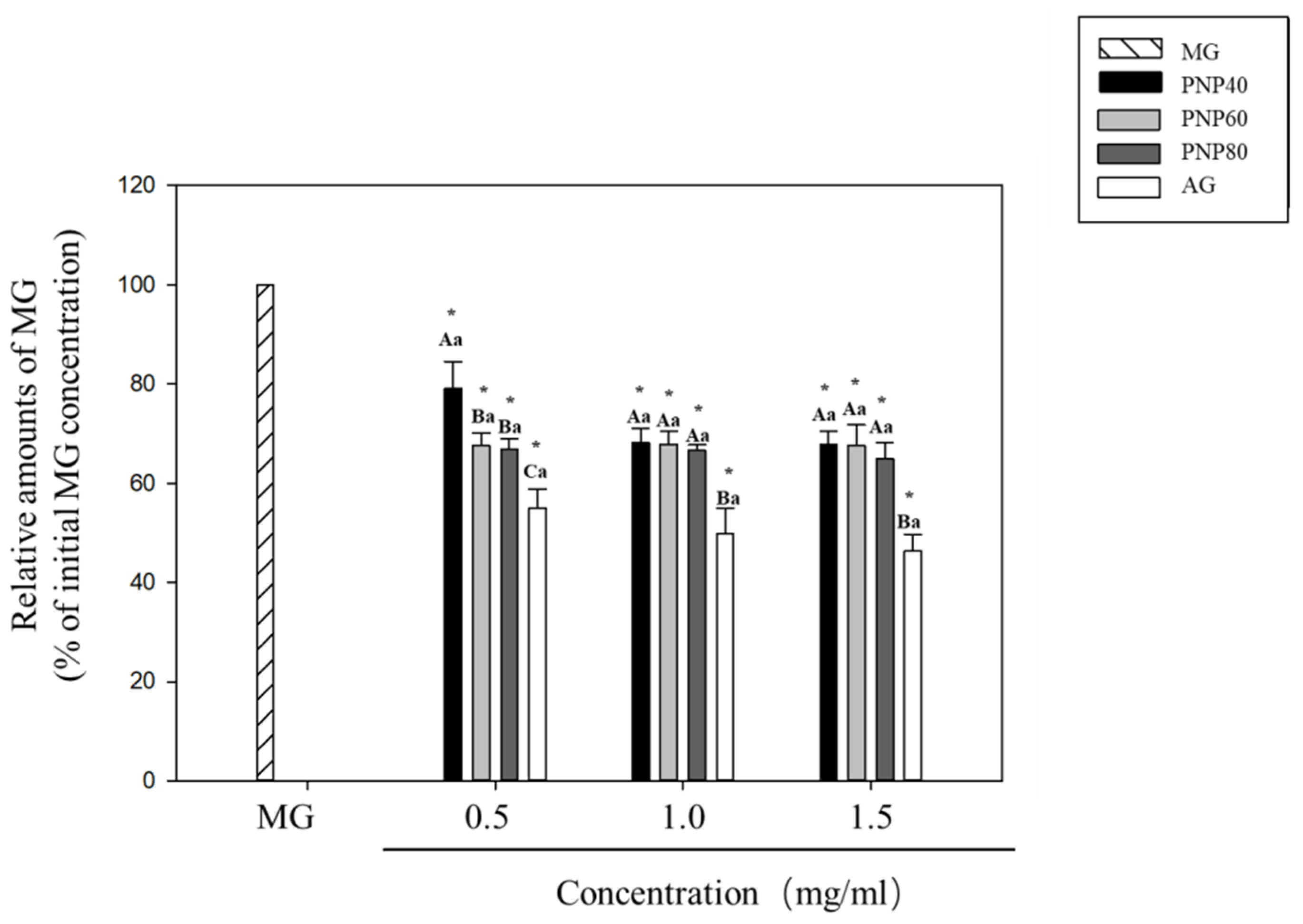
3.2. MG-Trapping Capacity of PNPs
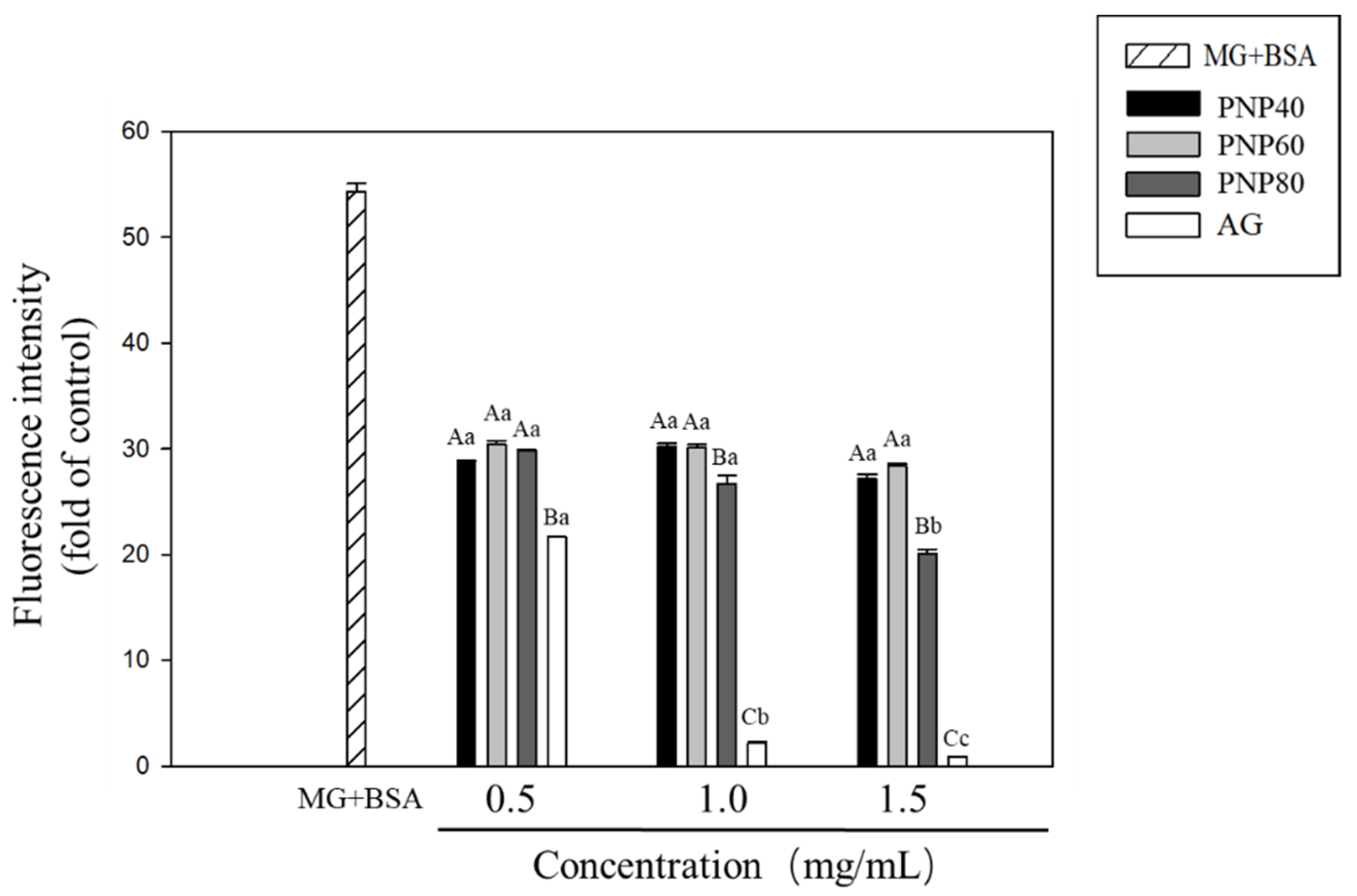
3.3. Inhibition Effects of PNPs on the Formation of the AGEs
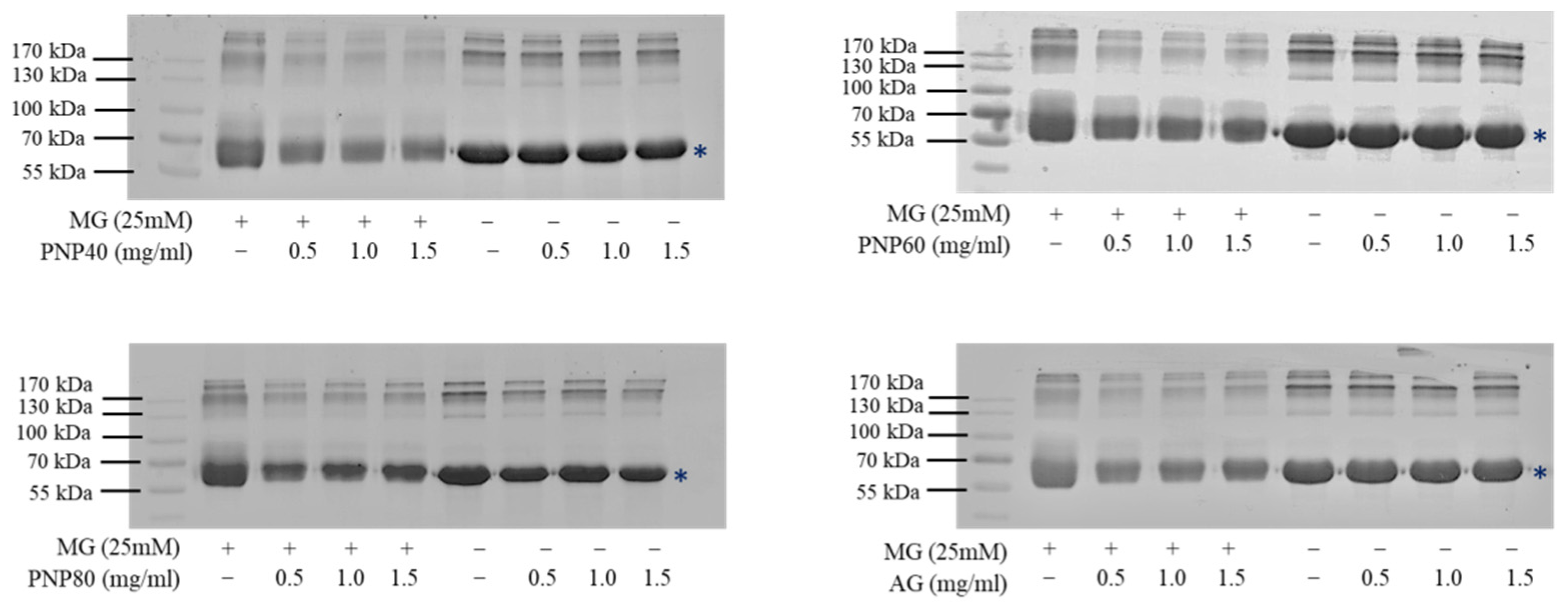
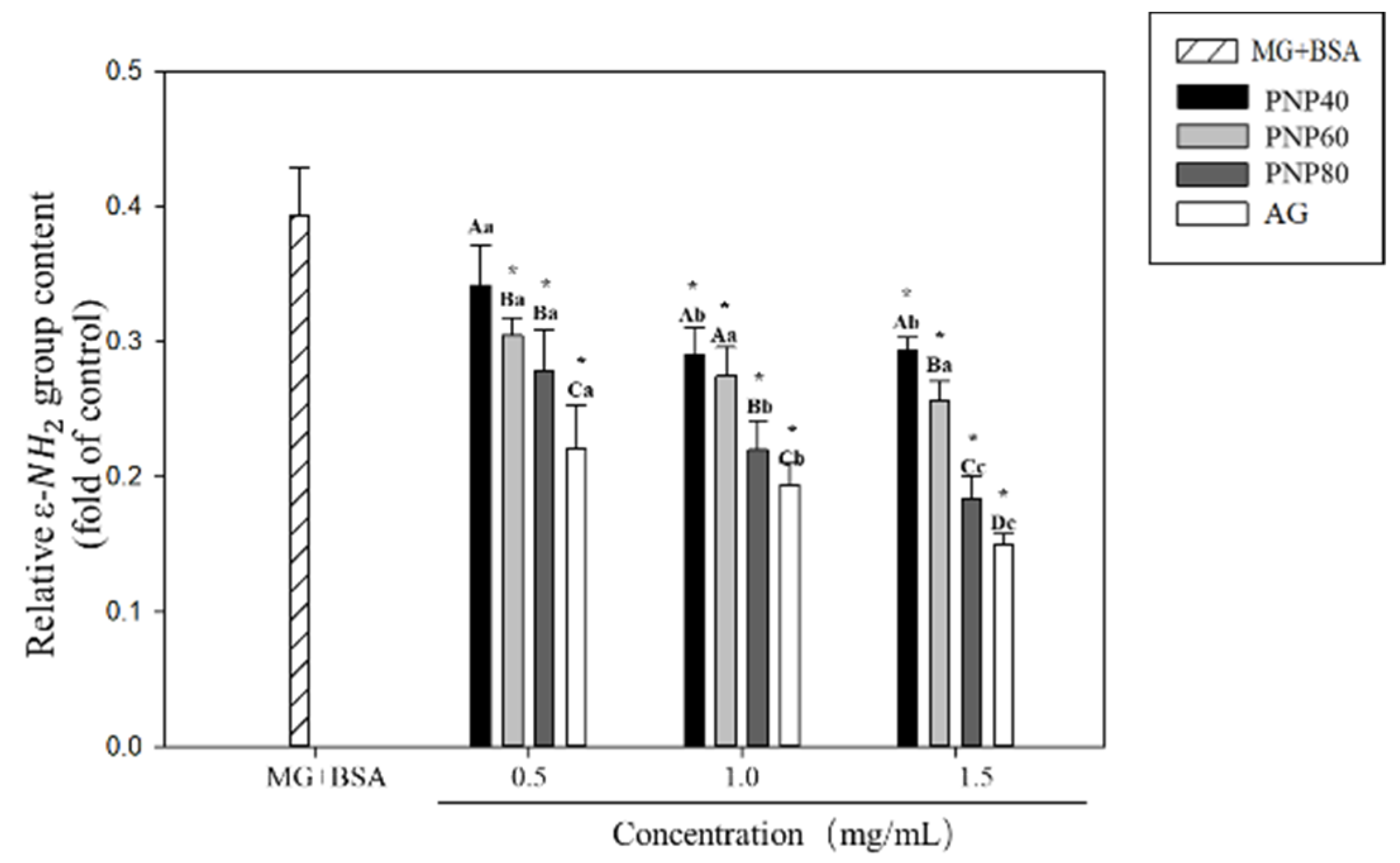
3.4. Inhibition Effects on the Formation of the Carbonyl Group and Decrease in the ε-NH2 Group
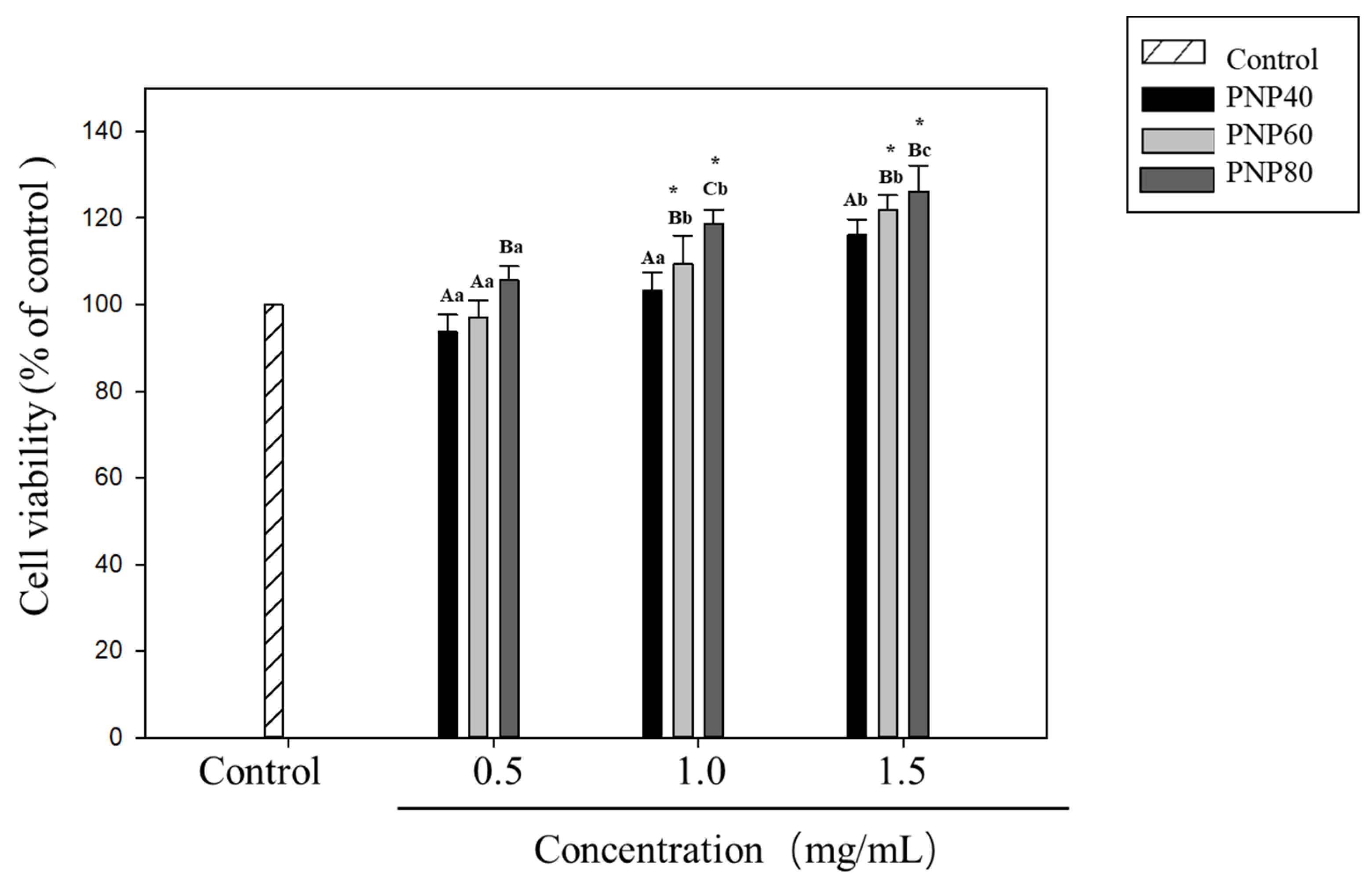
3.5. Effect of PNPs on Hs68 Cell Viability
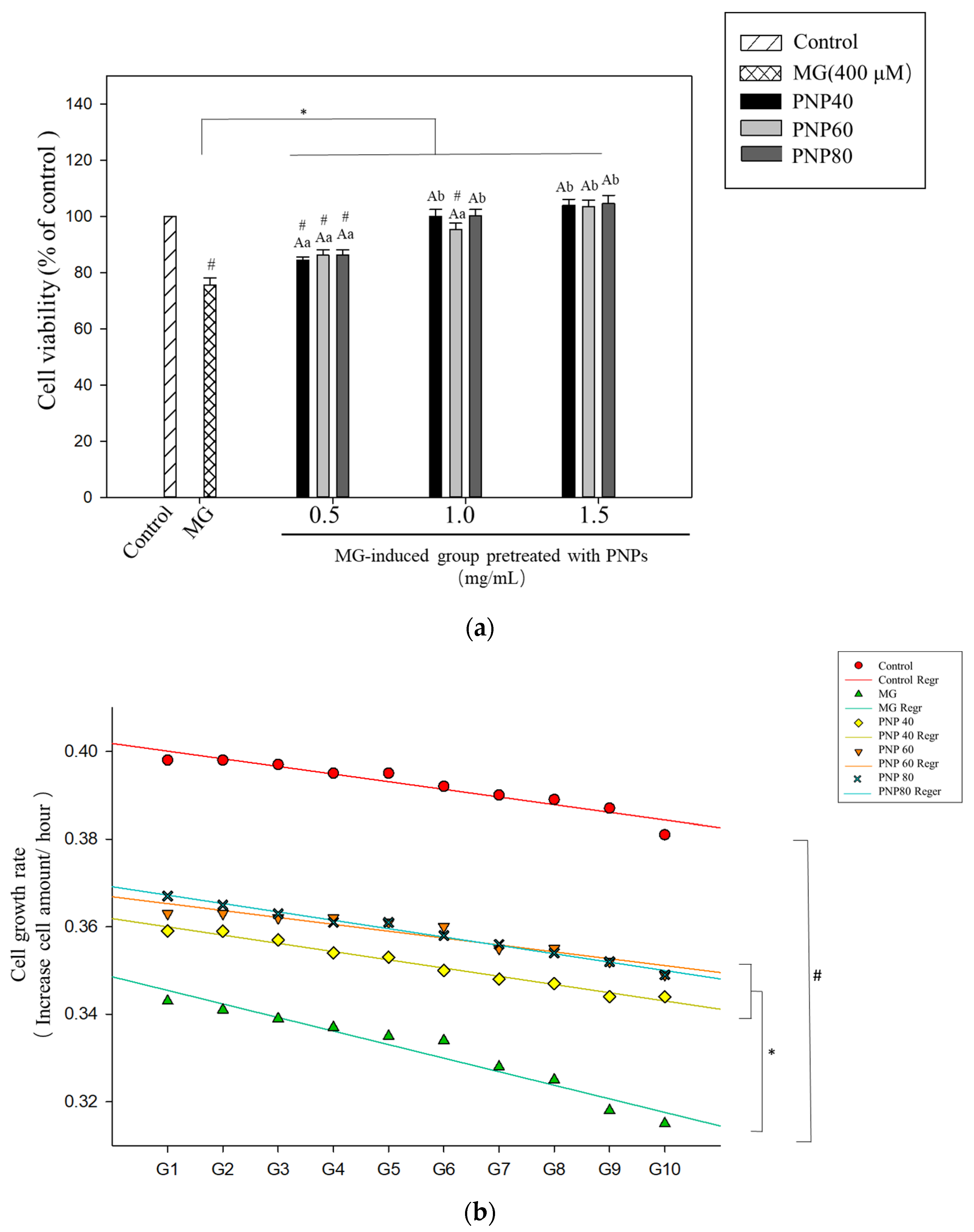
3.6. Effects of PNPs on the Viability of MG-Induced Hs68 Cells
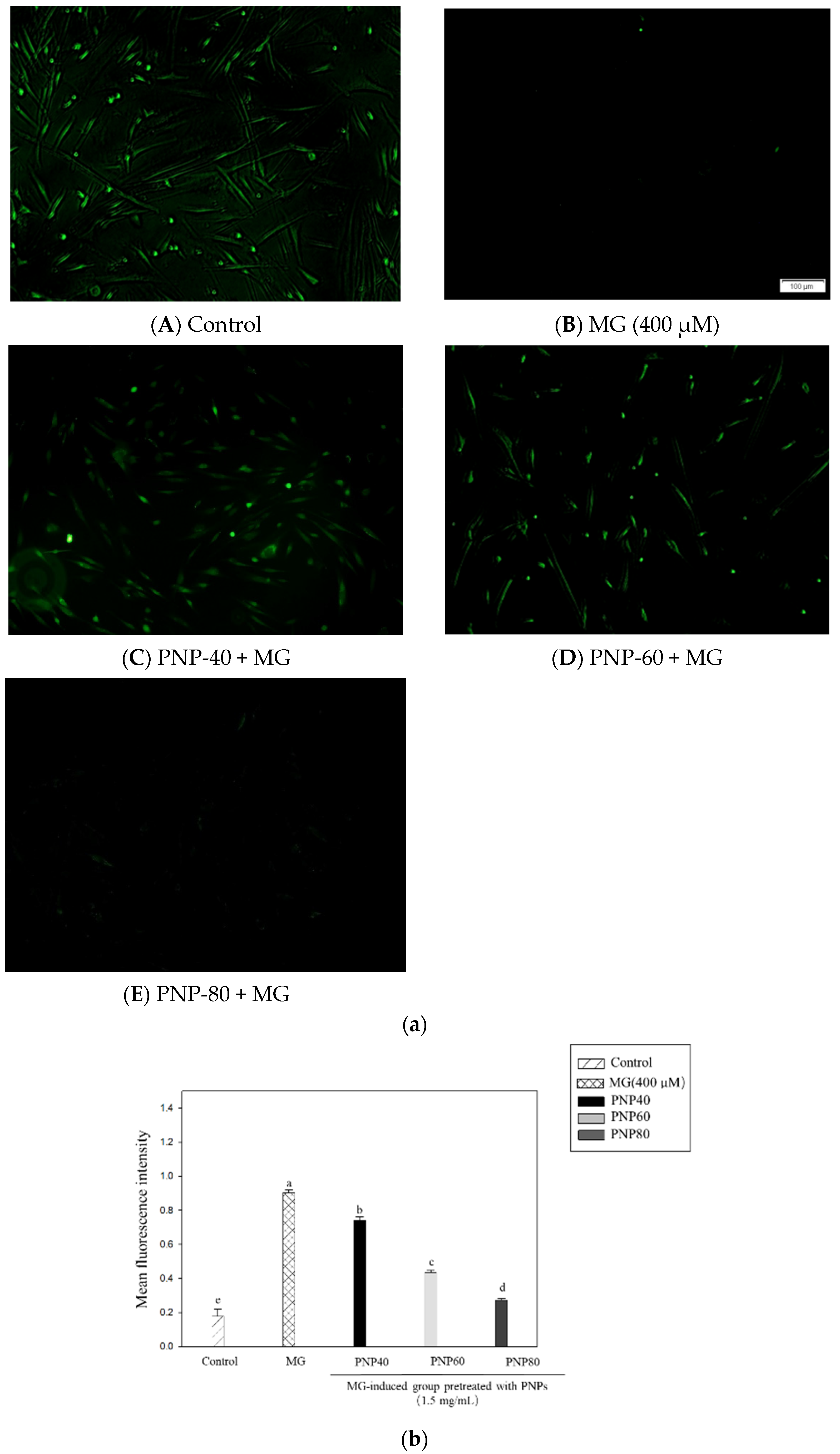
3.7. Effect of PNPs on MG-Induced ROS Production in Hs68 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cheng, A.-S.; Cheng, Y.-H.; Chiou, C.-H.; Chang, T.-L. Resveratrol upregulates Nrf2 expression to attenuate methylglyoxal-induced insulin resistance in Hep G2 cells. J. Agric. Food Chem. 2012, 60, 9180–9187. [Google Scholar] [CrossRef]
- Lin, J.A.; Wu, C.H.; Lu, C.C.; Hsia, S.M.; Yen, G.C. Glycative stress from advanced glycation end products (AGEs) and dicarbonyls: An emerging biological factor in cancer onset and progression. Mol. Nutr. Food Res. 2016, 60, 1850–1864. [Google Scholar] [CrossRef] [PubMed]
- Cianfruglia, L.; Perrelli, A.; Fornelli, C.; Magini, A.; Gorbi, S.; Salzano, A.M.; Antognelli, C.; Retta, F.; Benedetti, V.; Cassoni, P.; et al. KRIT1 Loss-Of-Function Associated with Cerebral Cavernous Malformation Disease Leads to Enhanced S-Glutathionylation of Distinct Structural and Regulatory Proteins. Antioxidants 2019, 8, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antognelli, C.; Perrelli, A.; Armeni, T.; Nicola Talesa, V.; Retta, S.F. Dicarbonyl Stress and S-Glutathionylation in Cerebrovascular Diseases: A Focus on Cerebral Cavernous Malformations. Antioxidants 2020, 9, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antognelli, C.; Trapani, E.; Delle Monache, S.; Perrelli, A.; Daga, M.; Pizzimenti, S.; Barrera, G.; Cassoni, P.; Angelucci, A.; Trabalzini, L.; et al. KRIT1 loss-of-function induces a chronic Nrf2-mediated adaptive homeostasis that sensitizes cells to oxidative stress: Implication for Cerebral Cavernous Malformation disease. Free Radic. Biol. Med. 2018, 115, 202–218. [Google Scholar] [CrossRef]
- Antognelli, C.; Trapani, E.; Delle Monache, S.; Perrelli, A.; Fornelli, C.; Retta, F.; Cassoni, P.; Talesa, V.N.; Retta, S.F. Data in support of sustained upregulation of adaptive redox homeostasis mechanisms caused by KRIT1 loss-of-function. Data Brief 2018, 16, 929–938. [Google Scholar] [CrossRef]
- Rinaldi, C.; Bramanti, P.; Famà, A.; Scimone, C.; Donato, L.; Antognelli, C.; Alafaci, C.; Tomasello, F.; D’Angelo, R.; Sidoti, A. Glyoxalase I A111E, Paraoxonase 1 Q192R and L55M Polymorphisms in Italian Patients with Sporadic Cerebral Cavernous Malformations: A Pilot Study. J. Biol. Regul. Homeost Agents 2015, 29, 493–500. [Google Scholar]
- Antognelli, C.; Mandarano, M.; Prosperi, E.; Sidoni, A.; Talesa, V.N. Glyoxalase-1-Dependent Methylglyoxal Depletion Sustains PD-L1 Expression in Metastatic Prostate Cancer Cells: A Novel Mechanism in Cancer Immunosurveillance Escape and a Potential Novel Target to Overcome PD-L1 Blockade Resistance. Cancers 2021, 13, 2965. [Google Scholar] [CrossRef]
- Semba, R.D.; Nicklett, E.J.; Ferrucci, L. Does accumulation of advanced glycation end products contribute to the aging phenotype? J. Gerontol. Ser. A Biomed. Sci. Med. Sci. 2010, 65, 963–975. [Google Scholar] [CrossRef] [Green Version]
- Lai-Cheong, J.E.; McGrath, J.A. Structure and function of skin, hair and nails. Medicine 2017, 45, 347–351. [Google Scholar] [CrossRef]
- Merecz-Sadowska, A.; Sitarek, P.; Kucharska, E.; Kowalczyk, T.; Zajdel, K.; Cegliński, T.; Zajdel, R. Antioxidant Properties of Plant-Derived Phenolic Compounds and Their Effect on Skin Fibroblast Cells. Antioxidants 2021, 10, 726. [Google Scholar] [CrossRef]
- Yagi, M.; Yonei, Y. Glycative stress and anti-aging: 7. Glycative stress and skin aging. Glycative Stress Res. 2018, 5, 50–54. [Google Scholar]
- Chou, C.-H.; Sung, T.-J.; Hu, Y.-N.; Lu, H.-Y.; Yang, L.-C.; Cheng, K.-C.; Lai, P.-S.; Hsieh, C.-W. Chemical analysis, moisture-preserving, and antioxidant activities of polysaccharides from Pholiota nameko by fractional precipitation. Int. J. Biol. Macromol. 2019, 131, 1021–1031. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.; Freitas, A.C.; Sousa, S.; Amorim, M.; Vasconcelos, M.W.; da Costa, J.P.; Silva, A.M.; Rocha-Santos, T.A.; Duarte, A.C.; Gomes, A.M. Chemical and structural characterization of Pholiota nameko extracts with biological properties. Food Chem. 2017, 216, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Liu, M.; Zhai, G.Y.; Ma, Z.; Wang, L.Q.; Jia, L. Antioxidant and anti-ageing activities of mycelia zinc polysaccharide from Pholiota nameko SW-03. J. Sci. Food Agric. 2015, 95, 3117–3126. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lu, X.; Zhang, S.; Lu, M.; Liu, H. Anti-inflammatory activity of polysaccharide from Pholiota nameko. Biochemistry 2008, 73, 669–675. [Google Scholar] [CrossRef]
- Li, H.; Zhang, M.; Ma, G. Hypolipidemic effect of the polysaccharide from Pholiota nameko. Nutrition 2010, 26, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Sung, T.-J.; Wang, Y.-Y.; Liu, K.-L.; Chou, C.-H.; Lai, P.-S.; Hsieh, C.-W. Pholiota nameko Polysaccharides Promotes Cell Proliferation and Migration and Reduces ROS Content in H2O2-Induced L929 Cells. Antioxidants 2020, 9, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Q.; Liu, J.; Dong, L.; Wang, X.; Zhang, X. Novel advances in inhibiting advanced glycation end product formation using natural compounds. Biomed. Pharmacother. 2021, 140, 111750. [Google Scholar] [CrossRef]
- Mohammed, A.S.A.; Naveed, M.; Jost, N. Polysaccharides; Classification, Chemical Properties, and Future Perspective Applications in Fields of Pharmacology and Biological Medicine (A Review of Current Applications and Upcoming Potentialities). J. Polym. Environ. 2021, 29, 2359–2371. [Google Scholar] [CrossRef]
- Fang, M.; Zou, T.; Yang, X.; Zhang, Z.; Cao, P.; Han, J.; Duan, Y.; Ruan, B.-F.; Li, Q.-S. Discovery of Novel Pterostilbene Derivatives That Might Treat Sepsis by Attenuating Oxidative Stress and Inflammation through Modulation of MAPKs/NF-κB Signaling Pathways. Antioxidants 2021, 10, 1333. [Google Scholar] [CrossRef]
- Antognelli, C.; Moretti, S.; Frosini, R.; Puxeddu, E.; Sidoni, A.; Talesa, V.N. Methylglyoxal Acts as a Tumor-Promoting Factor in Anaplastic Thyroid Cancer. Cells 2019, 8, 547. [Google Scholar] [CrossRef] [Green Version]
- Delle Monache, S.; Pulcini, F.; Frosini, R.; Mattei, V.; Talesa, V.N.; Antognelli, C. Methylglyoxal-Dependent Glycative Stress Is Prevented by the Natural Antioxidant Oleuropein in Human Dental Pulp Stem Cells through Nrf2/Glo1 Pathway. Antioxidants 2021, 10, 716. [Google Scholar] [CrossRef] [PubMed]
- Liping, S.; Xuejiao, S.; Yongliang, Z. Preparation, characterization and antiglycation activities of the novel polysaccharides from Boletus snicus. Int. J. Biol. Macromol. 2016, 92, 607–614. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, G.; Yu, Z.; Song, X.; Li, X.; Yang, Y.; Wang, L.; Liu, L.; Dai, J. Purification, characterization and antiglycation activity of a novel polysaccharide from black currant. Food Chem. 2016, 199, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Armbruster, D.A. Fructosamine: Structure, analysis, and clinical usefulness. Clin. Chem. 1987, 33, 2153–2163. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.; Baker, S.S.; Liu, W.; Desai, S.; Alkhouri, R.; Kozielski, R.; Mastrandrea, L.; Sarfraz, A.; Cai, W.; Vlassara, H. Effect of dietary advanced glycation end products on mouse liver. PLoS ONE 2012, 7, e35143. [Google Scholar] [CrossRef]
- Lo, C.Y.; Hsiao, W.T.; Chen, X.Y. Efficiency of trapping methylglyoxal by phenols and phenolic acids. J. Food Sci. 2011, 76, H90–H96. [Google Scholar] [CrossRef]
- Levine, R.L.; Garland, D.; Oliver, C.N.; Amici, A.; Climent, I.; Lenz, A.-G.; Ahn, B.-W.; Shaltiel, S.; Stadtman, E.R. [49] Determination of carbonyl content in oxidatively modified proteins. Methods Enzymol. 1990, 186, 464–478. [Google Scholar]
- Hashimoto, C.; Iwaihara, Y.; Chen, S.J.; Tanaka, M.; Watanabe, T.; Matsui, T. Highly-sensitive detection of free advanced glycation end-products by liquid chromatography-electrospray ionization-tandem mass spectrometry with 2,4,6-trinitrobenzene sulfonate derivatization. Anal. Chem. 2013, 85, 4289–4295. [Google Scholar] [CrossRef]
- Li, B.-Y.; Li, X.-L.; Cai, Q.; Gao, H.-Q.; Cheng, M.; Zhang, J.-H.; Wang, J.-F.; Yu, F.; Zhou, R.-H. Induction of lactadherin mediates the apoptosis of endothelial cells in response to advanced glycation end products and protective effects of grape seed procyanidin B2 and resveratrol. Apoptosis 2011, 16, 732–745. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Tarai, M.; Mishra, A.K. Unconventional steady-state fluorescence spectroscopy as an analytical technique for analyses of complex-multifluorophoric mixtures. TRAC Trends Anal. Chem. 2017, 97, 216–243. [Google Scholar] [CrossRef]
- Liu, C.; Guo, H.; Dain, J.A.; Wan, Y.; Gao, X.-H.; Chen, H.-D.; Seeram, N.P.; Ma, H. Cytoprotective effects of a proprietary red maple leaf extract and its major polyphenol, ginnalin A, against hydrogen peroxide and methylglyoxal induced oxidative stress in human keratinocytes. Food Funct. 2020, 11, 5105–5114. [Google Scholar] [CrossRef]
- Nishigaki, I.; Rajendran, P.; Venugopal, R.; Ekambaram, G.; Sakthisekaran, D.; Nishigaki, Y. Cytoprotective role of astaxanthin against glycated protein/iron chelate-induced toxicity in human umbilical vein endothelial cells. Phytother. Res. Int. J. Devoted Pharmacol. Toxicol. Eval. Nat. Prod. Deriv. 2010, 24, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Yaylayan, V. Mechanochemical generation of Schiff bases and Amadori products and utilization of diagnostic MS/MS fragmentation patterns in negative ionization mode for their analysis. Carbohydr. Res. 2020, 495, 108091. [Google Scholar] [CrossRef] [PubMed]
- Shumilina, J.; Kusnetsova, A.; Tsarev, A.; Janse van Rensburg, H.C.; Medvedev, S.; Demidchik, V.; Van den Ende, W.; Frolov, A. Glycation of plant proteins: Regulatory roles and interplay with sugar signalling? Int. J. Mol. Sci. 2019, 20, 2366. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.-W.; Hsieh, C.-L.; Wang, H.-Y.; Chen, H.-Y. Inhibitory effects of guava (Psidium guajava L.) leaf extracts and its active compounds on the glycation process of protein. Food Chem. 2009, 113, 78–84. [Google Scholar] [CrossRef]
- Berlett, B.S.; Stadtman, E.R. Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 1997, 272, 20313–20316. [Google Scholar] [CrossRef] [Green Version]
- Dalle-Donne, I.; Giustarini, D.; Colombo, R.; Rossi, R.; Milzani, A. Protein carbonylation in human diseases. Trends Mol. Med. 2003, 9, 169–176. [Google Scholar] [CrossRef]
- Zeng, J.; Dunlop, R.A.; Rodgers, K.J.; Davies, M.J. Evidence for inactivation of cysteine proteases by reactive carbonyls via glycation of active site thiols. Biochem. J. 2006, 398, 197–206. [Google Scholar] [CrossRef]
- Baynes, J.W. Chemical modification of proteins by lipids in diabetes. Clin. Chem. Lab. Med. 2003, 41, 1159–1165. [Google Scholar] [CrossRef] [PubMed]
- Potipiranun, T.; Adisakwattana, S.; Worawalai, W.; Ramadhan, R.; Phuwapraisirisan, P. Identification of pinocembrin as an anti-glycation agent and α-glucosidase inhibitor from fingerroot (Boesenbergia rotunda): The tentative structure–activity relationship towards MG-trapping activity. Molecules 2018, 23, 3365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blackburn, N.J.; Vulesevic, B.; McNeill, B.; Cimenci, C.E.; Ahmadi, A.; Gonzalez-Gomez, M.; Ostojic, A.; Zhong, Z.; Brownlee, M.; Beisswenger, P.J. Methylglyoxal-derived advanced glycation end products contribute to negative cardiac remodeling and dysfunction post-myocardial infarction. Basic Res. Cardiol. 2017, 112, 57. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Shamsi, A.; Khan, M.S.; Husain, F.M.; Bano, B. Methylglyoxal induced glycation and aggregation of human serum albumin: Biochemical and biophysical approach. Int. J. Biol. Macromol. 2018, 113, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.W.; Lee, J.; Park, Y.I. 7, 8-Dihydroxyflavone attenuates TNF-α-induced skin aging in Hs68 human dermal fibroblast cells via down-regulation of the MAPKs/Akt signaling pathways. Biomed. Pharmacother. 2017, 95, 1580–1587. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.-C.; Hu, M.-L. The limitations and validities of senescence associated-β-galactosidase activity as an aging marker for human foreskin fibroblast Hs68 cells. Exp. Gerontol. 2005, 40, 813–819. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.M.; Huang, D.Y.; Huang, Y.P.; Hsu, S.H.; Kang, L.Y.; Shen, C.M.; Lin, W.W. Methylglyoxal induces cell death through endoplasmic reticulum stress-associated ROS production and mitochondrial dysfunction. J. Cell. Mol. Med. 2016, 20, 1749–1760. [Google Scholar] [CrossRef] [Green Version]
- Fusté, N.; Guasch, M.; Guillen, P.; Anerillas, C.; Cemeli, T.; Pedraza, N.; Ferrezuelo, F.; Encinas, M.; Moralejo, M.; Garí, E. Barley β-glucan accelerates wound healing by favoring migration versus proliferation of human dermal fibroblasts. Carbohydr. Polym. 2019, 210, 389–398. [Google Scholar] [CrossRef]
- Choi, J.-S.; Kim, J.W.; Jung, G.-W.; Moon, S.-B.; Cho, H.-R.; Sung, S.H.; Jung, J.J.; Kwon, Y.S.; Ku, S.K.; Sohn, J.-H. Effect of a β-glucan from Aureobasidium on TGF-β 1-modulated in vitro dermal wound repair. Toxicol. Environ. Health Sci. 2016, 8, 12–18. [Google Scholar] [CrossRef]
- Moldogazieva, N.T.; Mokhosoev, I.M.; Mel’nikova, T.I.; Porozov, Y.B.; Terentiev, A.A. Oxidative stress and advanced lipoxidation and glycation end products (ALEs and AGEs) in aging and age-related diseases. Oxidative Med. Cell. Longev. 2019, 2019. [Google Scholar] [CrossRef] [Green Version]
- Kislinger, T.; Fu, C.; Huber, B.; Qu, W.; Taguchi, A.; Du Yan, S.; Hofmann, M.; Yan, S.F.; Pischetsrieder, M.; Stern, D. N ε-(carboxymethyl) lysine adducts of proteins are ligands for receptor for advanced glycation end products that activate cell signaling pathways and modulate gene expression. J. Biol. Chem. 1999, 274, 31740–31749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alikhani, M.; MacLellan, C.M.; Raptis, M.; Vora, S.; Trackman, P.C.; Graves, D.T. Advanced glycation end products induce apoptosis in fibroblasts through activation of ROS, MAP kinases, and the FOXO1 transcription factor. Am. J. Physiol.-Cell Physiol. 2007, 292, C850–C856. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, H.; Lin, T.-Y.; Lin, J.-A.; Cheng, K.-C.; Santoso, S.P.; Chou, C.-H.; Hsieh, C.-W. Effect of Pholiota nameko Polysaccharides Inhibiting Methylglyoxal-Induced Glycation Damage In Vitro. Antioxidants 2021, 10, 1589. https://doi.org/10.3390/antiox10101589
Lin H, Lin T-Y, Lin J-A, Cheng K-C, Santoso SP, Chou C-H, Hsieh C-W. Effect of Pholiota nameko Polysaccharides Inhibiting Methylglyoxal-Induced Glycation Damage In Vitro. Antioxidants. 2021; 10(10):1589. https://doi.org/10.3390/antiox10101589
Chicago/Turabian StyleLin, His, Ting-Yun Lin, Jer-An Lin, Kuan-Chen Cheng, Shella Permatasari Santoso, Chun-Hsu Chou, and Chang-Wei Hsieh. 2021. "Effect of Pholiota nameko Polysaccharides Inhibiting Methylglyoxal-Induced Glycation Damage In Vitro" Antioxidants 10, no. 10: 1589. https://doi.org/10.3390/antiox10101589





