Identification of Begomoviruses from Three Cryptic Species of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) in Nepal
Abstract
1. Introduction
2. Materials and Methods
2.1. Whitefly Samples
2.2. DNA Extraction and Polymerase Chain Reaction
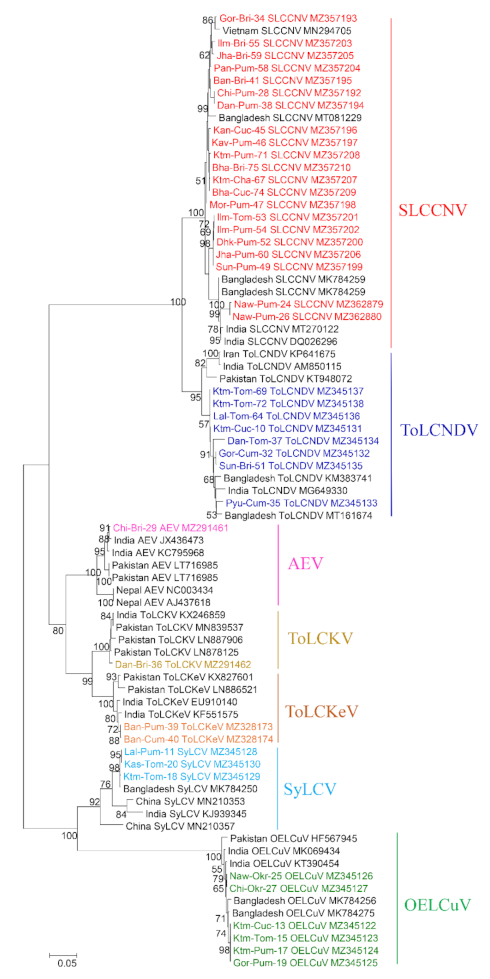
2.3. Sequence Alignment and Phylogenetic Analyses
3. Results
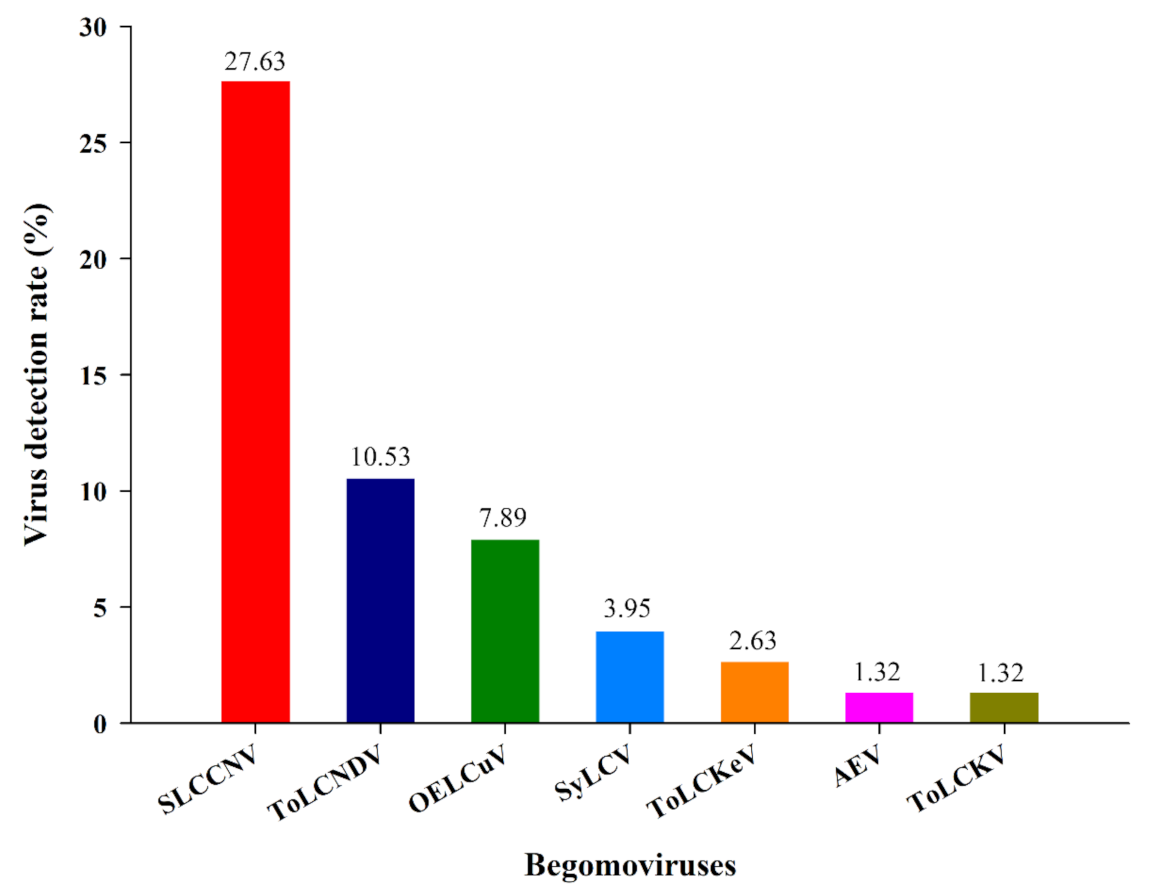
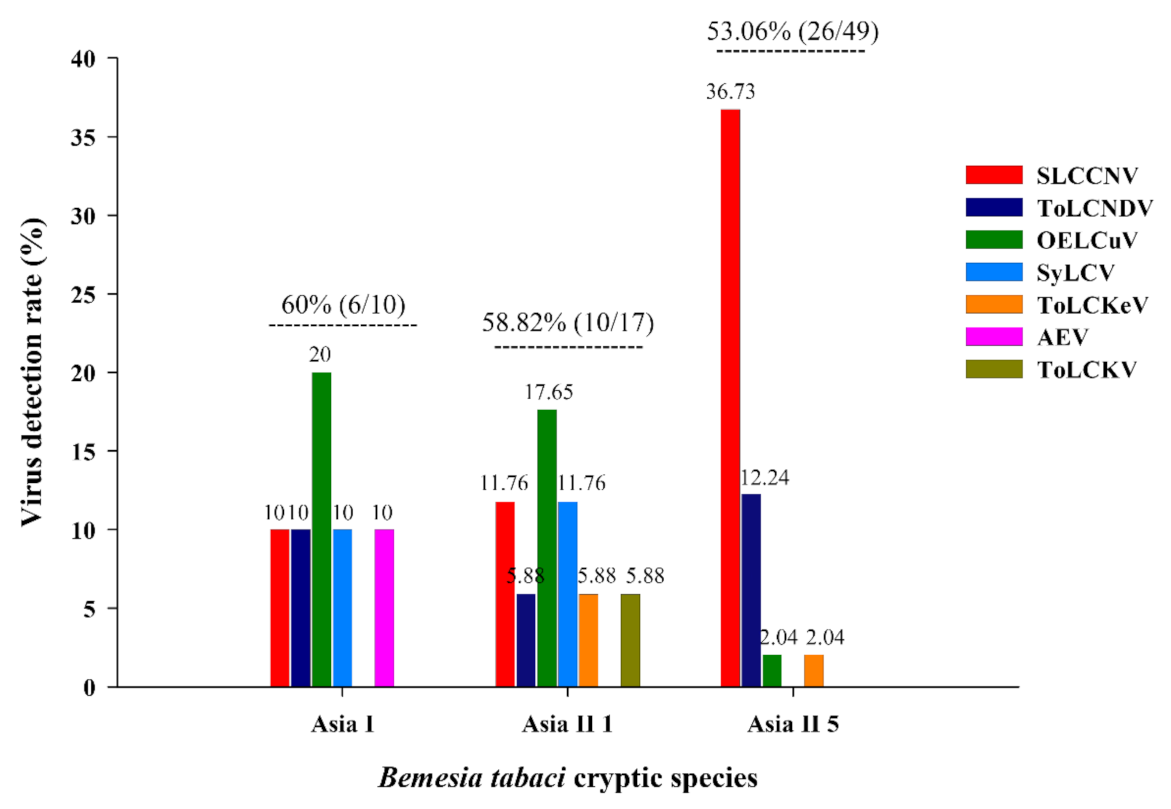
3.1. Identification of Begomovirus from Different Cryptic Species of Bemisia tabaci and Their Distribution
3.2. Identification of the Begomoviruses in B. tabaci Collected from Different Host Plants
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hogenhout, S.A.; Ammar, E.-D.; Whitfield, A.E.; Redinbaugh, M.G. Insect Vector Interactions with Persistently Transmitted Viruses. Annu. Rev. Phytopathol. 2008, 46, 327–359. [Google Scholar] [CrossRef]
- Navas-Castillo, J.; Fiallo-Olivé, E.; Sánchez-Campos, S. Emerging Virus Diseases Transmitted by Whiteflies. Annu. Rev. Phytopathol. 2011, 49, 219–248. [Google Scholar] [CrossRef] [PubMed]
- Krause-Sakate, R.; Watanabe, L.F.M.; Gorayeb, E.S.; Da Silva, F.B.; Alvarez, D.D.L.; Bello, V.H.; Nogueira, A.M.; De Marchi, B.R.; Vicentin, E.; Ribeiro-Junior, M.R.; et al. Population Dynamics of Whiteflies and Associated Viruses in South America: Research Progress and Perspectives. Insects 2020, 11, 847. [Google Scholar] [CrossRef] [PubMed]
- Nigam, D. Genomic Variation and Diversification in Begomovirus Genome in Implication to Host and Vector Adaptation. Plants 2021, 10, 1706. [Google Scholar] [CrossRef]
- Varma, A.; Mandal, B.; Singh, M.K. Global Emergence and Spread of Whitefly (Bemisia tabaci) Transmitted Geminiviruses; Thompson, W.M.O., Ed.; Springer: Dordrecht, The Netherlands, 2011; ISBN 9789400715240. [Google Scholar]
- Rubinstein, G.; Czosnek, H. Long-term association of tomato yellow leaf curl virus with its whitefly vector Bemisia tabaci: Effect on the insect transmission capacity, longevity and fecundity. J. Gen. Virol. 1997, 78, 2683–2689. [Google Scholar] [CrossRef]
- Kanakala, S.; Ghanim, M. Global genetic diversity and geographical distribution of Bemisia tabaci and its bacterial endosymbionts. PLoS ONE 2019, 14, e0213946. [Google Scholar] [CrossRef]
- Hu, J.; Zhang, X.; Jiang, Z.; Zhang, F.; Liu, Y.; Li, Z.; Zhang, Z. New putative cryptic species detection and genetic network analysis of Bemisia tabaci (Hempitera: Aleyrodidae) in China based on mitochondrial COI sequences. Mitochondrial DNA Part A DNA Mapping. Seq. Anal. 2018, 29, 474–484. [Google Scholar] [CrossRef]
- Firdaus, S.; Vosman, B.; Hidayati, N.; Supena, E.D.J.; Visser, R.G.; Van Heusden, A.W. The Bemisia tabaci species complex: Additions from different parts of the world. Insect Sci. 2013, 20, 723–733. [Google Scholar] [CrossRef]
- Esterhuizen, L.L.; Mabasa, K.G.; van Heerden, S.W.; Czosnek, H.; Brown, J.K.; van Heerden, H.; Rey, M.E.C. Genetic identification of members of the Bemisia tabaci cryptic species complex from South Africa reveals native and introduced haplotypes. J. Appl. Èntomol. 2013, 137, 122–135. [Google Scholar] [CrossRef]
- Chowda-Reddy, R.; Kirankumar, M.; Seal, S.E.; Muniyappa, V.; Valand, G.B.; Govindappa, M.R.; Colvin, J. Bemisia tabaci Phylogenetic Groups in India and the Relative Transmission Efficacy of Tomato leaf curl Bangalore virus by an Indigenous and an Exotic Population. J. Integr. Agric. 2012, 11, 235–248. [Google Scholar] [CrossRef]
- Alemandri, V.; De Barro, P.; Bejerman, N.; Caro, E.B.A.; Dumón, A.D.; Mattio, M.F.; Rodriguez, S.M.; Truol, G. Species within the Bemisia tabaci (Hemiptera: Aleyrodidae) Complex in Soybean and Bean Crops in Argentina. J. Econ. Èntomol. 2012, 105, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Boykin, L.M.; Armstrong, K.; Kubatko, L.; De Barro, P.J. Species Delimitation and Global Biosecurity. Evol. Bioinform. 2011, 8, EBO.S8532–37. [Google Scholar] [CrossRef]
- De Barro, P.J.; Liu, S.-S.; Boykin, L.; Dinsdale, A.B. Bemisia tabaci: A Statement of Species Status. Annu. Rev. Èntomol. 2011, 56, 1–19. [Google Scholar] [CrossRef]
- Dinsdale, A.; Cook, L.G.; Riginos, C.; Buckley, Y.M.; De Barro, P. Refined Global Analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) Mitochondrial Cytochrome Oxidase 1 to Identify Species Level Genetic Boundaries. Ann. Entomol. Soc. Am. 2010, 103, 196–208. [Google Scholar] [CrossRef]
- Rosell, R.C.; Bedford, I.D.; Frohlich, D.R.; Gill, R.J.; Brown, J.K.; Markham, P.G. Analysis of Morphological Variation in Distinct Populations of Bemisia tabaci (Homoptera: Aleyrodidae). Ann. Èntomol. Soc. Am. 1997, 90, 575–589. [Google Scholar] [CrossRef]
- Oliveira, M.; Henneberry, T.; Anderson, P. History, current status, and collaborative research projects for Bemisia tabaci. Crop. Prot. 2001, 20, 709–723. [Google Scholar] [CrossRef]
- Guo, T.; Guo, Q.; Cui, X.-Y.; Liu, Y.-Q.; Hu, J.; Liu, S.-S. Comparison of transmission of Papaya leaf curl China virus among four cryptic species of the whitefly Bemisia tabaci complex. Sci. Rep. 2015, 5, 15432. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Zhao, J.-J.; Zhang, T.; Li, F.-F.; Ghanim, M.; Zhou, X.-P.; Ye, G.-Y.; Liu, S.-S.; Wang, X.-W. Specific Cells in the Primary Salivary Glands of the Whitefly Bemisia tabaci Control Retention and Transmission of Begomoviruses. J. Virol. 2014, 88, 13460–13468. [Google Scholar] [CrossRef]
- Li, M.; Hu, J.; Xu, F.-C.; Liu, S.-S. Transmission of Tomato Yellow Leaf Curl Virus by two invasive biotypes and a Chinese indigenous biotype of the whitefly Bemisia tabaci. Int. J. Pest. Manag. 2010, 56, 275–280. [Google Scholar] [CrossRef]
- Pan, L.; Chen, Q.; Guo, T.; Wang, X.; Li, P.; Wang, X.; Liu, S. Differential efficiency of a begomovirus to cross the midgut of different species of whiteflies results in variation of virus transmission by the vectors. Sci. China Life Sci. 2018, 61, 1254–1265. [Google Scholar] [CrossRef]
- Bedford, I.D.; Briddon, R.W.; Brown, J.K.; Rosell, R.C.; Markham, P.G. Geminivirus transmission and biological characterisation of Bemisia tabaci (Gennadius) biotypes from different geographic regions. Ann. Appl. Biol. 1994, 125, 311–325. [Google Scholar] [CrossRef]
- Sánchez-Campos, S.; Navas-Castillo, J.; Camero, R.; Soria, C.; Díaz, J.A.; Moriones, E. Displacement of Tomato Yellow Leaf Curl Virus (TYLCV)-Sr by TYLCV-Is in Tomato Epidemics in Spain. Phytopathology 1999, 89, 1038–1043. [Google Scholar] [CrossRef]
- Maruthi, M.N.; Alam, S.N.; Kader, K.A.; Rekha, A.R.; Cork, A.; Colvin, J. Nucleotide Sequencing, Whitefly Transmission, and Screening Tomato for Resistance against Two Newly Described Begomoviruses in Bangladesh. Phytopathology 2005, 95, 1472–1481. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Islam, W.; Lin, W.; Islam, S.U.; Arif, M.; Li, X.; Yang, Y.; Ding, X.; Du, Z.; Wu, Z. Genetic diversity of begomoviruses in Pakistan captured through a vector based survey. Microb. Pathog. 2018, 118, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.S.; Ikegami, M.; Natsuaki, K.T. First report of Mungbean yellow mosaic India virus in Nepal. New Dis. Rep. 2012, 25, 30. [Google Scholar] [CrossRef]
- Shahid, M.S.; Pudashini, B.J.; Khatri-Chhetri, G.B.; Briddon, R.W.; Natsuaki, K.T. Molecular characterization of a distinct monopartite begomovirus associated with betasatellites and alphasatellites infecting Pisum sativum in Nepal. Virus Genes 2016, 53, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Bull, S.E.; Amin, I.; Idris, A.M.; Mansoor, S.; Bedford, I.D.; Dhawan, P.; Rishi, N.; Siwatch, S.S.; Abdel-Salam, A.M.; et al. Diversity of DNA β, a satellite molecule associated with some monopartite begomoviruses. Virology 2003, 312, 106–121. [Google Scholar] [CrossRef]
- Shahid, M.S.; Yoshida, S.; Khatri-Chhetri, G.B.; Briddon, R.; Natsuaki, K.T. Complete nucleotide sequence of a monopartite Begomovirus and associated satellites infecting Carica papaya in Nepal. Virus Genes 2013, 46, 581–584. [Google Scholar] [CrossRef]
- Ghimire, S.R.; Subedi, P.P.; Green, S.K. Status of Tomato Yellow Leaf Curl Virus in Tomato in the Western Hills of Nepal. Nepal Agric. Res. J. 2001, 4, 1–4. [Google Scholar] [CrossRef]
- Acharya, R.; Shrestha, Y.K.; Sharma, S.R.; Lee, K.-Y. Genetic diversity and geographic distribution of Bemisia tabaci species complex in Nepal. J. Asia-Pacific Èntomol. 2020, 23, 509–515. [Google Scholar] [CrossRef]
- Malik, A.H.; Briddon, R.W.; Mansoor, S. Infectious clones of Tomato leaf curl Palampur virus with a defective DNA B and their pseudo-recombination with Tomato leaf curl New Delhi virus. Virol. J. 2011, 8, 173. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, A.A.; Aravind, L.; Madden, T.L.; Shavirin, S.; Spouge, J.L.; Wolf, Y.I.; Koonin, E.V.; Altschul, S.F. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001, 29, 2994–3005. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution (N. Y.) 1985, 39, 783–791. [Google Scholar]
- Kon, T.; Dolores, L.M.; Bajet, N.B.; Hase, S.; Takahashi, H.; Ikegami, M. Molecular characterization of a strain of Squash leaf curl China virus from the Philippines. J. Phytopathol. 2003, 151, 535–539. [Google Scholar] [CrossRef]
- Revill, P.; Ha, C.V.; Porchun, S.C.; Vu, M.T.; Dale, J. The complete nucleotide sequence of two distinct geminiviruses infecting cucurbits in Vietnam. Arch. Virol. 2003, 148, 1523–1541. [Google Scholar] [CrossRef] [PubMed]
- Dolores, L.M.; Valdez, R.B. Identification of squash viruses and screening for resistance. Phytopathology 1988, 24, 43–53. [Google Scholar]
- Khatun, M.F.; Hwang, H.-S.; Shim, J.-K.; Kil, E.-J.; Lee, S.; Lee, K.-Y. Identification of begomoviruses from different cryptic species of Bemisia tabaci in Bangladesh. Microb. Pathog. 2020, 142, 104069. [Google Scholar] [CrossRef] [PubMed]
- Agnihotri, A.K.; Mishra, S.P. Temporal and spatial dynamics of tomato leaf curl disease at Chitrakoot region in India. J. Pharmacogn. Phytochem. 2018, SP1, 531–536. [Google Scholar]
- Zaidi, S.S.E.A.; Shafiq, M.; Amin, I.; Scheffler, B.; Scheffler, J.A.; Briddon, R.; Mansoor, S. Frequent Occurrence of Tomato Leaf Curl New Delhi Virus in Cotton Leaf Curl Disease Affected Cotton in Pakistan. PLoS ONE 2016, 11, e0155520. [Google Scholar] [CrossRef]
- Shelat, M.; Murari, S.; Sharma, M.C.; Subramanian, R.B.; Jummanah, J.; Jarullah, B. Prevalence and distribution of Tomato leaf curl virus in major agroclimatic zones of Gujarat. Adv. Biosci. Biotechnol. 2014, 5, 1–3. [Google Scholar] [CrossRef][Green Version]
- Rishi, N. Current status of begomoviruses in the Indian subcontinent. Indian Phytopathol. 2004, 57, 396–407. [Google Scholar]
- Pandey, P.; Mukhopadhya, S.; Naqvi, A.R.; Mukherjee, S.K.; Shekhawat, G.S.; Choudhury, N.R. Molecular characterization of two distinct monopartite begomoviruses infecting tomato in India. Virol. J. 2010, 7, 337. [Google Scholar] [CrossRef] [PubMed]
- Chatchawankanphanich, O.; Maxwell, D.P. Tomato leaf curl Karnataka virus from Bangalore, India, Appears to be a Recombinant Begomovirus. Phytopathology 2002, 92, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Esakky, R.; Palicherla, S.R. The emergence of okra enation leaf curl virus—an important begomovirus, infecting okra in several states across India. Arch. Phytopathol. Plant. Prot. 2019, 52, 234–238. [Google Scholar] [CrossRef]
- Mishra, G.P.; Singh, B.; Seth, T.; Singh, A.K.; Halder, J.; Krishnan, N.; Tiwari, S.K.; Singh, P.M. Biotechnological Advancements and Begomovirus Management in Okra (Abelmoschus esculentus L.): Status and Perspectives. Front. Plant. Sci. 2017, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Bananej, K.; Kraberger, S.; Varsani, A. Okra Enation Leaf Curl Virus in Papaya from Iran Displaying Severe Leaf Curl Symptoms. J. Plant. Pathol 2016, 98, 637–639. [Google Scholar]
- Kon, T.; Rojas, M.R.; Abdourhamane, I.K.; Gilbertson, R.L. Roles and interactions of begomoviruses and satellite DNAs associated with okra leaf curl disease in Mali, West Africa. J. Gen. Virol. 2009, 90, 1001–1013. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, G.A.M. Okra leaf curl virus: A monopartite begomovirus infecting okra crop in Saudi Arabia. Arab J. Biotechnol 2003, 6, 139–152. [Google Scholar]
- Singh, S.J. Assessment of losses in okra due to enation leaf curl virus. Indian J. Virol. 1996, 12, 51–53. [Google Scholar]
- Swanson, M.; Harrison, B. Serological relationships and epitope profiles of isolates of okra leaf curl geminivirus from Africa and the Middle East. Biochim. 1993, 75, 707–711. [Google Scholar] [CrossRef]
- Atiri, G.I. The occurrence and importance of okra mosaic virus in Nigerian weeds. Ann. Appl. Biol. 1984, 104, 261–265. [Google Scholar] [CrossRef]
- Hameed, U.; Zia-Ur-Rehman, M.; Herrmann, H.W.; Haider, M.S.; Brown, J.K. First report of Okra enation leaf curl virus and associated cotton leaf curl Multan beta satellite and cotton leaf curl Multan alpha satellite infecting cotton in Pakistan; a new member of the cotton leaf curl disease complex. Plant. Dis. 2014, 98, 1447–1448. [Google Scholar] [CrossRef] [PubMed]
- Saleem, H.M.; Bedford, I.D.; Evans, A.A.; Markham, P.G. Geminivirus transmission by different biotypes of the whitefly Bemisia tabaci (Gennadius). Pak. J. Zool. 2003, 35, 343–351. [Google Scholar]
- Tahir, M.; Amin, I.; Haider, M.S.; Mansoor, S.; Briddon, R.W. Ageratum enation virus—A Begomovirus of Weeds with the Potential to Infect Crops. Viruses 2015, 7, 647–665. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Gunapati, S.; Singh, S.P.; Gadre, R.; Sharma, N.C.; Tuli, R. Molecular characterization and pathogenicity of a carrot (Daucus carota) infecting begomovirus and associated betasatellite from India. Virus Res. 2013, 178, 478–485. [Google Scholar] [CrossRef]
- Kumar, Y.; Hallan, V.; Zaidi, A.A. Chilli leaf curl Palampur virus is a distinct begomovirus species associated with a betasatellite. Plant. Pathol. 2011, 60, 1040–1047. [Google Scholar] [CrossRef]
- Kumar, Y.; Bhardwaj, P.; Hallan, V.; Zaidi, A.A. Detection and characterization of Ageratum enation virus and a nanovirus-like satellite DNA1 from zinnia causing leaf curl symptoms in India. J. Gen. Plant. Pathol. 2010, 76, 395–398. [Google Scholar] [CrossRef]
- Raj, S.K.; Snehi, S.K.; Khan, M.S.; Tiwari, A.K.; Rao, G.P. Detection of Ageratum enation virus from cat’s whiskers (Cleome gynandra L.) with leaf curl symptoms in India. J. Gen. Plant. Pathol. 2010, 76, 292–294. [Google Scholar] [CrossRef]
- Swarnalatha, P.; Mamatha, M.; Manasa, M.; Singh, R.P.; Krishnareddy, M. Molecular identification of Ageratum enation virus (AEV) associated with leaf curl disease of tomato(Solanum lycopersicum) in India. Australas. Plant. Dis. Notes 2013, 8, 67–71. [Google Scholar] [CrossRef]
- Singh-Pant, P.; Pant, P.; Mukherjee, S.K.; Mazumdar-Leighton, S. Spatial and temporal diversity of begomoviral complexes in papayas with leaf curl disease. Arch. Virol. 2012, 157, 1217–1232. [Google Scholar] [CrossRef] [PubMed]

| S.N. | Samples Name | Collection Sites | Collection Dates | Plant Viruses Detected in B. tabaci | ||||
|---|---|---|---|---|---|---|---|---|
| Virus Name | Accession Numbers 1 | % Identity (NCBI) | Numbers of Identical Bases | Accession Numbers 2 | ||||
| 1 | Ktm-Cuc-10 | Kritipur, Kathmandu | 18 July 2017 | ToLCNDV | MZ345131 | 98.74 | 1022/1035 | AY428769 |
| 2 | Lal-Pum-11 | Lele, Lalitpur | 19 July 2017 | SyLCV | MZ345128 | 99.61 | 1034/1038 | MK784250 |
| 3 | Ktm-Cuc-13 | Syuchatar, Kathmandu | 28 July 2017 | OELCuV | MZ345122 | 99.33 | 1037/1044 | MK784275 |
| 4 | Ktm-Tom-15 | Syuchatar, Kathmandu | 28 July 2017 | OELCuV | MZ345123 | 99.33 | 1037/1044 | MK784275 |
| 5 | Ktm-Pum-17 | Dahachwok, Kathmandu | 12 August 2017 | OELCuV | MZ345124 | 99.33 | 1037/1044 | MK784275 |
| 6 | Ktm-Tom-18 | Dahachwok, Kathmandu | 12 August 2017 | SyLCV | MZ345129 | 99.81 | 1036/1038 | MK784250 |
| 7 | Gor-Pum-19 | Gorkha, Gorkha | 13 August 2017 | OELCuV | MZ345125 | 99.33 | 1037/1044 | MK784275 |
| 8 | Kas-Tom-20 | Pokhara, Kaski | 13 August 2017 | SyLCV | MZ345130 | 99.61 | 1034/1038 | MK784250 |
| 9 | Naw-Pum-24 | Daldale, Nawalparasi | 17 July 2019 | SLCCNV | MZ362879 | 97.98 | 1018/1039 | DQ026296 |
| 10 | Naw-Okr-25 | Gaidakot, Nawalparasi | 17 July 2019 | OELCuV | MZ345126 | 99.62 | 1039/1043 | KT390454 |
| 11 | Naw-Pum-26 | Gaidakot, Nawalparasi | 17 July 2019 | SLCCNV | MZ362880 | 98.27 | 1018/1039 | DQ026296 |
| 12 | Chi-Okr-27 | Tandi, Chitwan | 17 July 2019 | OELCuV | MZ345127 | 99.62 | 1040/1044 | KT390454 |
| 13 | Chi-Pum-28 | Tandi, Chitwan | 17 July 2019 | SLCCNV | MZ357192 | 98.07 | 1017/1037 | MN294705 |
| 14 | Chi-Bri-29 | Ramnagar, Chitwan | 25 July 2019 | AEV | MZ291461 | 99.42 | 1031/1037 | JX436473 |
| 15 | Gor-Cum-32 | Manakamana, Gorkha | 17 July 2019 | ToLCNDV | MZ345132 | 98.65 | 1021/1035 | KM383742 |
| 16 | Gor-Bri-34 | Ghyalchwok, Gorkha | 22 May 2019 | SLCCNV | MZ357193 | 98.94 | 1026/1037 | MN294705 |
| 17 | Pyu-Cum-35 | Khaira, Pyuthan | 15 August 2019 | ToLCNDV | MZ345133 | 97.97 | 1014/1035 | KM383742 |
| 18 | Dan-Bri-36 | Ghorahi, Dang | 23 July 2019 | ToLCKV | MZ291462 | 99.23 | 1030/1038 | LN878125 |
| 19 | Dan-Tom-37 | Ghorahi, Dang | 23 July 2019 | ToLCNDV | MZ345134 | 97.1 | 1005/1035 | KM383742 |
| 20 | Dan-Pum-38 | Ghorahi, Dang | 23 July 2019 | SLCCNV | MZ357194 | 98.07 | 1017/1037 | MN294705 |
| 21 | Ban-Pum-39 | Koholpur, Banke | 23 July 2019 | ToLCKeV | MZ328173 | 98.94 | 1031/1042 | KF551575 |
| 22 | Ban-Cum-40 | Koholpur, Banke | 23 July 2019 | ToLCKeV | MZ328174 | 98.94 | 1031/1042 | KF551575 |
| 23 | Ban-Bri-41 | Koholpur, Banke | 23 July 2019 | SLCCNV | MZ357195 | 98.36 | 1020/1037 | MT081229 |
| 24 | Kan-Cuc-45 | Mahendranagar, Kanchanpur | 24 July 2019 | SLCCNV | MZ357196 | 98.26 | 1019/1037 | MN294705 |
| 25 | Kav-Pum-46 | Banepa, Kavre | 28 July 2019 | SLCCNV | MZ357197 | 98.36 | 1020/1037 | MT081229 |
| 26 | Mor-Pum-47 | Biratnagar, Morang | 1 August 2019 | SLCCNV | MZ357198 | 98.36 | 1020/1037 | MT081229 |
| 27 | Sun-Pum-49 | Dharan, Sunsari | 1 August 2019 | SLCCNV | MZ357199 | 97.69 | 1013/1037 | EU573715 |
| 28 | Sun-Bri-51 | Dharan, Sunsari | 1 August 2019 | ToLCNDV | MZ345135 | 98.74 | 1022/1035 | KM383742 |
| 29 | Dhk-Pum-52 | Guthitar, Dhankuta | 1 August 2019 | SLCCNV | MZ357200 | 97.78 | 1014/1037 | EU573715 |
| 30 | Ilm-Tom-53 | Fikkal, Ilam | 2 August 2019 | SLCCNV | MZ357201 | 97.69 | 1013/1037 | EU573715 |
| 31 | Ilm-Pum-54 | Fikkal, Ilam | 2 August 2019 | SLCCNV | MZ357202 | 97.69 | 1013/1037 | EU573715 |
| 32 | Ilm-Bri-55 | Fikkal, Ilam | 2 August 2019 | SLCCNV | MZ357203 | 98.26 | 1019/1037 | MN294705 |
| 33 | Pan-Pum-58 | Lalikharka, Panchthar | 3 August 2019 | SLCCNV | MZ357204 | 98.26 | 1019/1037 | MN294705 |
| 34 | Jha-Bri-59 | Kakadvitta, Jhapa | 3 August 2019 | SLCCNV | MZ357205 | 97.97 | 1016/1037 | MN294705 |
| 35 | Jha-Pum-60 | Kakadvitta, Jhapa | 3 August 2019 | SLCCNV | MZ357206 | 97.69 | 1013/1037 | EU573715 |
| 36 | Lal-Tom-64 | Godamchaur, Lalitpur | 18 July 2019 | ToLCNDV | MZ345136 | 98.74 | 1022/1035 | AY428769 |
| 37 | Ktm-Cha-67 | Banasthali, Kathmandu | 6 June 2019 | SLCCNV | MZ357207 | 98.17 | 1018/1037 | MT081229 |
| 38 | Ktm-Tom-69 | Naikap, Kathmandu | 29 July 2019 | ToLCNDV | MZ345137 | 98.84 | 1023/1035 | AY428769 |
| 39 | Ktm-Pum-71 | New Baneshwor, Kathmandu | 15 June 2019 | SLCCNV | MZ357208 | 98.46 | 1021/1037 | MT081229 |
| 40 | Ktm-Tom-72 | Dahachwok, Kathmandu | 30 July 2019 | ToLCNDV | MZ345138 | 98.84 | 1023/1035 | AY428769 |
| 41 | Bha-Cuc-74 | Sanga, Bhaktapur | 28 July 2019 | SLCCNV | MZ357209 | 98.26 | 1019/1037 | MT081229 |
| 42 | Bha-Bri-75 | Sanga, Bhaktapur | 28 July 2019 | SLCCNV | MZ357210 | 98.26 | 1019/1037 | MT081229 |
| S.N. | Begomovirus | B. tabaci Cryptic Species (Number of Virus Detected Samples) | Host Plants for B. tabaci |
|---|---|---|---|
| 1 | Squash leaf curl China virus (SLCCNV) | Asia I (1), Asia II 1 (2), Asia II 5 (18) | Cucurbita pepo, Cucumis sativus, Sechium edule, Solanum lycopersicum, Solanum melongena |
| 2 | Tomato leaf curl New Delhi virus (ToLCNDV) | Asia I (1), Asia II 1 (1), Asia II 5 (6) | Cucumis sativus, Solanum lycopersicum, Solanum melongena |
| 3 | Okra enation leaf curl virus (OELCuV) | Asia I (2), Asia II 1 (3), Asia II 5 (1) | Abelmoschus esculentus, Cucurbita pepo, Cucumis sativus, Solanum lycopersicum |
| 4 | Synedrella leaf curl virus (SyLCV) | Asia I (1), Asia II 1 (2) | Cucurbita pepo, Solanum lycopersicum |
| 5 | Tomato leaf curl Kerala virus (ToLCKeV) | Asia II 1 (1), Asia II 5 (1) | Cucurbita pepo, Cucumis sativus |
| 6 | Ageratum enation virus (AEV) | Asia I (1) | Solanum melongena |
| 7 | Tomato leaf Curl Karnataka virus (ToLCKV) | Asia II 1 (1) | Solanum melongena |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Acharya, R.; Shrestha, Y.K.; Khatun, M.F.; Lee, K.-Y. Identification of Begomoviruses from Three Cryptic Species of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) in Nepal. Agronomy 2021, 11, 2032. https://doi.org/10.3390/agronomy11102032
Acharya R, Shrestha YK, Khatun MF, Lee K-Y. Identification of Begomoviruses from Three Cryptic Species of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) in Nepal. Agronomy. 2021; 11(10):2032. https://doi.org/10.3390/agronomy11102032
Chicago/Turabian StyleAcharya, Rajendra, Yam Kumar Shrestha, Mst Fatema Khatun, and Kyeong-Yeoll Lee. 2021. "Identification of Begomoviruses from Three Cryptic Species of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) in Nepal" Agronomy 11, no. 10: 2032. https://doi.org/10.3390/agronomy11102032
APA StyleAcharya, R., Shrestha, Y. K., Khatun, M. F., & Lee, K.-Y. (2021). Identification of Begomoviruses from Three Cryptic Species of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) in Nepal. Agronomy, 11(10), 2032. https://doi.org/10.3390/agronomy11102032








