LncRNA MALAT1 Facilitates Ovarian Cancer Progression through Promoting Chemoresistance and Invasiveness in the Tumor Microenvironment
Abstract
:1. Introduction
2. Results
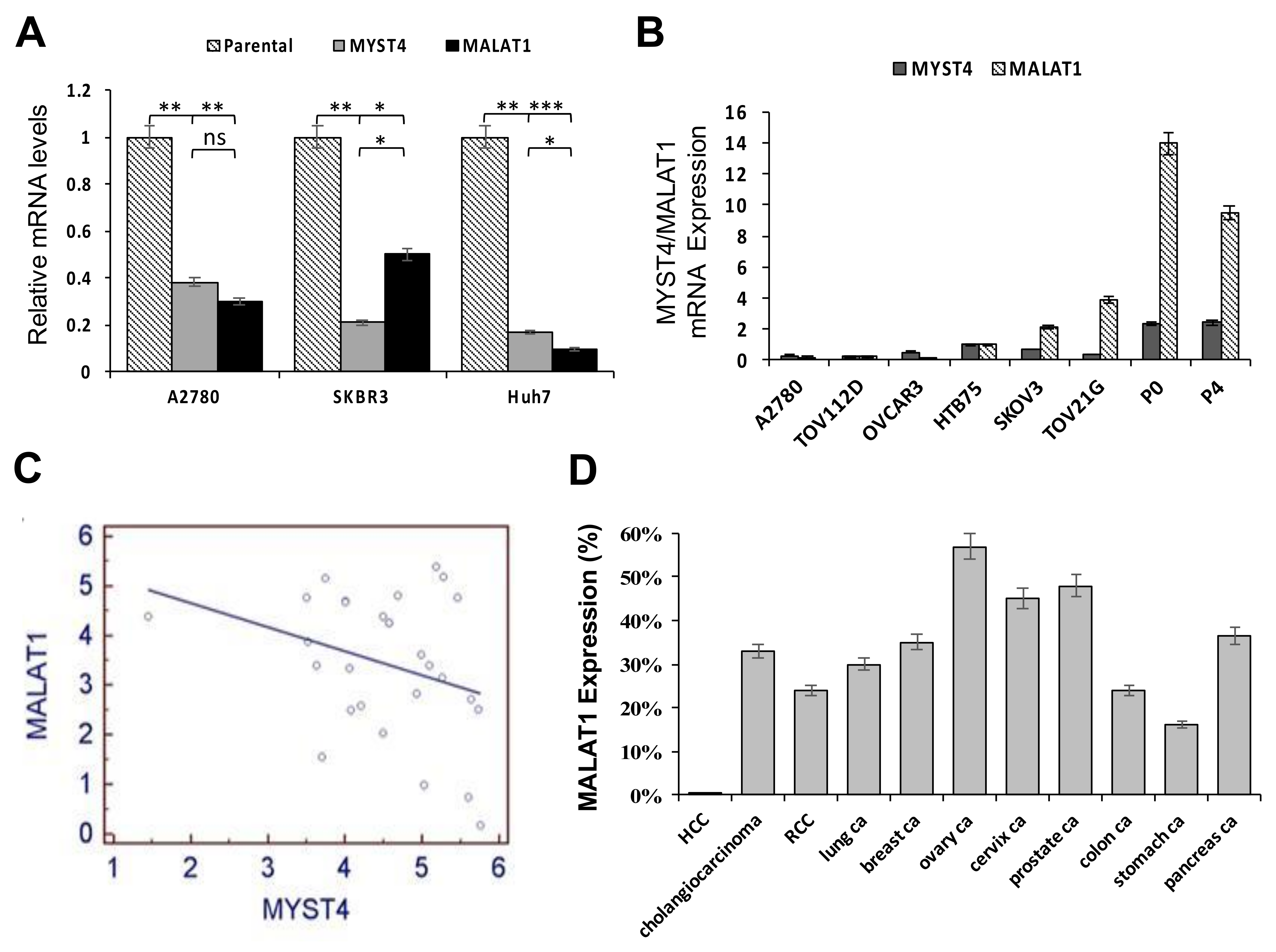
2.1. Relative Expressions between MYST4 and MALAT1, and Levels of MALAT1 in Different Clinical Cancer Tissues
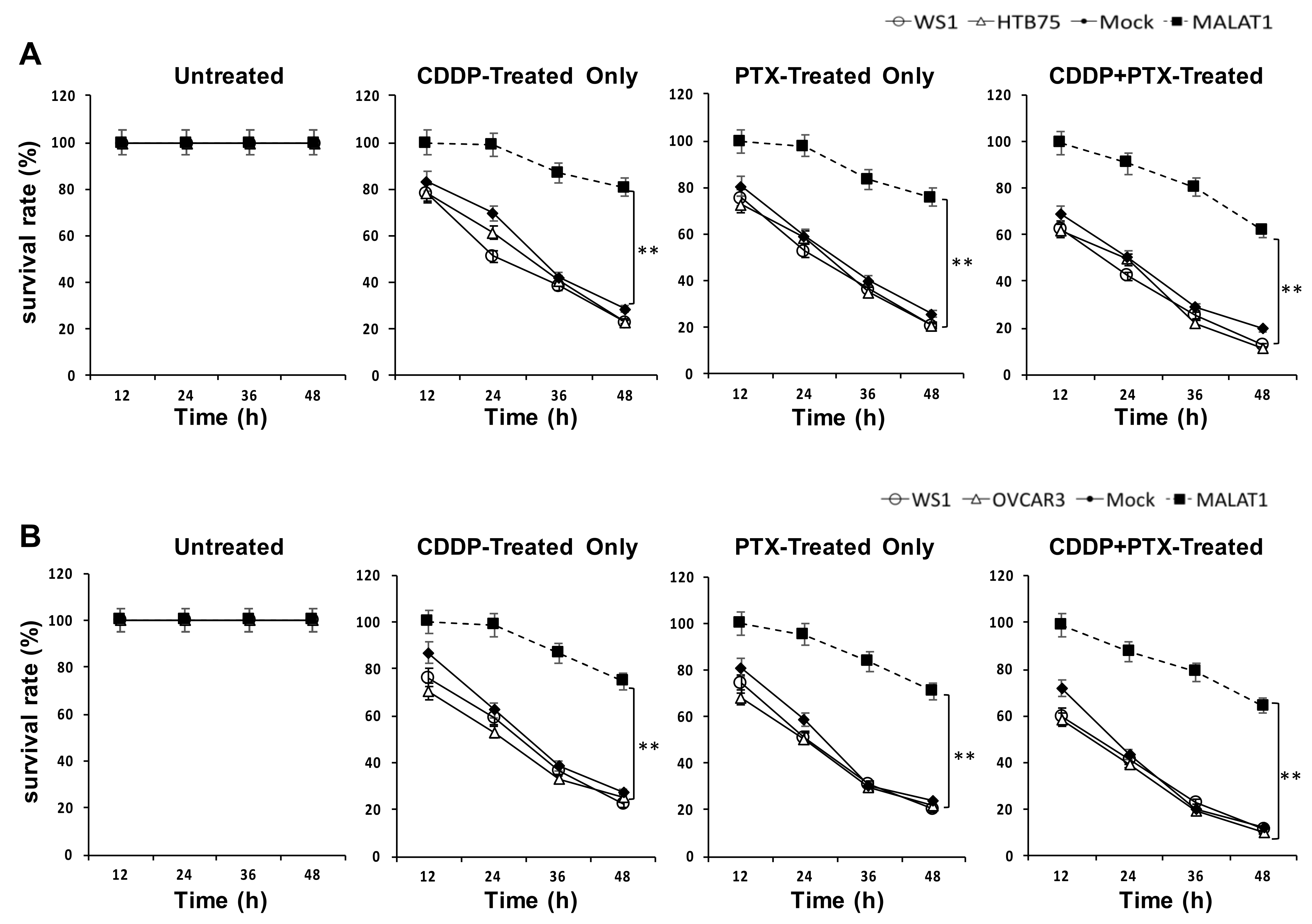
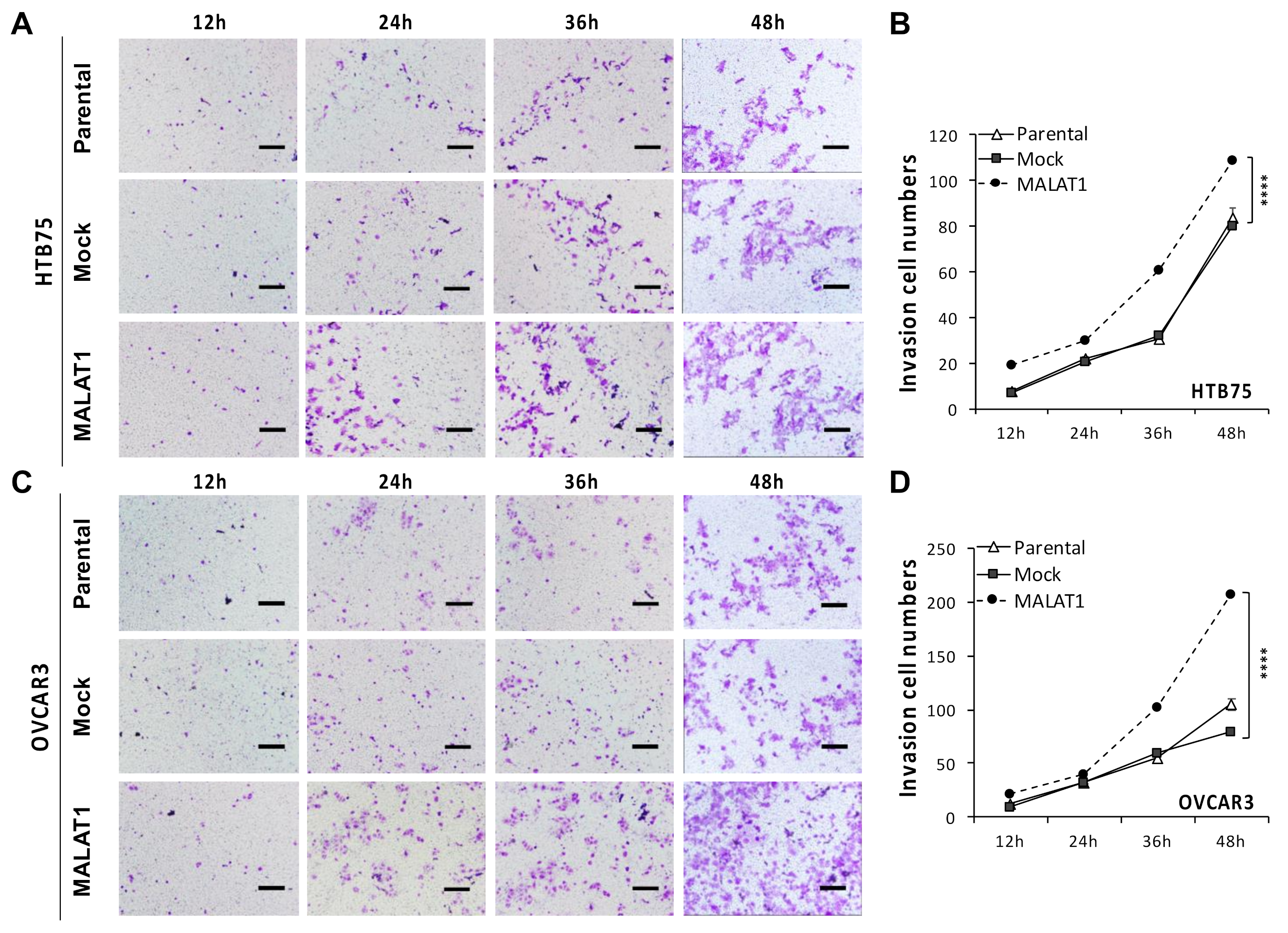
2.2. MALATI Upregulation in EOCs Stimulates Drug Resistance, Cell Migration, and Cell Invasion In Vitro
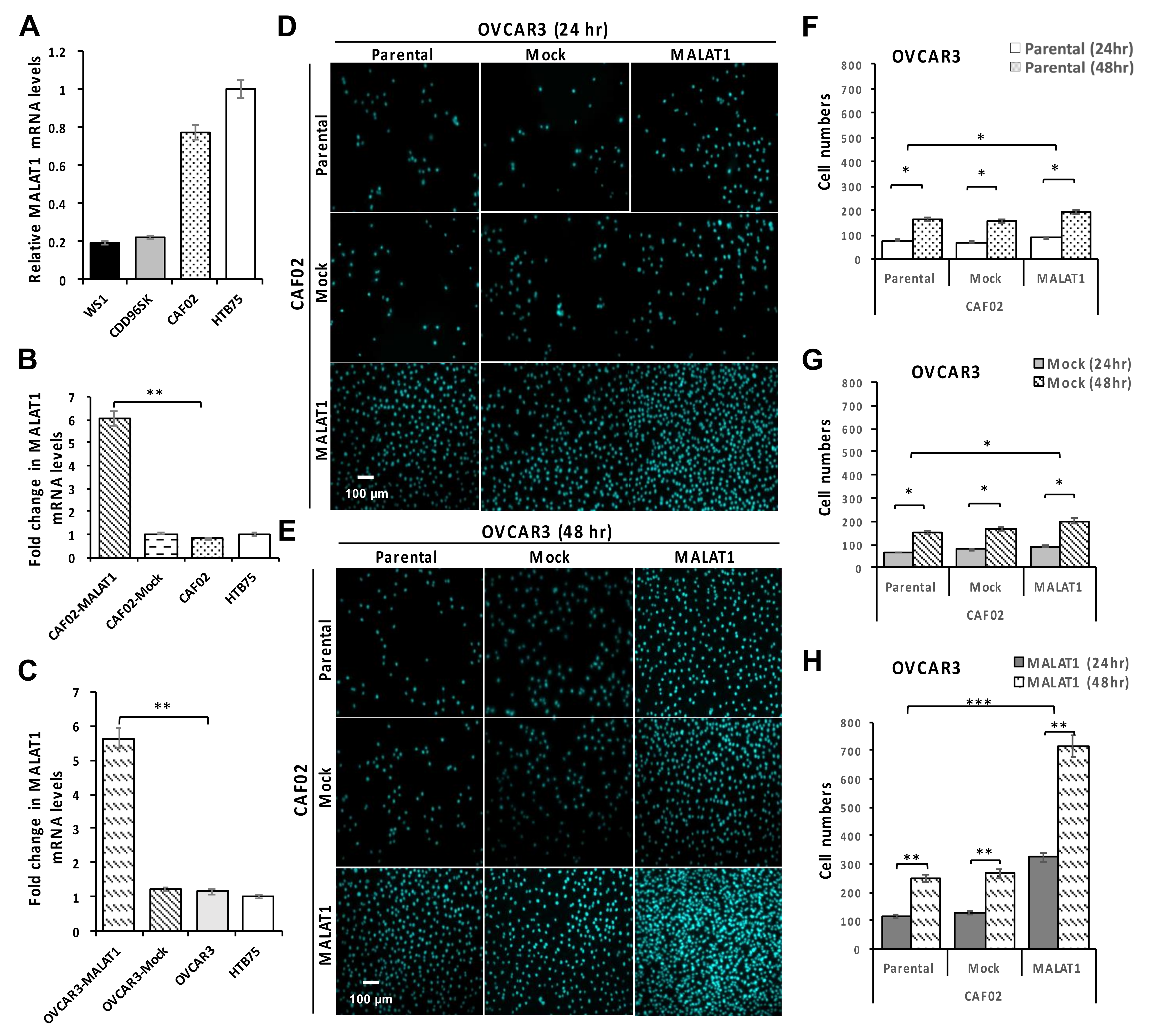
2.3. Overexpression of MALAT1 in CAF02 Fibroblasts Stimulates the In Vitro Invasive Ability of OVCAR3 Cells
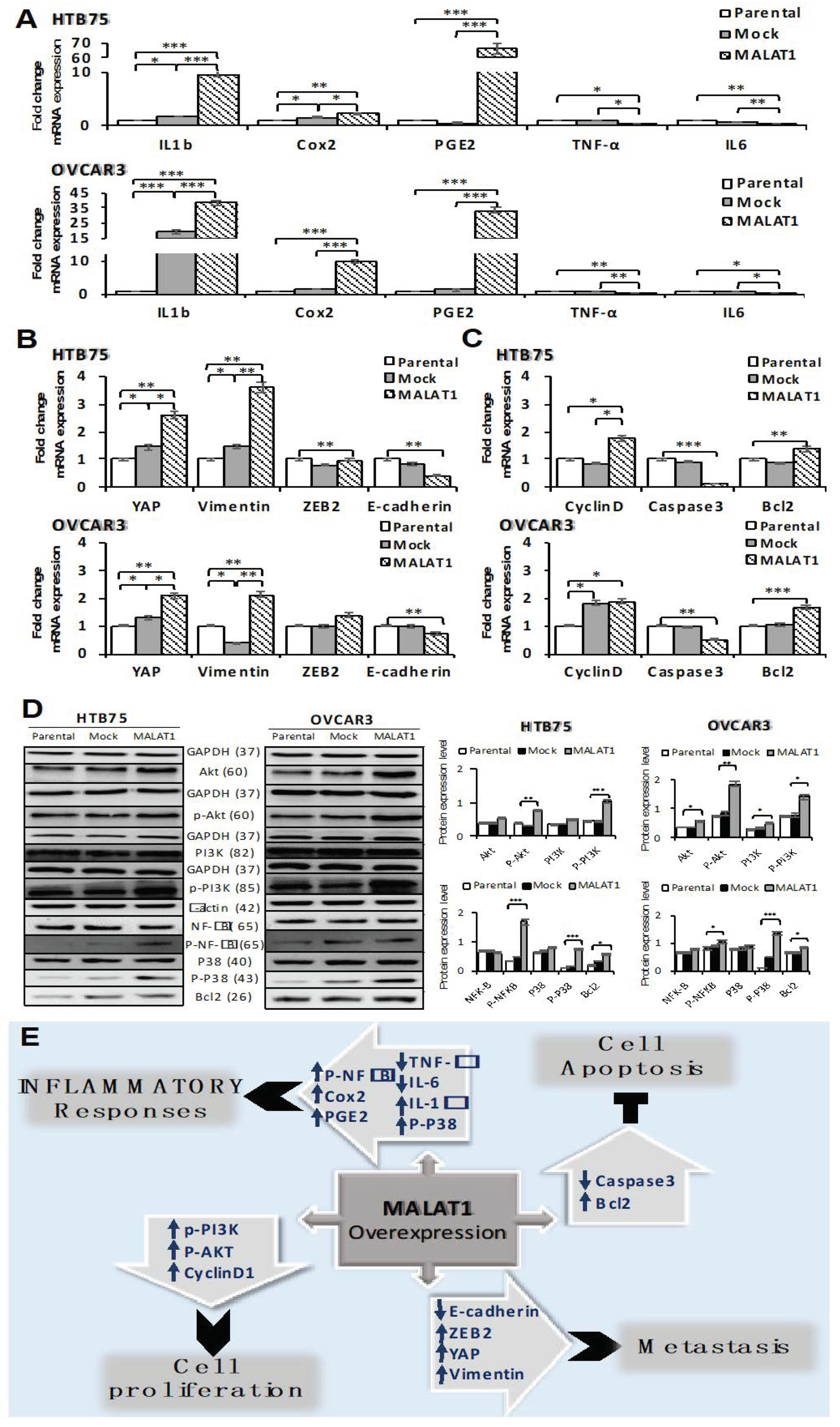
2.4. Overexpression of MALAT1 Modulates Tumor Cell Survival by Triggering Functional Molecules Involved in Different Signal Transduction Pathways
3. Discussion
4. Materials and Methods
4.1. Clinical Samples
4.2. In Situ Hybridization
4.3. Cell Lines and Cultivation
4.4. Lentiviral Production
4.5. MYST4 shRNA Knockdown
4.6. In Vitro Overexpression of MALAT1
4.7. Real-Time Reverse-Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
4.8. Drug-Resistance Cell Viability Assay
4.9. Wound-Healing Assay
4.10. Transwell-Invasion Assay
4.11. Western Blotting
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, J.; Chu, M. Targeting of IL-6-Relevant Long Noncoding RNA Profiles in Inflammatory and Tumorous Disease. Inflammation 2019, 42, 1139–1146. [Google Scholar] [CrossRef]
- Zhu, K.; Ren, Q.; Zhao, Y. lncRNA MALAT1 overexpression promotes proliferation, migration and invasion of gastric cancer by activating the PI3K/AKT pathway. Oncol. Lett. 2019, 17, 5335–5342. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Yu, Z.; Yang, H.; Lin, Y. Increased MALAT1 expression predicts poor prognosis in esophageal cancer patients. Biomed. Pharmacother. 2016, 83, 8–13. [Google Scholar] [CrossRef]
- Carlevaro-Fita, J.; Lanzós, A.; Feuerbach, L.; Hong, C.; Mas-Ponte, D.; Pedersen, J.S.; PCAWG Drivers and Functional Interpretation Group; Johnson, R.; PCAWG Consortium Cancer. LncRNA Census reveals evidence for deep functional conservation of long noncoding RNAs in tumorigenesis. Commun. Biol. 2020, 3, 56. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhao, Y.; Wang, Y.-M. LncRNA MALAT1 Promotes Survival of Epithelial Ovarian Cancer Cells by Downregulating miR-145-5p. Cancer Manag. Res. 2020, 12, 11359–11369. [Google Scholar] [CrossRef]
- Zhang, B.; Mao, Y.S.; Diermeier, S.D.; Novikova, I.V.; Nawrocki, E.P.; Jones, T.A.; Lazar, Z.; Tung, C.-S.; Luo, W.; Eddy, S.R.; et al. Identification and Characterization of a Class of MALAT1-like Genomic Loci. Cell Rep. 2017, 19, 1723–1738. [Google Scholar] [CrossRef] [Green Version]
- Lin, Q.; Guan, W.; Ren, W.; Zhang, L.; Zhang, J.; Xu, G. MALAT1 affects ovarian cancer cell behavior and patient survival. Oncol. Rep. 2018, 39, 2644–2652. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, M.K.; Doxtater, K.; Keramatnia, F.; Zacheaus, C.; Yallapu, M.; Jaggi, M.; Chauhan, S.C. Role of lncRNAs in ovarian cancer: Defining new biomarkers for therapeutic purposes. Drug Discov. Today 2018, 23, 1635–1643. [Google Scholar] [CrossRef]
- Zhang, J.; Han, C.; Song, K.; Chen, W.; Ungerleider, N.; Yao, L.; Ma, W.; Wu, T. The long-noncoding RNA MALAT1 regulates TGF-β/Smad signaling through formation of a lncRNA-protein complex with Smads, SETD2 and PPM1A in hepatic cells. PLoS ONE 2020, 15, e0228160. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Guo, L.; Li, Y.; Zhou, Q.; Li, Z. MALAT1 is an oncogenic long non-coding RNA associated with tumor invasion in non-small cell lung cancer regulated by DNA methylation. Int. J. Clin. Exp. Pathol. 2015, 8, 15903–15910. [Google Scholar]
- Pang, E.-J.; Yang, R.; Fu, X.-B.; Liu, Y.-F. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumor Biol. 2015, 36, 2403–2407. [Google Scholar] [CrossRef]
- Paniri, A.; Akhavan-Niaki, H. Emerging role of IL-6 and NLRP3 inflammasome as potential therapeutic targets to combat COVID-19: Role of lncRNAs in cytokine storm modulation. Life Sci. 2020, 257, 118114. [Google Scholar] [CrossRef]
- Liu, C.L.; Sheu, J.J.-C.; Lin, H.-P.; Jeng, Y.-M.; Chang, C.Y.-Y.; Chen, C.-M.; Cheng, J.; Mao, T.-L. The overexpression of MYST4 in human solid tumors is associated with increased aggressiveness and decreased overall survival. Int. J. Clin. Exp. Pathol. 2019, 12, 431–442. [Google Scholar]
- Zhao, M.; Wang, S.; Li, Q.; Ji, Q.; Guo, P.; Liu, X. MALAT1: A long non-coding RNA highly associated with human cancers. Oncol. Lett. 2018, 16, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Wang, X.; Chen, L.-P.; Li, M.; Li, M.; Hu, Y.-H.; Ding, W.-H.; Wang, X. Long non-coding RNA MALAT1 regulates ovarian cancer cell proliferation, migration and apoptosis through Wnt/β-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3703–3712. [Google Scholar]
- Braga, E.A.; Fridman, M.V.; Moscovtsev, A.A.; Filippova, E.A.; Dmitriev, A.A.; Kushlinskii, N.E. LncRNAs in Ovarian Cancer Progression, Metastasis, and Main Pathways: CeRNA and Alternative Mechanisms. Int. J. Mol. Sci. 2020, 21, 8855. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Yi, F.; Han, X.; Du, Q.; Liang, Z. MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett. 2013, 587, 3175–3181. [Google Scholar] [CrossRef] [Green Version]
- Hewitson, J.P.; West, K.A.; James, K.R.; Rani, G.F.; Dey, N.; Romano, A.; Brown, N.; Teichmann, S.A.; Kaye, P.M.; Lagos, D. Malat1 Suppresses Immunity to Infection through Promoting Expression of Maf and IL-10 in Th Cells. J. Immunol. 2020, 204, 2949–2960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, L.; Zhang, G.; Cheng, Z.; Wang, X.; Jia, L.; Jing, X.; Wang, H.; Zhang, R.; Liu, M.; Jiang, T.; et al. Knockdown of LncRNA MALAT1 contributes to the suppression of inflammatory responses by up-regulating miR-146a in LPS-induced acute lung injury. Connect. Tissue Res. 2018, 59, 581–592. [Google Scholar] [CrossRef]
- Li, H.; Shi, H.; Ma, N.; Zi, P.; Liu, Q.; Sun, R. BML-111 alleviates acute lung injury through regulating the expression of lncRNA MALAT1. Arch. Biochem. Biophys. 2018, 649, 15–21. [Google Scholar] [CrossRef]
- Zhang, X.; Tang, X.; Liu, K.; Hamblin, M.H.; Yin, K.-J. Long Noncoding RNA Malat1 Regulates Cerebrovascular Pathologies in Ischemic Stroke. J. Neurosci. 2017, 37, 1797–1806. [Google Scholar] [CrossRef] [PubMed]
- Yong, H.; Wu, G.; Chen, J.; Liu, X.; Bai, Y.; Tang, N.; Liu, L.; Wei, J. lncRNA MALAT1 Accelerates Skeletal Muscle Cell Apoptosis and Inflammatory Response in Sepsis by Decreasing BRCA1 Expression by Recruiting EZH2. Mol. Ther. - Nucleic Acids 2020, 19, 97–108. [Google Scholar] [CrossRef]
- Gast, M.; Rauch, B.H.; Nakagawa, S.; Haghikia, A.; Jasina, A.; Haas, J.; Nath, N.; Jensen, L.; Stroux, A.; Böhm, A.; et al. Immune system-mediated atherosclerosis caused by deficiency of long non-coding RNA MALAT1 in ApoE-/-mice. Cardiovasc. Res. 2019, 115, 302–314. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, X.; Hu, X.; Zhou, W.; Zhang, P.; Zhang, J.; Yang, S.; Liu, Y. The effects of lncRNA MALAT1 on proliferation, invasion and migration in colorectal cancer through regulating SOX9. Mol. Med. 2018, 24, 1–15. [Google Scholar] [CrossRef]
- Chou, J.; Wang, B.; Zheng, T.; Li, X.; Zheng, L.; Hu, J.; Zhang, Y.; Xing, Y.; Xi, T. MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42. Biochem. Biophys. Res. Commun. 2016, 472, 262–269. [Google Scholar] [CrossRef]
- Ji, Q.; Cai, G.; Liu, X.; Zhang, Y.; Wang, Y.; Zhou, L.; Sui, H.; Li, Q. MALAT1 regulates the transcriptional and translational levels of proto-oncogene RUNX2 in colorectal cancer metastasis. Cell Death Dis. 2019, 10, 1–17. [Google Scholar] [CrossRef]
- Lei, R.; Xue, M.; Zhang, L.; Lin, Z. Long noncoding RNA MALAT1-regulated microRNA 506 modulates ovarian cancer growth by targeting iASPP. OncoTargets Ther. 2016, 10, 35–46. [Google Scholar] [CrossRef] [Green Version]
- Tao, F.; Tian, X.; Ruan, S.; Shen, M.; Zhang, Z. miR-211 sponges lncRNA MALAT1 to suppress tumor growth and progression through inhibiting PHF19 in ovarian carcinoma. FASEB J. 2018, 32, 6330–6343. [Google Scholar] [CrossRef]
- Wu, X.; Wang, Y.; Zhong, W.; Cheng, H.; Tian, Z. The Long Non-Coding RNA MALAT1 Enhances Ovarian Cancer Cell Stemness by Inhibiting YAP Translocation from Nucleus to Cytoplasm. Med Sci. Monit. 2020, 26, 26. [Google Scholar] [CrossRef]
- Bai, L.; Wang, A.; Zhang, Y.; Xu, X.; Zhang, X. Knockdown of MALAT1 enhances chemosensitivity of ovarian cancer cells to cisplatin through inhibiting the Notch1 signaling pathway. Exp. Cell Res. 2018, 366, 161–171. [Google Scholar] [CrossRef]
- Kim, J.; Piao, H.-L.; Kim, B.-J.; Yao, F.; Han, Z.; Wang, Y.; Xiao, Z.; Siverly, A.N.; Lawhon, S.E.; Ton, B.N.; et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 2018, 50, 1705–1715. [Google Scholar] [CrossRef]
- Vogell, A.; Evans, M.L. Cancer Screening in Women. Obstet. Gynecol. Clin. N. Am. 2019, 46, 485–499. [Google Scholar]
- Luan, C.; Li, Y.; Liu, Z.; Zhao, C. Long Noncoding RNA MALAT1 Promotes the Development of Colon Cancer by Regulating miR-101-3p/STC1 Axis. OncoTargets Ther. 2020, 13, 3653–3665. [Google Scholar] [CrossRef]
- Pei, C.; Gong, X.; Zhang, Y. LncRNA MALAT-1 promotes growth and metastasis of epithelial ovarian cancer via sponging microrna-22. Am. J. Transl. Res. 2020, 12, 6977–6987. [Google Scholar] [PubMed]
- Wei, S.; Wang, K.; Huang, X.; Zhao, Z.; Zhao, Z. LncRNA MALAT1 contributes to non-small cell lung cancer progression via modulating miR-200a-3p/programmed death-ligand 1 axis. Int. J. Immunopathol. Pharmacol. 2019, 33, 2058738419859699. [Google Scholar] [CrossRef] [Green Version]
- Xiao, H.; Zhu, Q.; Zhou, J. Long non-coding RNA MALAT1 interaction with miR-429 regulates the proliferation and EMT of lung adenocarcinoma cells through RhoA. Int. J. Clin. Exp. Pathol. 2019, 12, 419–430. [Google Scholar] [PubMed]
- Stone, J.K.; Kim, J.-H.; Vukadin, L.; Richard, A.; Giannini, H.K.; Lim, S.-T.S.; Tan, M.; Ahn, E.-Y.E. Hypoxia induces cancer cell-specific chromatin interactions and increases MALAT1 expression in breast cancer cells. J. Biol. Chem. 2019, 294, 11213–11224. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, X.; Qiao, L.; Liu, W.; Xu, C.; Wang, X. MALAT1 regulates miR-34a expression in melanoma cells. Cell Death Dis. 2019, 10, 389. [Google Scholar] [CrossRef]
- Mao, X.; Su, Z.; Mookhtiar, A.K. Long non-coding RNA: A versatile regulator of the nuclear factor-κB signalling circuit. Immunology 2017, 150, 379–388. [Google Scholar] [CrossRef] [Green Version]
- Hirata, H.; Hinoda, Y.; Shahryari, V.; Deng, G.; Nakajima, K.; Tabatabai, Z.L.; Ishii, N.; Dahiya, R. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res. 2015, 75, 1322–1331. [Google Scholar] [CrossRef] [Green Version]
- Zhao, G.; Su, Z.; Song, D.; Mao, Y.; Mao, X. The long noncoding RNA MALAT1 regulates the lipopolysaccharide-induced inflammatory response through its interaction with NF-kappaB. FEBS Lett. 2016, 590, 2884–2895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Zhang, H.; Zheng, Y.; Jin, X.; Liu, M.; Li, S.; Zhao, Q.; Liu, X.; Wang, Y.; Shi, M.; et al. The Long Noncoding RNA MALAT1 Induces Tolerogenic Dendritic Cells and Regulatory T Cells via miR155/Dendritic Cell-Specific Intercellular Adhesion Molecule-3 Grabbing Nonintegrin/IL10 Axis. Front. Immunol. 2018, 9, 1847. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Song, Y.; Liu, F.; Liu, D.; Miao, H.; Ren, J.; Xu, J.; Ding, L.; Hu, Y.; Wang, Z.; et al. Long Non-Coding RNA MALAT1 Promotes Proliferation, Angiogenesis, and Immunosuppressive Properties of Mesenchymal Stem Cells by Inducing VEGF and IDO. J. Cell. Biochem. 2017, 118, 2780–2791. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.L.; Chen, Y.J.; Fan, M.H.; Liao, Y.J.; Mao, T.L. Characteristics of CD133-Sustained Chemoresistant Cancer Stem-Like Cells in Human Ovarian Carcinoma. Int. J. Mol. Sci. 2020, 21, 6467. [Google Scholar] [CrossRef]
- Liu, S.; Jiang, X.; Li, W.; Cao, D.; Shen, K.; Yang, J. Inhibition of the long non-coding RNA MALAT1 suppresses tumorigenicity and induces apoptosis in the human ovarian cancer SKOV3 cell line. Oncol. Lett. 2016, 11, 3686–3692. [Google Scholar] [CrossRef]
- Colvin, E.K.; Howell, V.M.; Mok, S.C.; Samimi, G.; Vafaee, F. Expression of long noncoding RNAs in cancer-associated fibroblasts linked to patient survival in ovarian cancer. Cancer Sci. 2020, 111, 1805–1817. [Google Scholar] [CrossRef]
- Yang, C.-Y.; Fan, M.H.; Miao, C.H.; Liao, Y.J.; Yuan, R.-H.; Liu, C.L. Engineering Chimeric Antigen Receptor T Cells against Immune Checkpoint Inhibitors PD-1/PD-L1 for Treating Pancreatic Cancer. Mol. Ther. - Oncolytics 2020, 17, 571–585. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Primer Sequences | GenBank No. |
|---|---|---|
| MYST4 F | 5′ AACCT GTTCC AGAGC CAATG 3′ | XM_867653 |
| MYST4 R | 5′ TGTAC TTTTG CAGGT GATTG 3′ | |
| MALAT1 F | 5′ GGTTG AGATG AAGCT TCTT 3′ | NR002819.4 |
| MALAT1 R | 5′ GCACT TCTTG TGTTC TCTT 3′ | |
| TNF-α F | 5′ TACTG ATACA TACAA CCACG 3′ | NT_167244.2 |
| TNF-α R | 5′ ATTTT CAGGT CCCCA GCAGG 3′ | |
| IL-6 F | 5′ TTGAG GGAAA AGAGT AGAGG 3′ | NC_000007.14 |
| IL-6 R | 5′ TAGGC TTTCT ATGGT GGTGG 3′ | |
| YAP F | 5′ ATCAGATCGTGCACGTCC 3′ | AB567721.1 |
| YAP R | 5′ ATCAGTACTGGCCTGTCG 3′ | |
| Cox2 F | 5′ AGTGCGATTGTACCCGG 3′ | AY462100.1 |
| Cox2 R | 5′ TTGCATTTCGAAGGAAGG 3′ | |
| PGE2 F | 5′ AACCCTGTGCGCAGGGC 3′ | NM_198938.3 |
| PGE2 R | 5′ TTGCCATTTGGCCTCCC 3′ | |
| IL-1β F | 5′ AAGCTGAGGAAGATGCTGG 3′ | NM_000576.3 |
| IL-1β R | 5′ TTGCTGTGAGTCCCGGAGC 3′ | |
| Cyclin-D1 F | 5′ AAGGCGGAGGAGACCTGC 3′ | NM_053056.3 |
| Cyclin-D1 R | 5′ ATGGCCAGCGGGAAGACC 3′ | |
| Caspase-3 F | 5′ TACCT GTGGC TGTGT ATCC 3′ | NC_000004.12 |
| Caspase-3 R | 5′ AAACT CACTG AAGTG CTGC 3′ | |
| Bcl2 F | 5′ TTGTC ATCAG CTCGC TCTCC 3′ | XR_935248.3 |
| Bcl2 R | 5′ TATCC GGCAC ATGGA GGCGG 3′ | |
| ZEB2 F | 5’ AAGTG AGAAG GGGTG TCC 3’ | NR149354.1 |
| ZEB2 R | 5’ TTTCC TGGGC TGGAT AGC 3’ | |
| E-Cadherin F | 5′ TTTCC CTCAA GGCTG CAGG 3′ | NC_000074.7 |
| E-Cadherin R | 5′ ATGCC AGGAG GCCGC TCTC 3′ | |
| Vimentin F | 5′ TAAACCGCTAGGAGCCC 3′ | NM_003380.5 |
| Vimentin R | 5′ ATGGCTGCGGAGGGTGG 3′ | |
| S26 F | 5’ CCGTG CCTCC AAGAT GACAA AG 3’ | NM001029.5 |
| S26 R | 5’ GTTCG GTCCT TGCGG GCTTC AC 3’ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, T.-L.; Fan, M.-H.; Dlamini, N.; Liu, C.-L. LncRNA MALAT1 Facilitates Ovarian Cancer Progression through Promoting Chemoresistance and Invasiveness in the Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 10201. https://doi.org/10.3390/ijms221910201
Mao T-L, Fan M-H, Dlamini N, Liu C-L. LncRNA MALAT1 Facilitates Ovarian Cancer Progression through Promoting Chemoresistance and Invasiveness in the Tumor Microenvironment. International Journal of Molecular Sciences. 2021; 22(19):10201. https://doi.org/10.3390/ijms221910201
Chicago/Turabian StyleMao, Tsui-Lien, Ming-Huei Fan, Nhlanhla Dlamini, and Chao-Lien Liu. 2021. "LncRNA MALAT1 Facilitates Ovarian Cancer Progression through Promoting Chemoresistance and Invasiveness in the Tumor Microenvironment" International Journal of Molecular Sciences 22, no. 19: 10201. https://doi.org/10.3390/ijms221910201
APA StyleMao, T.-L., Fan, M.-H., Dlamini, N., & Liu, C.-L. (2021). LncRNA MALAT1 Facilitates Ovarian Cancer Progression through Promoting Chemoresistance and Invasiveness in the Tumor Microenvironment. International Journal of Molecular Sciences, 22(19), 10201. https://doi.org/10.3390/ijms221910201







