Targeting Amino Acid Metabolic Reprogramming via L-Type Amino Acid Transporter 1 (LAT1) for Endocrine-Resistant Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients Cohorts and Sample Selection
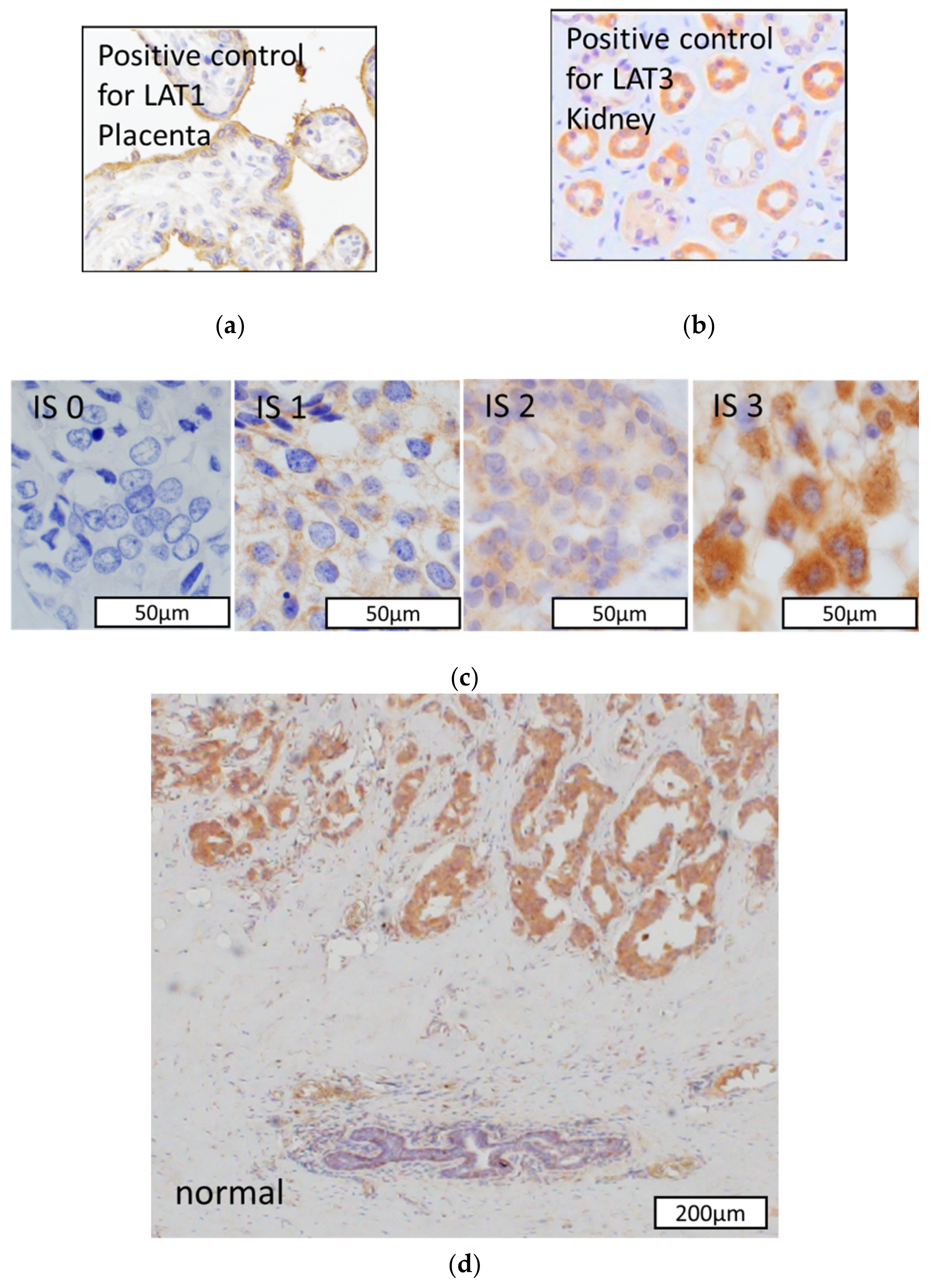
2.2. Immunohistochemistry
2.3. Scoring of Immunohistoreactivity
2.4. Cell Lines and Culture
2.5. Western Blotting
2.6. Cell Proliferation Assay
2.7. Metabolic Analysis
2.8. Amino Acid Uptake with Radiotracers
2.9. Statistical Analysis
3. Results
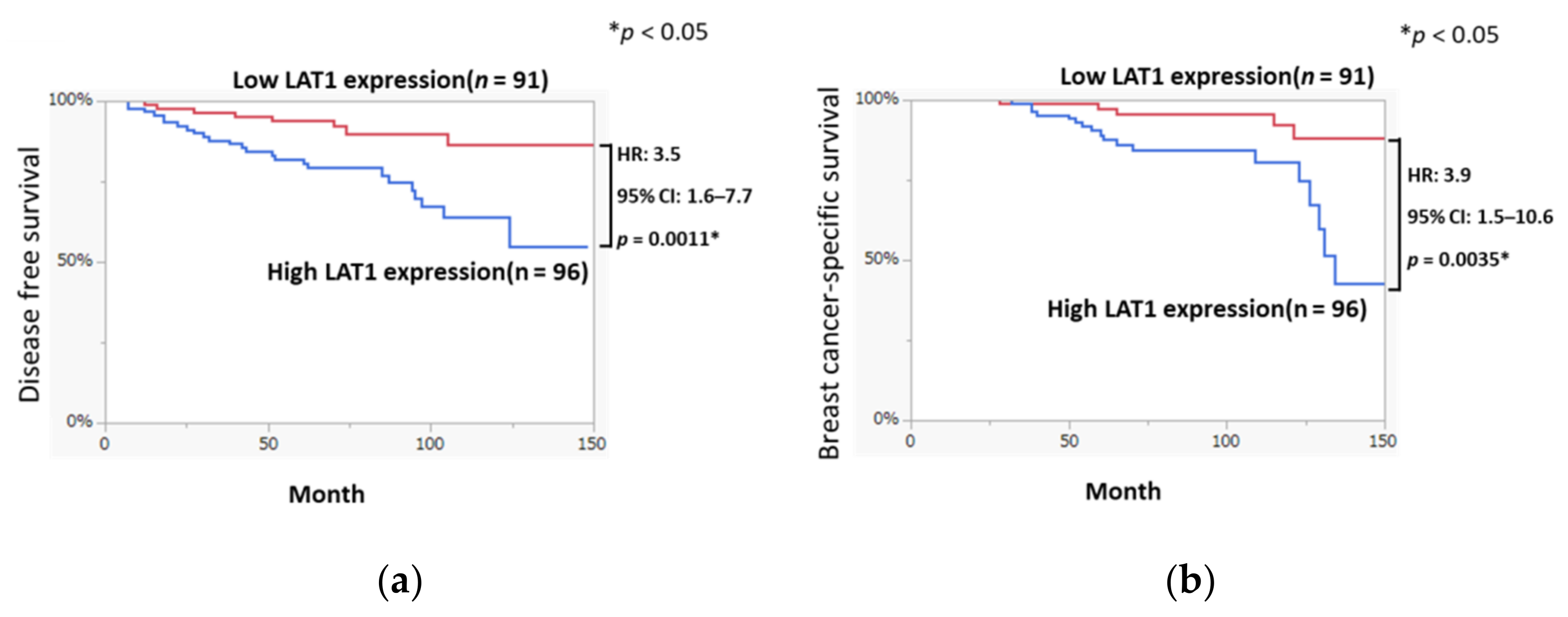
3.1. LAT1 Expression Was Associated with Disease-Free Survival and Breast Cancer-Specific Survival in Breast Cancer
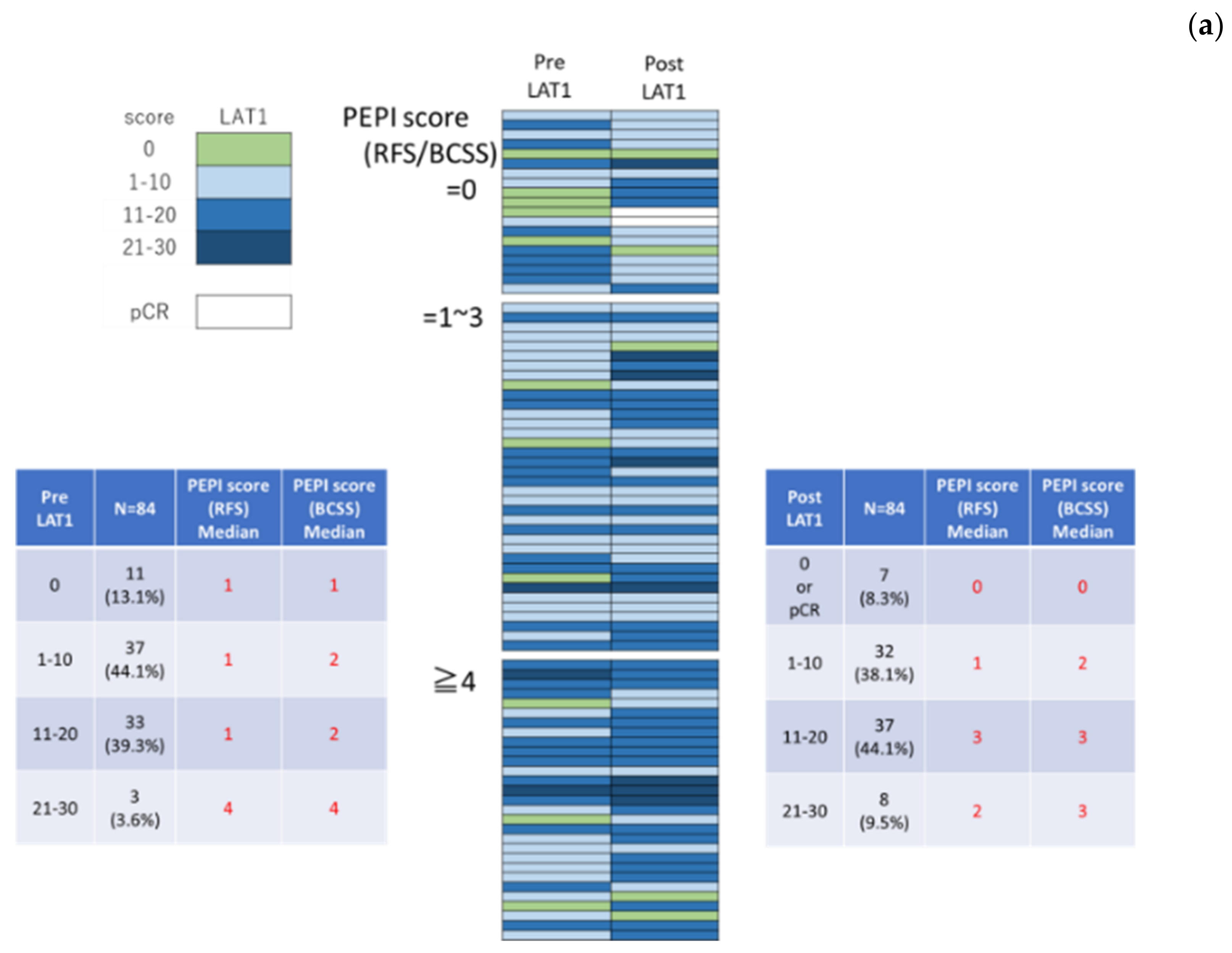
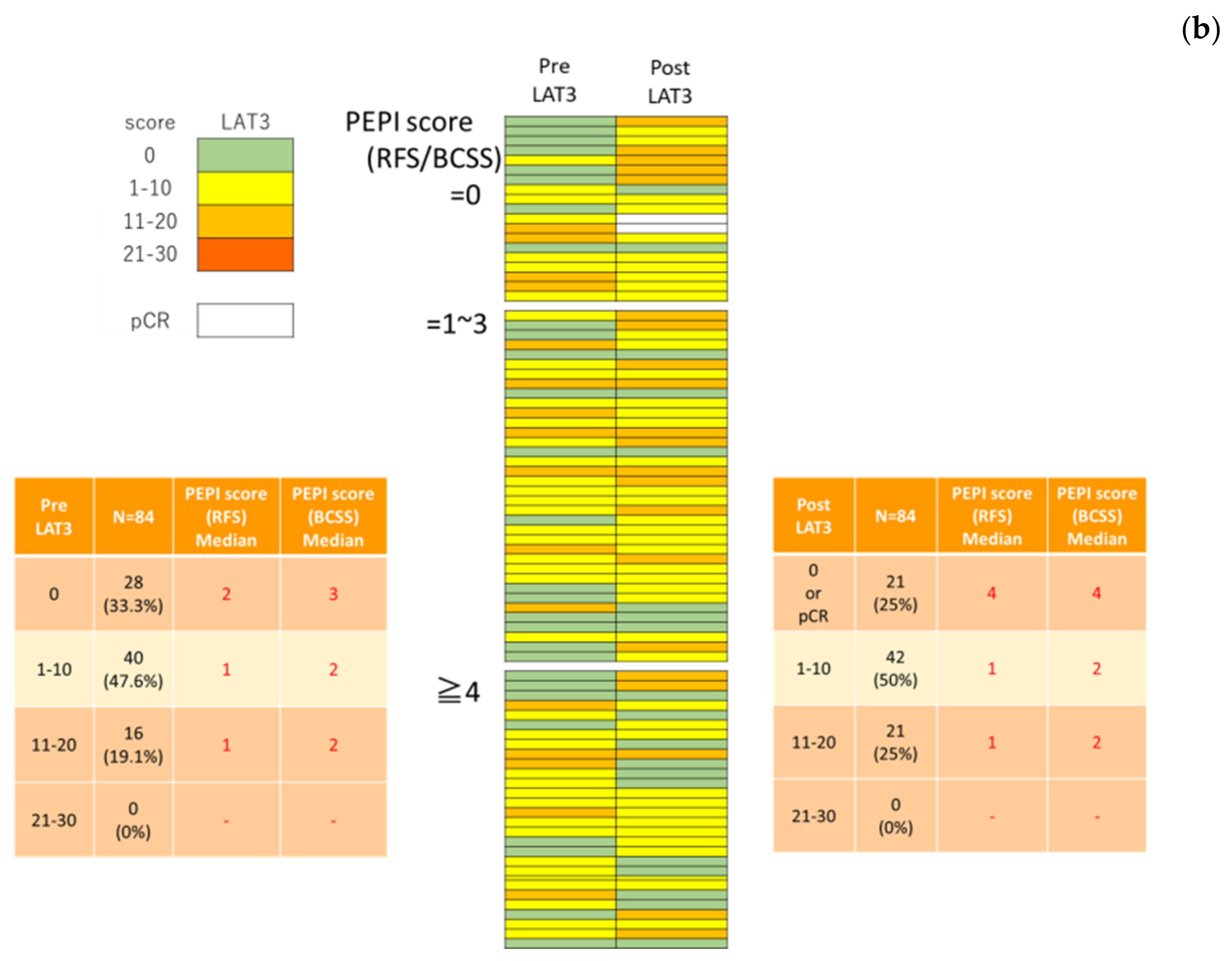
3.2. LAT1 Expression Was Altered by Neoadjuvant Hormone Therapy
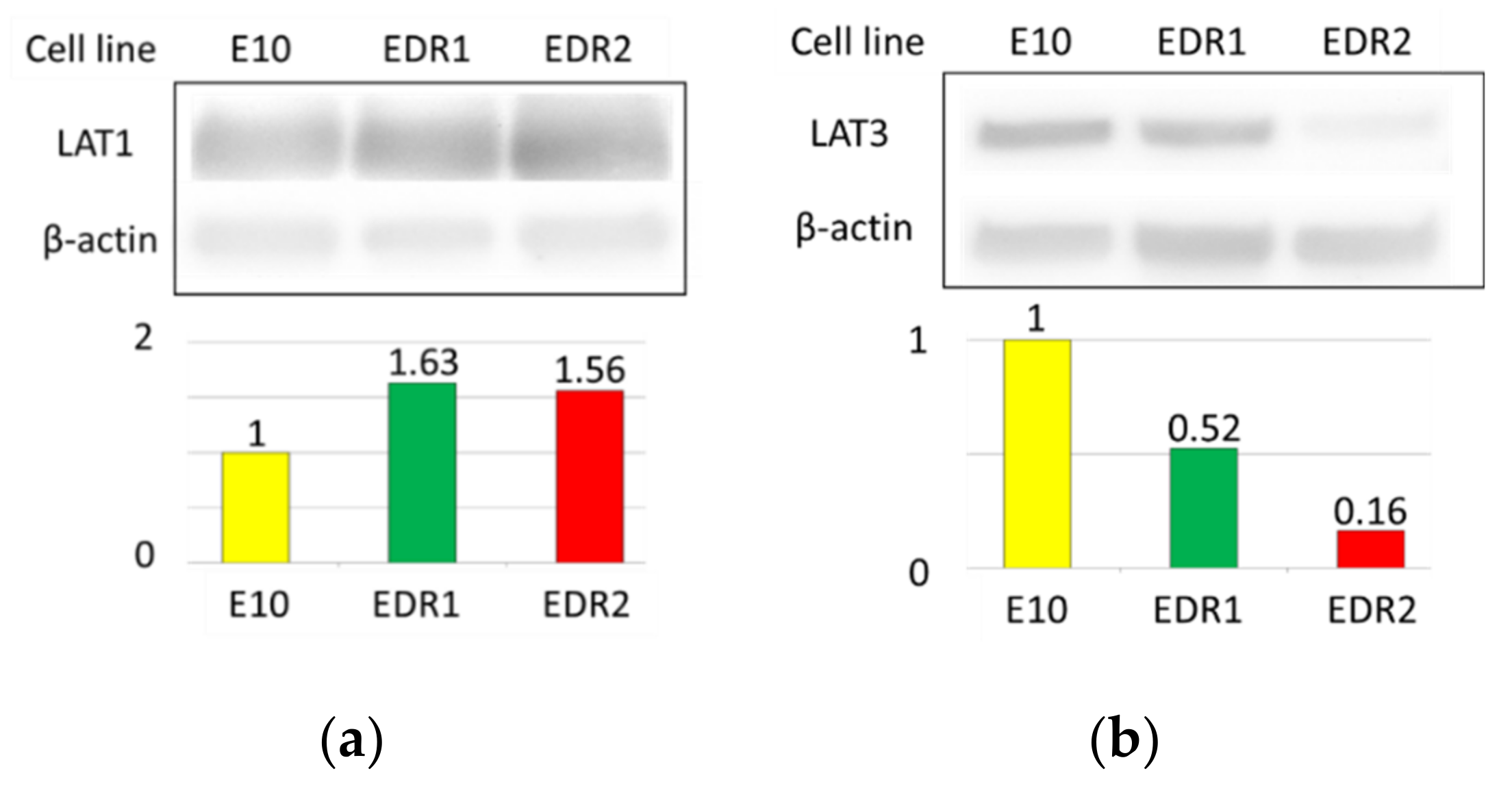
3.3. LAT1 Was Overexpressed in Estrogen Deprivation-Resistant Cells (Aromatase Inhibitor-Resistant Breast Carcinoma Model Cells)
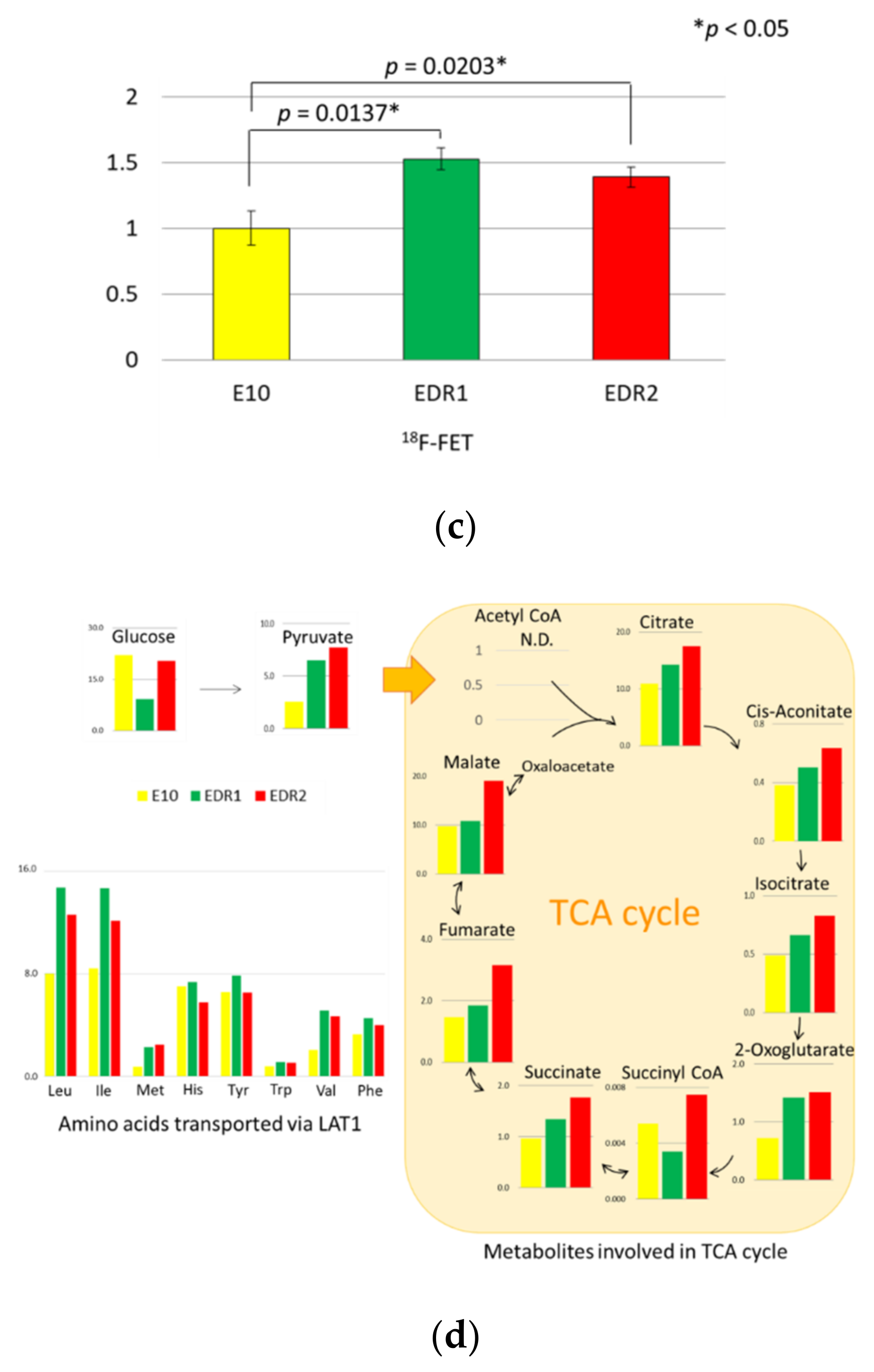
3.4. Amino Acids via LAT1 Are Enhanced in Breast Carcinoma Cells
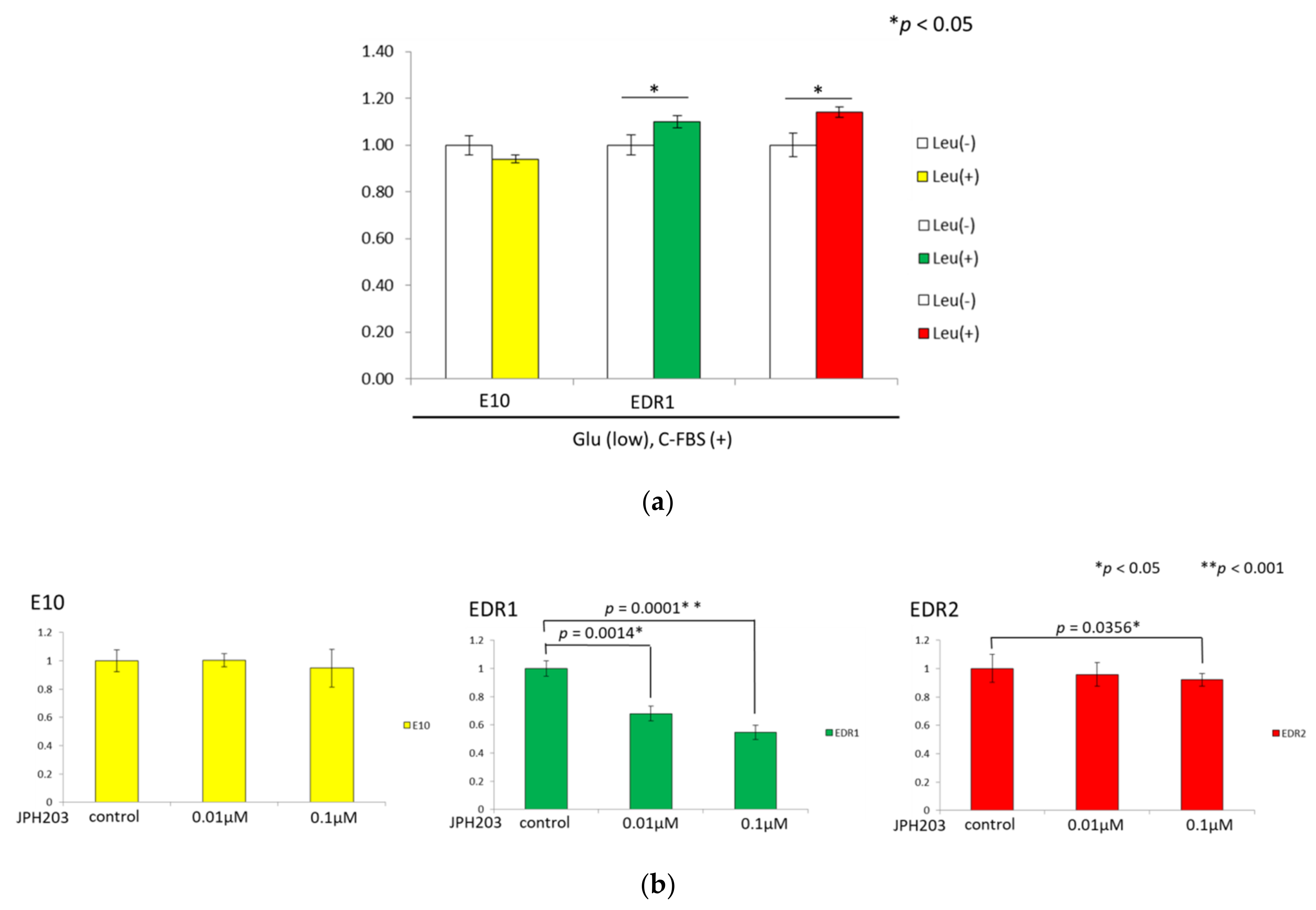
3.5. Leucine Uptake Reliefs Nutrient Stress in Estrogen Deprivation-Resistant Cells
3.6. JPH203 Inhibits the Proliferation of Estrogen Deprivation-Resistant Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iida, M.; Tsuboi, K.; Niwa, T.; Ishida, T.; Hayashi, S.I. Compensatory role of insulin-like growth factor 1 receptor in estrogen receptor signaling pathway and possible therapeutic target for hormone therapy-resistant breast cancer. Breast Cancer 2019, 26, 272–281. [Google Scholar] [CrossRef]
- Qin, C.; Yang, G.; Yang, J.; Ren, B.; Wang, H.; Chen, G.; Zhao, F.; You, L.; Wang, W.; Zhao, Y. Metabolism of pancreatic cancer: Paving the way to better anticancer strategies. Mol. Cancer 2020, 19, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harada-Shoji, N.; Soga, T.; Tada, H.; Miyashita, M.; Harada, M.; Watanabe, G.; Hamanaka, Y.; Sato, A.; Suzuki, T.; Suzuki, A.; et al. A metabolic profile of routine needle biopsies identified tumor type specific metabolic signatures for breast cancer stratification: A pilot study. Metabolomics 2019, 15, 147. [Google Scholar] [CrossRef] [PubMed]
- Eagle, H. Nutrition needs of mammalian cells in tissue culture. Science 1955, 122, 501–504. [Google Scholar] [CrossRef]
- Sato, M.; Harada-Shoji, N.; Toyohara, T.; Soga, T.; Itoh, M.; Miyashita, M.; Tada, H.; Amari, M.; Anzai, N.; Furumoto, S.; et al. L-type amino acid transporter 1 is associated with chemoresistance in breast cancer via the promotion of amino acid metabolism. Sci. Rep. 2021, 11, 589. [Google Scholar] [CrossRef]
- Wang, Q.; Holst, J. L-type amino acid transport and cancer: Targeting the mTORC1 pathway to inhibit neoplasia. Am. J. Cancer Res. 2015, 5, 1281–1294. [Google Scholar]
- Liang, Z.; Cho, H.T.; Williams, L.; Zhu, A.; Liang, K.; Huang, K.; Wu, H.; Jiang, C.; Hong, S.; Crowe, R.; et al. Potential biomarker of L-type amino acid transporter 1 in breast cancer progression. Nucl. Med. Mol. Imaging 2011, 45, 93–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanai, Y.; Segawa, H.; Miyamoto, K.; Uchino, H.; Takeda, E.; Endou, H. Expression cloning and characterization of a transporter for large neutral amino acids activated by the heavy chain of 4F2 antigen (CD98). J. Biol. Chem. 1998, 273, 23629–23632. [Google Scholar] [CrossRef] [Green Version]
- Hafliger, P.; Charles, R.P. The L-type amino acid transporter LAT1-an emerging target in cancer. Int. J. Mol. Sci. 2019, 20, 2428. [Google Scholar] [CrossRef] [Green Version]
- Saito, Y.; Li, L.; Coyaud, E.; Luna, A.; Sander, C.; Raught, B.; Asara, J.M.; Brown, M.; Muthuswamy, S.K. LLGL2 rescues nutrient stress by promoting leucine uptake in ER(+) breast cancer. Nature 2019, 569, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Baselga, J.; Campone, M.; Piccart, M.; Burris, H.A.; Rugo, H.S.; Sahmoud, T.; Noguchi, S.; Gnant, M.; Pritchard, K.I.; Lebrun, F.; et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N. Engl. J. Med. 2012, 366, 520–529. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Bailey, C.G.; Ng, C.; Tiffen, J.; Thoeng, A.; Minhas, V.; Lehman, M.L.; Hendy, S.C.; Buchanan, G.; Nelson, C.C.; et al. Androgen receptor and nutrient signaling pathways coordinate the demand for increased amino acid transport during prostate cancer progression. Cancer Res. 2011, 71, 7525–7536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.K.; Moran, A.M.; Bailey, C.G.; Rasko, J.E.J.; Holst, J.; Wang, Q. EGF-activated PI3K/Akt signalling coordinates leucine uptake by regulating LAT3 expression in prostate cancer. Cell Commun. Signal. 2019, 17, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okano, N.; Naruge, D.; Kawai, K.; Kobayashi, T.; Nagashima, F.; Endou, H.; Furuse, J. First-in-human phase I study of JPH203, an L-type amino acid transporter 1 inhibitor, in patients with advanced solid tumors. Investig. New Drugs 2020, 38, 1495–1506. [Google Scholar] [CrossRef]
- Horii, R.; Akiyama, F. Histological assessment of therapeutic response in breast cancer. Breast Cancer 2016, 23, 540–545. [Google Scholar] [CrossRef]
- Nosaka, K.; Makishima, K.; Sakabe, T.; Yurugi, Y.; Wakahara, M.; Kubouchi, Y.; Horie, Y.; Umekita, Y. Upregulation of glucose and amino acid transporters in micropapillary carcinoma. Histol. Histopathol. 2019, 34, 1009–1014. [Google Scholar] [PubMed]
- Fujiki, N.; Konno, H.; Kaneko, Y.; Gohno, T.; Hanamura, T.; Imami, K.; Ishihama, Y.; Nakanishi, K.; Niwa, T.; Seino, Y.; et al. Estrogen Response element-GFP (ERE-GFP) introduced MCF-7 cells demonstrated the coexistence of multiple estrogen-deprivation resistant mechanisms. J. Steroid Biochem. Mol. Biol. 2014, 139, 61–72. [Google Scholar] [CrossRef]
- Tsuboi, K.; Kaneko, Y.; Nagatomo, T.; Fujii, R.; Hanamura, T.; Gohno, T.; Yamaguchi, Y.; Niwa, T.; Hayashi, S.I. Different epigenetic mechanisms of ERα implicated in the fate of fulvestrant-resistant breast cancer. J. Steroid Biochem. Mol. Biol. 2017, 167, 115–125. [Google Scholar] [CrossRef]
- Iwabuchi, E.; Miki, Y.; Ono, K.; Onodera, Y.; Suzuki, T.; Hirakawa, H.; Ishida, T.; Ohuchi, N.; Sasano, H. In situ detection of estrogen receptor dimers in breast carcinoma cells in archival materials using proximity ligation assay (PLA). J. Steroid Biochem. Mol. Biol. 2017, 165, 159–169. [Google Scholar] [CrossRef]
- Shibahara, Y.; Miki, Y.; Onodera, Y.; Hata, S.; Chan, M.S.; Yiu, C.C.; Loo, T.Y.; Nakamura, Y.; Akahira, J.; Ishida, T.; et al. Aromatase inhibitor treatment of breast cancer cells increases the expression of let-7f, a micro RNA targeting CYP19A1. J. Pathol. 2012, 227, 357–366. [Google Scholar] [CrossRef]
- Hirayama, A.; Kami, K.; Sugimoto, M.; Sugawara, M.; Toki, N.; Onozuka, H.; Kinoshita, T.; Saito, N.; Ochiai, A.; Tomita, M.; et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009, 69, 4918–4925. [Google Scholar] [CrossRef] [Green Version]
- Habermeier, A.; Graf, J.; Sandhöfer, B.F.; Boissel, J.P.; Roesch, F.; Closs, E.I. System L amino acid transporter LAT1 accumulates O-(2-fluoroethyl)-L-tyrosine (FET). Amino Acids 2015, 47, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Salisbury, T.B.; Arthur, S. The regulation and function of the L-type amino acid transporter 1 (LAT1) in cancer. Int. J. Mol. Sci. 2018, 19, 2373. [Google Scholar] [CrossRef] [Green Version]
- Furuya, M.; Horiguchi, J.; Nakajima, H.; Kanai, Y.; Oyama, T. Correlation of L-type amino acid transporter 1 and CD98 expression with triple negative breast cancer prognosis. Cancer Sci. 2012, 103, 382–819. [Google Scholar] [CrossRef]
- Sevigny, C.M.; Sengupta, S.; Luo, Z.; Liu, X.; Hu, R.; Zhang, Z.; Jin, L.; Pearce, D.; Demas, D.; Shajahan-Haq, A.N.; et al. SLCs contribute to endocrine resistance in breast cancer: Role of SLC7A5 (LAT1). bioRxiv 2019. [Google Scholar] [CrossRef]
- Ananieva, E.A.; Wilkinson, A.C. Branched-chain amino acid metabolism in cancer. Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 64–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ananieva, E. Targeting amino acid metabolism in cancer growth and anti-tumor immune response. World J. Biol. Chem. 2015, 6, 281–289. [Google Scholar] [CrossRef]
- Higuchi, K.; Sakamoto, S.; Ando, K.; Maimaiti, M.; Takeshita, N.; Okunushi, K.; Reien, Y.; Imamura, Y.; Sazuka, T.; Nakamura, K.; et al. Characterization of the expression of LAT1 as a prognostic indicator and a therapeutic target in renal cell carcinoma. Sci. Rep. 2019, 9, 16776. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Sakamoto, S.; Matsushima, J.; Kimura, T.; Ueda, T.; Mizokami, A.; Kanai, Y.; Ichikawa, T. Up-regulation of LAT1 during antiandrogen therapy contributes to progression in prostate cancer cells. J. Urol. 2016, 195, 1588–1597. [Google Scholar] [CrossRef]
- Kongpracha, P.; Nagamori, S.; Wiriyasermkul, P.; Tanaka, Y.; Kaneda, K.; Okuda, S.; Ohgaki, R.; Kanai, Y. Structure-activity relationship of a novel series of inhibitors for cancer type transporter L-type amino acid transporter1 (LAT1). J. Pharmacol. Sci. 2017, 133, 96–103. [Google Scholar] [CrossRef]
- Häfliger, P.; Graff, J.; Rubin, M.; Stooss, A.; Dettmer, M.S.; Altmann, K.H.; Gertsch, J.; Charles, R.P. The LAT1 inhibitor JPH203 reduces growth of thyroid carcinoma in a fully immunocompetent mouse model. J. Exp. Clin. Cancer Res. 2018, 37, 234. [Google Scholar] [CrossRef] [PubMed]
- Yun, D.W.; Lee, S.A.; Park, M.G.; Kim, J.S.; Yu, S.K.; Park, M.R.; Kim, S.G.; Oh, J.S.; Kim, C.S.; Kim, H.J.; et al. JPH203, an L-type amino acid transporter 1-selective compound, induces apoptosis of YD-38 human oral cancer cells. J. Pharmacol. Sci. 2014, 124, 208–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaira, K.; Oriuchi, N.; Takahashi, T.; Nakagawa, K.; Ohde, Y.; Okumura, T.; Murakami, H.; Shukuya, T.; Kenmotsu, H.; Naito, T.; et al. LAT1 expression is closely associated with hypoxic markers and mTOR in resected non-small cell lung cancer. Am. J. Transl. Res. 2011, 3, 468–478. [Google Scholar] [PubMed]
| n = 187 | % | Positive | % | ||
|---|---|---|---|---|---|
| Age, Median (years) | 55 | LAT1 | |||
| (27–88) | pTis | 1 | 50.0 | ||
| pT1 | 66 | 48.9 | |||
| pT | pT2 | 16 | 66.7 | ||
| pTis | 2 | 1.1 | pT3 | 0 | 0 |
| pT1 | 135 | 72.2 | pT4 | 13 | 59.1 |
| pT2 | 24 | 12.8 | Total | 96 | 51.3 |
| pT3 | 4 | 2.1 | |||
| pT4 | 22 | 11.8 | |||
| pN | |||||
| pN0 | 127 | 67.9 | |||
| pN1< | 60 | 32.1 | |||
| Ki-67, Median (%) | 10 | ||||
| (1–53) | |||||
| Premenopausal | 67 | 35.8 | |||
| Postmenopausal | 120 | 64.2 |
| n = 84 | % | Positive | |||
|---|---|---|---|---|---|
| Age, Median (years) | 71.5 | Pre LAT1 | |||
| (53–90) | cTis | 1 | 50.0 | ||
| cT1 | 12 | 32.4 | |||
| cT | cT2 | 14 | 63.6 | ||
| cTis | 2 | 2.4 | cT3 | 1 | 25.0 |
| cT1 | 37 | 44.1 | cT4 | 8 | 42.1 |
| cT2 | 22 | 26.2 | Total | 36 | 42.9 |
| cT3 | 4 | 4.8 | |||
| cT4 | 19 | 22.6 | Pre LAT3 | ||
| cTis | 0 | 0 | |||
| cN | cT1 | 23 | 62.2 | ||
| cN0 | 59 | 70.2 | cT2 | 14 | 63.6 |
| cN1< | 25 | 29.8 | cT3 | 4 | 100 |
| cT4 | 15 | 78.9 | |||
| Pre Ki-67, Median (%) | 13.0 | Total | 56 | 66.7 | |
| (0.1–80) | |||||
| Medicine of NAH | |||||
| ANA | 35 | 41.7 | |||
| LET | 46 | 54.8 | |||
| EXE | 3 | 3.6 |
| Pre LAT1 | Post LAT1 | Pre LAT3 | Post LAT3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Total Score Mean | p-Value | Total Score Mean | p-Value | Total Score Mean | p-Value | Total Score Mean | p-Value | ||
| Total | 10.0 (0–30) | 12.5 (0–30) | 6.5 (0–30) | 8.2 (0–30) | |||||
| pStage 0–I | 8.9 | 0.0778 | 9.7 | 0.0003 * | 6.6 | 0.4783 | 9.3 | 0.0474 * | |
| II–IV | 11.0 | 15.4 | 6.5 | 7.0 | |||||
| pT is–1 | 9.6 | 0.2528 | 11.1 | 0.0096 * | 6.4 | 0.3835 | 9.1 | 0.0386 * | |
| 2–4 | 10.6 | 15.1 | 6.8 | 6.6 | |||||
| pN − | 9.4 | 0.1655 | 11.4 | 0.0365 * | 6.5 | 0.4981 | 9.0 | 0.0519 | |
| + | 11.0 | 14.6 | 6.6 | 6.7 | |||||
| Histological Grade | 1 | 9.4 | 0.2606 | 10.4 | 0.0058 * | 5.8 | 0.1661 | 9.0 | 0.1254 |
| (HG) | 2–3 | 10.4 | 14.6 | 7.1 | 7.4 | ||||
| Post Ki-67/ Pre Ki-67 | <1.00 | 9.5 | 0.1332 | 12.0 | 0.1251 | 6.8 | 0.2144 | 8.5 | 0.258 |
| 1.00≤ | 11.5 | 14.3 | 5.7 | 7.4 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shindo, H.; Harada-Shoji, N.; Ebata, A.; Sato, M.; Soga, T.; Miyashita, M.; Tada, H.; Kawai, M.; Kosaka, S.; Onuki, K.; et al. Targeting Amino Acid Metabolic Reprogramming via L-Type Amino Acid Transporter 1 (LAT1) for Endocrine-Resistant Breast Cancer. Cancers 2021, 13, 4375. https://doi.org/10.3390/cancers13174375
Shindo H, Harada-Shoji N, Ebata A, Sato M, Soga T, Miyashita M, Tada H, Kawai M, Kosaka S, Onuki K, et al. Targeting Amino Acid Metabolic Reprogramming via L-Type Amino Acid Transporter 1 (LAT1) for Endocrine-Resistant Breast Cancer. Cancers. 2021; 13(17):4375. https://doi.org/10.3390/cancers13174375
Chicago/Turabian StyleShindo, Haruhiko, Narumi Harada-Shoji, Akiko Ebata, Miku Sato, Tomoyoshi Soga, Minoru Miyashita, Hiroshi Tada, Masaaki Kawai, Shinkichi Kosaka, Koji Onuki, and et al. 2021. "Targeting Amino Acid Metabolic Reprogramming via L-Type Amino Acid Transporter 1 (LAT1) for Endocrine-Resistant Breast Cancer" Cancers 13, no. 17: 4375. https://doi.org/10.3390/cancers13174375
APA StyleShindo, H., Harada-Shoji, N., Ebata, A., Sato, M., Soga, T., Miyashita, M., Tada, H., Kawai, M., Kosaka, S., Onuki, K., Usami, S., Furumoto, S., Hayashi, S., Abe, T., Suzuki, T., Ishida, T., & Sasano, H. (2021). Targeting Amino Acid Metabolic Reprogramming via L-Type Amino Acid Transporter 1 (LAT1) for Endocrine-Resistant Breast Cancer. Cancers, 13(17), 4375. https://doi.org/10.3390/cancers13174375







