The Liquid Biopsy for Lung Cancer: State of the Art, Limitations and Future Developments
Abstract
:Simple Summary
Abstract
1. Introduction
1.1. Lung Cancer Screening and Surveillance/Response Assessment
1.2. Liquid Biopsy in Lca Diagnosis and Molecular Assessment
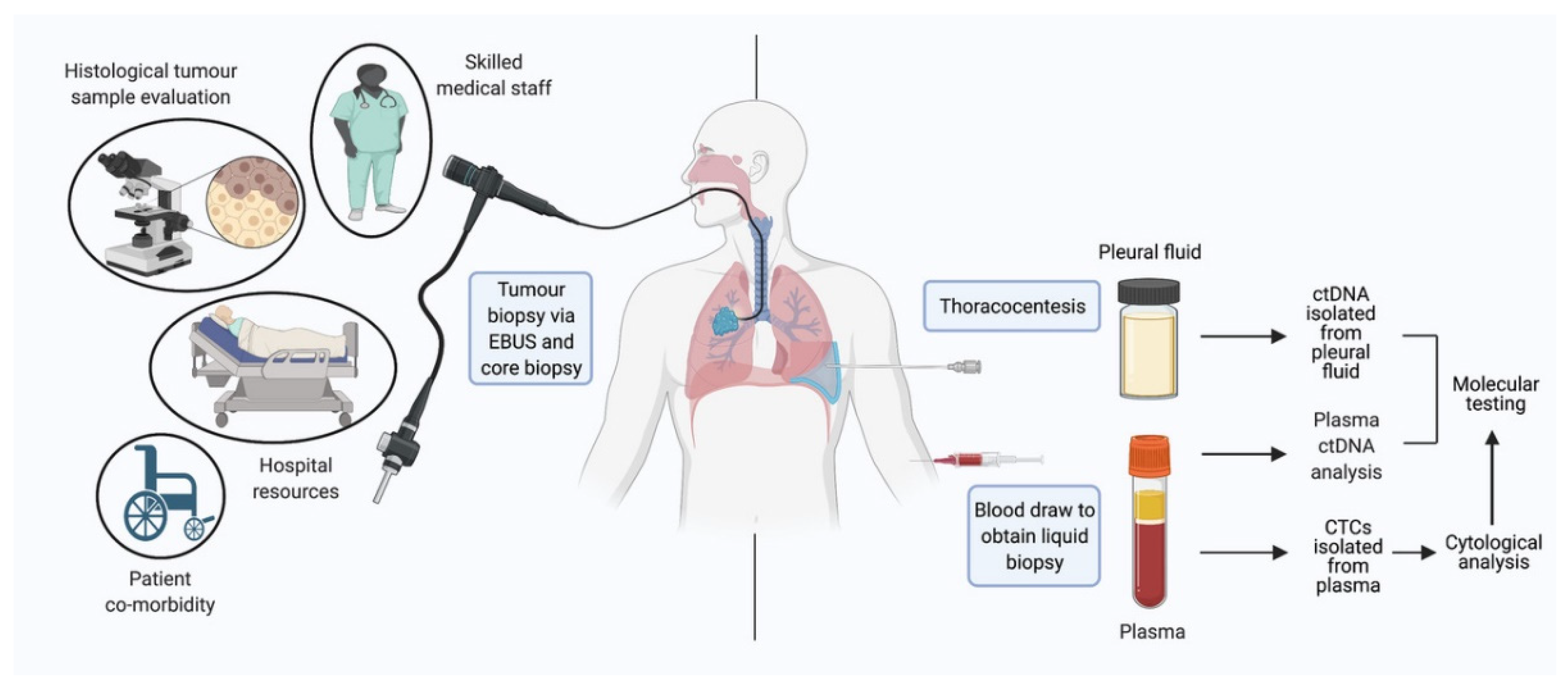
2. Circulating Tumor Cells
2.1. Capture/Isolation
2.2. Clinical Use
3. Circulating Tumor DNA
3.1. Methodology Considerations
3.2. Molecular Testing
3.3. Clinical Use
3.4. ctDNA in Other Fluids—Pleural and CSF
3.5. ctDNA Limitations
3.6. Minimal Residual Disease
4. Future Clinical Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Globocan. Cancer Fact Sheets, Lung Cancer 2018. Available online: http://gco.iarc.fr/today/fact-sheets-cancers (accessed on 16 October 2020).
- American Lung Association. Lung Cancer Fact Sheet. Available online: https://www.lung.org/lung-health-diseases/lung-disease-lookup/lung-cancer/resource-library/lung-cancer-fact-sheet (accessed on 24 November 2020).
- Arbour, K.C.; Riely, G.J. Systemic therapy for locally advanced and metastatic non-small cell lung cancer: A review. JAMA 2019, 322, 764–774. [Google Scholar] [CrossRef]
- Barlesi, F.; Mazieres, J.; Merlio, J.P.; Debieuvre, D.; Mosser, J.; Lena, H.; Ouafik, L.; Besse, B.; Rouquette, I.; Westeel, V.; et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: Results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet 2016, 387, 1415–1426. [Google Scholar] [CrossRef]
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.B.; Barlesi, F.; Lolkema, M.P.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F.; et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020, 31, 1491–1505. [Google Scholar] [CrossRef]
- Morgensztern, D.; Ng, S.H.; Gao, F.; Govindan, R. Trends in stage distribution for patients with non-small cell lung cancer: A National Cancer Database survey. J. Thorac. Oncol. 2010, 5, 29–33. [Google Scholar] [CrossRef] [Green Version]
- Fintelmann, F.J.; Troschel, F.M.; Kuklinski, M.W.; McDermott, S.; Petranovic, M.; Digumarthy, S.R.; Sharma, A.; Troschel, A.S.; Price, M.C.; Hariri, L.P.; et al. Safety and success of repeat lung needle biopsies in patients with epidermal growth factor receptor-mutant lung cancer. Oncologist 2019, 24, 1570–1576. [Google Scholar] [CrossRef] [Green Version]
- Heerink, W.J.; de Bock, G.H.; de Jonge, G.J.; Groen, H.J.; Vliegenthart, R.; Oudkerk, M. Complication rates of CT-guided transthoracic lung biopsy: Meta-analysis. Eur. Radiol. 2017, 27, 138–148. [Google Scholar] [CrossRef] [Green Version]
- Chouaid, C.; Dujon, C.; Do, P.; Monnet, I.; Madroszyk, A.; Le Caer, H.; Auliac, J.B.; Berard, H.; Thomas, P.; Lena, H.; et al. Feasibility and clinical impact of re-biopsy in advanced non small-cell lung cancer: A prospective multicenter study in a real-world setting (GFPC study 12-01). Lung Cancer 2014, 86, 170–173. [Google Scholar] [CrossRef]
- Gregg, J.P.; Li, T.; Yoneda, K.Y. Molecular testing strategies in non-small cell lung cancer: Optimizing the diagnostic journey. Transl. Lung Cancer Res. 2019, 8, 286–301. [Google Scholar] [CrossRef] [Green Version]
- Jamal-Hanjani, M.; Wilson, G.A.; McGranahan, N.; Birkbak, N.J.; Watkins, T.B.K.; Veeriah, S.; Shafi, S.; Johnson, D.H.; Mitter, R.; Rosenthal, R.; et al. Tracking the evolution of non–small-cell lung cancer. N. Engl. J. Med. 2017, 376, 2109–2121. [Google Scholar] [CrossRef] [Green Version]
- de Bruin, E.C.; McGranahan, N.; Mitter, R.; Salm, M.; Wedge, D.C.; Yates, L.; Jamal-Hanjani, M.; Shafi, S.; Murugaesu, N.; Rowan, A.J.; et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014, 346, 251. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Fujimoto, J.; Zhang, J.; Wedge, D.C.; Song, X.; Zhang, J.; Seth, S.; Chow, C.W.; Cao, Y.; Gumbs, C.; et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 2014, 346, 256. [Google Scholar] [CrossRef] [Green Version]
- Imamura, F.; Uchida, J.; Kukita, Y.; Kumagai, T.; Nishino, K.; Inoue, T.; Kimura, M.; Oba, S.; Kato, K. Monitoring of treatment responses and clonal evolution of tumor cells by circulating tumor DNA of heterogeneous mutant EGFR genes in lung cancer. Lung Cancer 2016, 94, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.C.; Reis-Filho, J.S. Genetic heterogeneity and cancer drug resistance. Lancet Oncol. 2012, 13, e178–e185. [Google Scholar] [CrossRef]
- Siravegna, G.; Mussolin, B.; Buscarino, M.; Corti, G.; Cassingena, A.; Crisafulli, G.; Ponzetti, A.; Cremolini, C.; Amatu, A.; Lauricella, C.; et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 2015, 21, 795–801. [Google Scholar] [CrossRef] [Green Version]
- Dexter, D.L.; Leith, J.T. Tumor heterogeneity and drug resistance. J. Clin. Oncol. 1986, 4, 244–257. [Google Scholar] [CrossRef]
- Lovly, C.M.; Salama, A.K.S.; Salgia, R. Tumor heterogeneity and therapeutic resistance. Am. Soc. Clin. Oncol. Educ. Book 2016, 36, e585–e593. [Google Scholar] [CrossRef]
- Schneider, B.J.; Ismaila, N.; Aerts, J.; Chiles, C.; Daly, M.E.; Detterbeck, F.C.; Hearn, J.W.D.; Katz, S.I.; Leighl, N.B.; Levy, B.; et al. Lung cancer surveillance after definitive curative-intent therapy: ASCO guideline. J. Clin. Oncol. 2019, 38, 753–766. [Google Scholar] [CrossRef]
- de Koning, H.J.; van der Aalst, C.M.; de Jong, P.A.; Scholten, E.T.; Nackaerts, K.; Heuvelmans, M.A.; Lammers, J.J.; Weenink, C.; Yousaf-Khan, U.; Horeweg, N.; et al. Reduced lung-cancer mortality with volume CT screening in a randomized trial. N. Engl. J. Med. 2020, 382, 503–513. [Google Scholar] [CrossRef]
- Orme, N.M.; Fletcher, J.G.; Siddiki, H.A.; Harmsen, W.S.; O’Byrne, M.M.; Port, J.D.; Tremaine, W.J.; Pitot, H.C.; McFarland, E.G.; Robinson, M.E.; et al. Incidental findings in imaging research: Evaluating incidence, benefit, and burden. Arch. Intern. Med. 2010, 170, 1525–1532. [Google Scholar] [CrossRef]
- Chapman, C.J.; Healey, G.F.; Murray, A.; Boyle, P.; Robertson, C.; Peek, L.J.; Allen, J.; Thorpe, A.J.; Hamilton-Fairley, G.; Parsy-Kowalska, C.B.; et al. EarlyCDT(R)-Lung test: Improved clinical utility through additional autoantibody assays. Tumour Biol. 2012, 33, 1319–1326. [Google Scholar] [CrossRef] [Green Version]
- Kearney, P.; Hunsucker, S.W.; Li, X.J.; Porter, A.; Springmeyer, S.; Mazzone, P. An integrated risk predictor for pulmonary nodules. PLoS ONE 2017, 12, e0177635. [Google Scholar] [CrossRef] [Green Version]
- Vachani, A.; Whitney, D.H.; Parsons, E.C.; Lenburg, M.; Ferguson, J.S.; Silvestri, G.A.; Spira, A. Clinical utility of a bronchial genomic classifier in patients with suspected lung cancer. Chest 2016, 150, 210–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostrin, E.J.; Sidransky, D.; Spira, A.; Hanash, S.M. Biomarkers for lung cancer screening and detection. Cancer Epidemiol. Biomark. Prev. 2020, 29, 2411–2415. [Google Scholar] [CrossRef]
- Hofman, V.; Bonnetaud, C.; Ilie, M.I.; Vielh, P.; Vignaud, J.M.; Fléjou, J.F.; Lantuejoul, S.; Piaton, E.; Mourad, N.; Butori, C.; et al. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with lung cancer is a new prognostic biomarker. Clin. Cancer Res. 2011, 17, 827–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayarri-Lara, C.; Ortega, F.G.; Cueto Ladrón de Guevara, A.; Puche, J.L.; Ruiz Zafra, J.; de Miguel-Pérez, D.; Ramos, A.S.; Giraldo-Ospina, C.F.; Navajas Gómez, J.A.; Delgado-Rodriguez, M.; et al. Circulating tumor cells identify early recurrence in patients with non-small cell lung cancer undergoing radical resection. PLoS ONE 2016, 11, e0148659. [Google Scholar] [CrossRef]
- Benešová, L.; Hálková, T.; Ptáčková, R.; Semyakina, A.; Menclová, K.; Pudil, J.; Ryska, M.; Levý, M.; Šimša, J.; Pazdírek, F.; et al. Significance of postoperative follow-up of patients with metastatic colorectal cancer using circulating tumor DNA. World J. Gastroenterol. 2019, 25, 6939–6948. [Google Scholar] [CrossRef]
- Lee, B.; Lipton, L.; Cohen, J.; Tie, J.; Javed, A.A.; Li, L.; Goldstein, D.; Burge, M.; Cooray, P.; Nagrial, A.; et al. Circulating tumor DNA as a potential marker of adjuvant chemotherapy benefit following surgery for localized pancreatic cancer. Ann. Oncol. 2019, 30, 1472–1478. [Google Scholar] [CrossRef]
- Massihnia, D.; Pizzutilo, E.G.; Amatu, A.; Tosi, F.; Ghezzi, S.; Bencardino, K.; Di Masi, P.; Righetti, E.; Patelli, G.; Scaglione, F.; et al. Liquid biopsy for rectal cancer: A systematic review. Cancer Treat. Rev. 2019, 79, 101893. [Google Scholar] [CrossRef]
- Suzuki, C.; Jacobsson, H.; Hatschek, T.; Torkzad, M.R.; Bodén, K.; Eriksson-Alm, Y.; Berg, E.; Fujii, H.; Kubo, A.; Blomqvist, L. Radiologic measurements of tumor response to treatment: Practical approaches and limitations. Radiographics 2008, 28, 329–344. [Google Scholar] [CrossRef] [Green Version]
- Jia, W.; Gao, Q.; Han, A.; Zhu, H.; Yu, J. The potential mechanism, recognition and clinical significance of tumor pseudoprogression after immunotherapy. Cancer Biol. Med. 2019, 16, 655–670. [Google Scholar]
- Katz, S.I.; Hammer, M.; Bagley, S.J.; Aggarwal, C.; Bauml, J.M.; Thompson, J.C.; Nachiappan, A.C.; Simone, C.B.; Langer, C.J. Radiologic pseudoprogression during anti-PD-1 therapy for advanced non-small cell lung cancer. J. Thorac. Oncol. 2018, 13, 978–986. [Google Scholar] [CrossRef] [Green Version]
- Sestini, S.; Boeri, M.; Marchiano, A.; Pelosi, G.; Galeone, C.; Verri, C.; Suatoni, P.; Sverzellati, N.; La Vecchia, C.; Sozzi, G.; et al. Circulating microRNA signature as liquid-biopsy to monitor lung cancer in low-dose computed tomography screening. Oncotarget 2015, 6, 32868–32877. [Google Scholar] [CrossRef] [Green Version]
- Syrigos, K.; Fiste, O.; Charpidou, A.; Grapsa, D. Circulating tumor cells count as a predictor of survival in lung cancer. Crit. Rev. Oncol Hematol. 2018, 125, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Vendrell, J.A.; Mau-Them, F.T.; Béganton, B.; Godreuil, S.; Coopman, P.; Solassol, J. Circulating cell free tumor DNA detection as a routine tool for lung cancer patient management. Int. J. Mol. Sci. 2017, 18, 264. [Google Scholar] [CrossRef]
- Sozzi, G.; Conte, D.; Mariani, L.; Lo Vullo, S.; Roz, L.; Lombardo, C.; Pierotti, M.A.; Tavecchio, L. Analysis of circulating tumor DNA in plasma at diagnosis and during follow-up of lung cancer patients. Cancer Res. 2001, 61, 4675. [Google Scholar]
- Krishnamurthy, N.; Spencer, E.; Torkamani, A.; Nicholson, L. Liquid biopsies for cancer: Coming to a patient near you. J. Clin. Med. 2017, 6, 3. [Google Scholar] [CrossRef] [Green Version]
- Dawson, S.-J.; Tsui, D.W.Y.; Murtaza, M.; Biggs, H.; Rueda, O.M.; Chin, S.-F.; Dunning, M.J.; Gale, D.; Forshew, T.; Mahler-Araujo, B.; et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013, 368, 1199–1209. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Forbes, M.E.; Bitting, R.L.; O’Neill, S.S.; Chou, P.C.; Topaloglu, U.; Miller, L.D.; Hawkins, G.A.; Grant, S.C.; DeYoung, B.R.; et al. Incorporating blood-based liquid biopsy information into cancer staging: Time for a TNMB system? Ann. Oncol. 2018, 29, 311–323. [Google Scholar] [CrossRef] [Green Version]
- Loveland, P.; Christie, M.; Hammerschlag, G.; Irving, L.; Steinfort, D. Diagnostic yield of pleural fluid cytology in malignant effusions: An Australian tertiary centre experience. Intern. Med. J. 2018, 48, 1318–1324. [Google Scholar] [CrossRef]
- Brun, C.; Gay, P.; Cottier, M.; Karpathiou, G.; Patoir, A.; Tiffet, O.; Barral, F.G.; Vergnon, J.M.; Froudarakis, M.E. Comparison of cytology, chest computed and positron emission tomography findings in malignant pleural effusion from lung cancer. J. Thorac. Dis. 2018, 10, 6903–6911. [Google Scholar] [CrossRef] [PubMed]
- Glantz, M.J.; Cole, B.F.; Glantz, L.K.; Cobb, J.; Mills, P.; Lekos, A.; Walters, B.C.; Recht, L.D. Cerebrospinal fluid cytology in patients with cancer: Minimizing false-negative results. Cancer 1998, 82, 733–739. [Google Scholar] [CrossRef]
- Guo, Z.; Xie, Z.; Shi, H.; Du, W.; Peng, L.; Han, W.; Duan, F.; Zhang, X.; Chen, M.; Duan, J.; et al. Malignant pleural effusion supernatant is an alternative liquid biopsy specimen for comprehensive mutational profiling. Thorac. Cancer 2019, 10, 823–831. [Google Scholar] [CrossRef]
- Villatoro, S.; Mayo-de-Las-Casas, C.; Jordana-Ariza, N.; Viteri-Ramirez, S.; Garzon-Ibanez, M.; Moya-Horno, I.; Garcia-Pelaez, B.; Gonzalez-Cao, M.; Malapelle, U.; Balada-Bel, A.; et al. Prospective detection of mutations in cerebrospinal fluid, pleural effusion, and ascites of advanced cancer patients to guide treatment decisions. Mol. Oncol. 2019, 13, 2633–2645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeo, C.D.; Kim, J.W.; Kim, K.H.; Ha, J.H.; Rhee, C.K.; Kim, S.J.; Kim, Y.K.; Park, C.K.; Lee, S.H.; Park, M.S.; et al. Detection and comparison of EGFR mutations in matched tumor tissues, cell blocks, pleural effusions, and sera from patients with NSCLC with malignant pleural effusion, by PNA clamping and direct sequencing. Lung Cancer 2013, 81, 207–212. [Google Scholar] [CrossRef]
- Meng, S.; Tripathy, D.; Frenkel, E.P.; Shete, S.; Naftalis, E.Z.; Huth, J.F.; Beitsch, P.D.; Leitch, M.; Hoover, S.; Euhus, D.; et al. Circulating tumor cells in patients with breast cancer dormancy. Clin. Cancer Res. 2004, 10, 8152–8162. [Google Scholar] [CrossRef] [Green Version]
- Nagrath, S.; Sequist, L.V.; Maheswaran, S.; Bell, D.W.; Irimia, D.; Ulkus, L.; Smith, M.R.; Kwak, E.L.; Digumarthy, S.; Muzikansky, A.; et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450, 1235–1239. [Google Scholar] [CrossRef] [Green Version]
- Lou, J.; Ben, S.; Yang, G.; Liang, X.; Wang, X.; Ni, S.; Han, B. Quantification of rare circulating tumor cells in non-small cell lung cancer by ligand-targeted PCR. PLoS ONE 2013, 8, e80458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, T.; Zhu, D.; Yang, Y.; Yuan, G.; Xie, H.; Shen, R. The application of nano-enrichment in CTC detection and the clinical significance of CTCs in non-small cell lung cancer (NSCLC) treatment. PLoS ONE 2019, 14, e0219129. [Google Scholar]
- Freidin, M.B.; Tay, A.; Freydina, D.V.; Chudasama, D.; Nicholson, A.G.; Rice, A.; Anikin, V.; Lim, E. An assessment of diagnostic performance of a filter-based antibody-independent peripheral blood circulating tumour cell capture paired with cytomorphologic criteria for the diagnosis of cancer. Lung Cancer 2014, 85, 182–185. [Google Scholar] [CrossRef]
- Jin, X.R.; Zhu, L.Y.; Qian, K.; Feng, Y.G.; Zhou, J.H.; Wang, R.W.; Bai, L.; Deng, B.; Liang, N.; Tan, Q.Y. Circulating tumor cells in early stage lung adenocarcinoma: A case series report and literature review. Oncotarget 2017, 8, 23130–23141. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Tian, X.; Gao, L.; Jiang, X.; Fu, R.; Zhang, T.; Ren, T.; Hu, P.; Wu, Y.; Zhao, P.; et al. Clinical significance of circulating tumor cells and tumor markers in the diagnosis of lung cancer. Cancer Med. 2019, 8, 3782–3792. [Google Scholar] [CrossRef] [PubMed]
- Poggiana, C.; Rossi, E.; Zamarchi, R. Possible role of circulating tumor cells in early detection of lung cancer. J. Thorac. Dis. 2020, 12, 3821–3835. [Google Scholar] [CrossRef] [PubMed]
- Crosbie, P.A.; Shah, R.; Krysiak, P.; Zhou, C.; Morris, K.; Tugwood, J.; Booton, R.; Blackhall, F.; Dive, C. Circulating tumor cells detected in the tumor-draining pulmonary vein are associated with disease recurrence after surgical resection of NSCLC. J. Thorac. Oncol. 2016, 11, 1793–1797. [Google Scholar] [CrossRef] [Green Version]
- Janning, M.; Kobus, F.; Babayan, A.; Wikman, H.; Velthaus, J.L.; Bergmann, S.; Schatz, S.; Falk, M.; Berger, L.A.; Bottcher, L.M.; et al. Determination of PD-L1 expression in circulating tumor cells of NSCLC patients and correlation with response to PD-1/PD-L1 inhibitors. Cancers 2019, 11, 835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krebs, M.G.; Sloane, R.; Priest, L.; Lancashire, L.; Hou, J.M.; Greystoke, A.; Ward, T.H.; Ferraldeschi, R.; Hughes, A.; Clack, G.; et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J. Clin. Oncol. 2011, 29, 1556–1563. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, C.R.; Blackhall, F.H.; Carmel, A.; Fernandez-Gutierrez, F.; Gazzaniga, P.; Groen, H.J.M.; Hiltermann, T.J.N.; Krebs, M.G.; Loges, S.; Lopez-Lopez, R.; et al. EPAC-lung: Pooled analysis of circulating tumour cells in advanced non-small cell lung cancer. Eur. J. Cancer 2019, 117, 60–68. [Google Scholar] [CrossRef] [Green Version]
- Manjunath, Y.; Upparahalli, S.V.; Suvilesh, K.N.; Avella, D.M.; Kimchi, E.T.; Staveley-O’Carroll, K.F.; Li, G.; Kaifi, J.T. Circulating tumor cell clusters are a potential biomarker for detection of non-small cell lung cancer. Lung Cancer 2019, 134, 147–150. [Google Scholar] [CrossRef]
- Murlidhar, V.; Reddy, R.M.; Fouladdel, S.; Zhao, L.; Ishikawa, M.K.; Grabauskiene, S.; Zhang, Z.; Lin, J.; Chang, A.C.; Carrott, P.; et al. Poor prognosis indicated by venous circulating tumor cell clusters in early-stage lung cancers. Cancer Res. 2017, 77, 5194–5206. [Google Scholar] [CrossRef] [Green Version]
- Tamminga, M.; de Wit, S.; van de Wauwer, C.; van den Bos, H.; Swennenhuis, J.F.; Klinkenberg, T.J.; Hiltermann, T.J.N.; Andree, K.C.; Spierings, D.C.J.; Lansdorp, P.M.; et al. Analysis of released circulating tumor cells during surgery for non-small cell lung cancer. Clin. Cancer Res. 2020, 26, 1656–1666. [Google Scholar] [CrossRef]
- Xu, M.; Zhao, H.; Chen, J.; Liu, W.; Li, E.; Wang, Q.; Zhang, L. An integrated microfluidic chip and its clinical application for circulating tumor cell isolation and single-cell analysis. Cytom. A 2020, 97, 46–53. [Google Scholar] [CrossRef]
- Andree, K.C.; van Dalum, G.; Terstappen, L.W. Challenges in circulating tumor cell detection by the CellSearch system. Mol. Oncol. 2016, 10, 395–407. [Google Scholar] [CrossRef] [Green Version]
- Barr, J.; Chudasama, D.; Rice, A.; Karteris, E.; Anikin, V. Lack of association between Screencell-detected circulating tumour cells and long-term survival of patients undergoing surgery for non-small cell lung cancer: A pilot clinical study. Mol. Clin. Oncol. 2020, 12, 191–195. [Google Scholar] [CrossRef] [Green Version]
- Chudasama, D.; Rice, A.; Anikin, V.; Soppa, G.; Dalal, P. Circulating tumour cells in patients with malignant lung tumors undergoing radio-frequency ablation. Anticancer Res. 2015, 35, 2823–2826. [Google Scholar]
- Alama, A.; Truini, A.; Coco, S.; Genova, C.; Grossi, F. Prognostic and predictive relevance of circulating tumor cells in patients with non-small-cell lung cancer. Drug Discov. Today 2014, 19, 1671–1676. [Google Scholar] [CrossRef]
- Zhou, J.; Kulasinghe, A.; Bogseth, A.; O’Byrne, K.; Punyadeera, C.; Papautsky, I. Isolation of circulating tumor cells in non-small-cell-lung-cancer patients using a multi-flow microfluidic channel. Microsyst. Nanoeng. 2019, 5, 8. [Google Scholar] [CrossRef] [Green Version]
- Sollier, E.; Go, D.E.; Che, J.; Gossett, D.R.; O’Byrne, S.; Weaver, W.M.; Kummer, N.; Rettig, M.; Goldman, J.; Nickols, N.; et al. Size-selective collection of circulating tumor cells using Vortex technology. Lab Chip 2014, 14, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Andree, K.C.; Mentink, A.; Zeune, L.L.; Terstappen, L.; Stoecklein, N.H.; Neves, R.P.; Driemel, C.; Lampignano, R.; Yang, L.; Neubauer, H.; et al. Toward a real liquid biopsy in metastatic breast and prostate cancer: Diagnostic LeukApheresis increases CTC yields in a European prospective multicenter study (CTCTrap). Int. J. Cancer 2018, 143, 2584–2591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, X.L.; Xiao, Z.L.; Liu, L.; Liu, X.X.; Nie, W.; Li, P.; Chen, N.Y.; Wei, Y.Q. Meta-analysis of circulating tumor cells as a prognostic marker in lung cancer. Asian Pac. J. Cancer Prev. 2012, 13, 1137–1144. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Qin, A.; Zhang, K.; Ren, H.; Liu, S.; Liu, X.; Pan, X.; Yu, G. Circulating tumor cells predict prognosis following tyrosine kinase inhibitor treatment in EGFR-mutant non-small cell lung cancer patients. Oncol. Res. 2017, 25, 1601–1606. [Google Scholar] [CrossRef]
- Chemi, F.; Rothwell, D.G.; McGranahan, N.; Gulati, S.; Abbosh, C.; Pearce, S.P.; Zhou, C.; Wilson, G.A.; Jamal-Hanjani, M.; Birkbak, N.; et al. Pulmonary venous circulating tumor cell dissemination before tumor resection and disease relapse. Nat. Med. 2019, 25, 1534–1539. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Tanaka, F.; Yoneda, K.; Takuwa, T.; Matsumoto, S.; Okumura, Y.; Kondo, N.; Tsujimura, T.; Nakano, T.; Hasegawa, S. Positive correlation between postoperative tumor recurrence and changes in circulating tumor cell counts in pulmonary venous blood (pvCTC) during surgical manipulation in non-small cell lung cancer. J. Thorac. Dis. 2018, 10, 298–306. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, M.; Tanaka, F.; Yoneda, K.; Takuwa, T.; Matsumoto, S.; Okumura, Y.; Kondo, N.; Tsubota, N.; Tsujimura, T.; Tabata, C.; et al. Significant increase in circulating tumour cells in pulmonary venous blood during surgical manipulation in patients with primary lung cancer. Interact. Cardiovasc. Thorac. Surg. 2014, 18, 775–783. [Google Scholar] [CrossRef] [Green Version]
- Chudasama, D.; Burnside, N.; Beeson, J.; Karteris, E.; Rice, A.; Anikin, V. Perioperative detection of circulating tumour cells in patients with lung cancer. Oncol. Lett. 2017, 14, 1281–1286. [Google Scholar] [CrossRef] [Green Version]
- Funaki, S.; Sawabata, N.; Nakagiri, T.; Shintani, Y.; Inoue, M.; Kadota, Y.; Minami, M.; Okumura, M. Novel approach for detection of isolated tumor cells in pulmonary vein using negative selection method: Morphological classification and clinical implications. Eur. J. Cardiothorac. Surg. 2011, 40, 322–327. [Google Scholar] [CrossRef]
- Ilie, M.; Hofman, V.; Long-Mira, E.; Selva, E.; Vignaud, J.M.; Padovani, B.; Mouroux, J.; Marquette, C.H.; Hofman, P. “Sentinel” circulating tumor cells allow early diagnosis of lung cancer in patients with chronic obstructive pulmonary disease. PLoS ONE 2014, 9, e111597. [Google Scholar] [CrossRef] [Green Version]
- Marquette, C.H.; Boutros, J.; Benzaquen, J.; Ferreira, M.; Pastre, J.; Pison, C.; Padovani, B.; Bettayeb, F.; Fallet, V.; Guibert, N.; et al. Circulating tumour cells as a potential biomarker for lung cancer screening: A prospective cohort study. Lancet Respir. Med. 2020, 8, 709–716. [Google Scholar] [CrossRef]
- Maheswaran, S.; Sequist, L.V.; Nagrath, S.; Ulkus, L.; Brannigan, B.; Collura, C.V.; Inserra, E.; Diederichs, S.; Iafrate, A.J.; Bell, D.W.; et al. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl. J. Med. 2008, 359, 366–377. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Nong, J.; Wang, J.; Yan, Z.; Yi, L.; Gao, X.; Liu, Z.; Zhang, H.; Zhang, S. Isolation of circulating tumor cells and detection of EGFR mutations in patients with non-small-cell lung cancer. Oncol. Lett. 2019, 17, 3799–3807. [Google Scholar] [CrossRef]
- Pailler, E.; Adam, J.; Barthelemy, A.; Oulhen, M.; Auger, N.; Valent, A.; Borget, I.; Planchard, D.; Taylor, M.; Andre, F.; et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J. Clin. Oncol. 2013, 31, 2273–2281. [Google Scholar] [CrossRef] [Green Version]
- Pailler, E.; Auger, N.; Lindsay, C.R.; Vielh, P.; Islas-Morris-Hernandez, A.; Borget, I.; Ngo-Camus, M.; Planchard, D.; Soria, J.C.; Besse, B.; et al. High level of chromosomal instability in circulating tumor cells of ROS1-rearranged non-small-cell lung cancer. Ann. Oncol. 2015, 26, 1408–1415. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.R.; El Messaoudi, S.; Gahan, P.B.; Anker, P.; Stroun, M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016, 35, 347–376. [Google Scholar] [CrossRef] [Green Version]
- Corcoran, R.B.; Chabner, B.A. Application of Cell-free DNA Analysis to Cancer Treatment. N. Engl. J. Med. 2018, 379, 1754–1765. [Google Scholar] [CrossRef] [Green Version]
- Perez-Barrios, C.; Nieto-Alcolado, I.; Torrente, M.; Jimenez-Sanchez, C.; Calvo, V.; Gutierrez-Sanz, L.; Palka, M.; Donoso-Navarro, E.; Provencio, M.; Romero, A. Comparison of methods for circulating cell-free DNA isolation using blood from cancer patients: Impact on biomarker testing. Transl. Lung Cancer Res. 2016, 5, 665–672. [Google Scholar] [CrossRef] [Green Version]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef]
- Herbreteau, G.; Vallee, A.; Knol, A.C.; Theoleyre, S.; Quereux, G.; Khammari, A.; Dreno, B.; Denis, M.G. Circulating tumour DNA: Analytical aspects and clinical applications for metastatic melanoma patients. Ann. Biol. Clin. 2017, 75, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Montalvo, L.; Chrebtow, V.; Busch, M.P. Quantitation of genomic DNA in plasma and serum samples: Higher concentrations of genomic DNA found in serum than in plasma. Transfusion 2001, 41, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Nikolaev, S.; Lemmens, L.; Koessler, T.; Blouin, J.L.; Nouspikel, T. Circulating tumoral DNA: Preanalytical validation and quality control in a diagnostic laboratory. Anal. Biochem. 2018, 542, 34–39. [Google Scholar] [CrossRef]
- Kang, Q.; Henry, N.L.; Paoletti, C.; Jiang, H.; Vats, P.; Chinnaiyan, A.M.; Hayes, D.F.; Merajver, S.D.; Rae, J.M.; Tewari, M. Comparative analysis of circulating tumor DNA stability In K3EDTA, Streck, and CellSave blood collection tubes. Clin. Biochem. 2016, 49, 1354–1360. [Google Scholar] [CrossRef]
- Chae, Y.K.; Oh, M.S. Detection of minimal residual disease using ctDNA in lung cancer: Current evidence and future directions. J. Thorac. Oncol. 2019, 14, 16–24. [Google Scholar] [CrossRef] [Green Version]
- Rolfo, C.; Mack, P.C.; Scagliotti, G.V.; Baas, P.; Barlesi, F.; Bivona, T.G.; Herbst, R.S.; Mok, T.S.; Peled, N.; Pirker, R.; et al. Liquid biopsy for advanced Non-Small Cell Lung Cancer (NSCLC): A statement paper from the IASLC. J. Thorac. Oncol. 2018, 13, 1248–1268. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Diehl, F.; Dressman, D.; Vogelstein, B.; Kinzler, K.W. BEAMing up for detection and quantification of rare sequence variants. Nat. Methods 2006, 3, 95–97. [Google Scholar] [CrossRef]
- Samorodnitsky, E.; Jewell, B.M.; Hagopian, R.; Miya, J.; Wing, M.R.; Lyon, E.; Damodaran, S.; Bhatt, D.; Reeser, J.W.; Datta, J.; et al. Evaluation of hybridization capture versus amplicon-based methods for whole-exome sequencing. Hum. Mutat. 2015, 36, 903–914. [Google Scholar] [CrossRef] [Green Version]
- Simon, R.; Roychowdhury, S. Implementing personalized cancer genomics in clinical trials. Nat. Rev. Drug. Discov. 2013, 12, 358–369. [Google Scholar] [CrossRef]
- Schenk, D.; Song, G.; Ke, Y.; Wang, Z. Amplification of overlapping DNA amplicons in a single-tube multiplex PCR for targeted next-generation sequencing of BRCA1 and BRCA2. PLoS ONE 2017, 12, e0181062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.H.; Wu, N.C.; Sun, R. A benchmark study on error-correction by read-pairing and tag-clustering in amplicon-based deep sequencing. BMC Genom. 2016, 17, 108. [Google Scholar] [CrossRef] [Green Version]
- Bruno, R.; Fontanini, G. Next generation sequencing for gene fusion analysis in lung cancer: A literature review. Diagnostics 2020, 10, 521. [Google Scholar] [CrossRef] [PubMed]
- Esposito Abate, R.; Frezzetti, D.; Maiello, M.R.; Gallo, M.; Camerlingo, R.; De Luca, A.; De Cecio, R.; Morabito, A.; Normanno, N. Next generation sequencing-based profiling of cell free DNA in patients with advanced non-small cell lung cancer: Advantages and pitfalls. Cancers 2020, 12, 3804. [Google Scholar] [CrossRef] [PubMed]
- Plagnol, V.; Woodhouse, S.; Howarth, K.; Lensing, S.; Smith, M.; Epstein, M.; Madi, M.; Smalley, S.; Leroy, C.; Hinton, J.; et al. Analytical validation of a next generation sequencing liquid biopsy assay for high sensitivity broad molecular profiling. PLoS ONE 2018, 13, e0193802. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Cao, Y.; MacLeay, A.; Lennerz, J.K.; Baig, A.; Frazier, R.P.; Lee, J.; Hu, K.; Pacula, M.; Meneses, E.; et al. Clinical validation of a cell-free DNA gene panel. J. Mol. Diagn. 2019, 21, 632–645. [Google Scholar] [CrossRef]
- Muller, J.N.; Falk, M.; Talwar, J.; Neemann, N.; Mariotti, E.; Bertrand, M.; Zacherle, T.; Lakis, S.; Menon, R.; Gloeckner, C.; et al. Concordance between comprehensive cancer genome profiling in plasma and tumor specimens. J. Thorac. Oncol. 2017, 12, 1503–1511. [Google Scholar] [CrossRef] [Green Version]
- Remon, J.; Lacroix, L.; Jovelet, C.; Caramella, C.; Howarth, K.; Plagnol, V.; Rosenfeld, N.; Morris, C.; Mezquita, L.; Pannet, C.; et al. Real-world utility of an amplicon-based next-generation sequencing liquid biopsy for broad molecular profiling in patients with advanced non-small-cell lung cancer. JCO Precis. Oncol. 2019, 3, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Schrock, A.B.; Welsh, A.; Chung, J.H.; Pavlick, D.; Bernicker, E.H.; Creelan, B.C.; Forcier, B.; Ross, J.S.; Stephens, P.J.; Ali, S.M.; et al. Hybrid capture-based genomic profiling of circulating tumor DNA from patients with advanced non-small cell lung cancer. J. Thorac. Oncol. 2019, 14, 255–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oxnard, G.R.; Thress, K.S.; Alden, R.S.; Lawrance, R.; Paweletz, C.P.; Cantarini, M.; Yang, J.C.; Barrett, J.C.; Janne, P.A. Association between plasma genotyping and outcomes of treatment with Osimertinib (AZD9291) in advanced non-small-cell lung cancer. J. Clin. Oncol. 2016, 34, 3375–3382. [Google Scholar] [CrossRef] [Green Version]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Newman, A.M.; Lovejoy, A.F.; Klass, D.M.; Kurtz, D.M.; Chabon, J.J.; Scherer, F.; Stehr, H.; Liu, C.L.; Bratman, S.V.; Say, C.; et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 2016, 34, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Lin, D.; Wu, C.; Li, X.; Zhao, C.; Zheng, L.; Chuai, S.; Fei, K.; Zhou, C.; Hirsch, F.R. Intratumoral heterogeneity of ALK-rearranged and ALK/EGFR coaltered lung adenocarcinoma. J. Clin. Oncol. 2015, 33, 3701–3709. [Google Scholar] [CrossRef]
- Chaudhuri, A.A.; Chabon, J.J.; Lovejoy, A.F.; Newman, A.M.; Stehr, H.; Azad, T.D.; Khodadoust, M.S.; Esfahani, M.S.; Liu, C.L.; Zhou, L.; et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 2017, 7, 1394–1403. [Google Scholar] [CrossRef] [Green Version]
- Cho, M.S.; Park, C.H.; Lee, S.; Park, H.S. Clinicopathological parameters for circulating tumor DNA shedding in surgically resected non-small cell lung cancer with EGFR or KRAS mutation. PLoS ONE 2020, 15, e0230622. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.S.; Jiang, B.Y.; Yang, J.J.; Zhang, X.C.; Zhang, Z.; Ye, J.Y.; Zhong, W.Z.; Tu, H.Y.; Chen, H.J.; Wang, Z.; et al. Unique genetic profiles from cerebrospinal fluid cell-free DNA in leptomeningeal metastases of EGFR-mutant non-small-cell lung cancer: A new medium of liquid biopsy. Ann. Oncol. 2018, 29, 945–952. [Google Scholar] [CrossRef]
- Papadopoulou, E.; Tsoulos, N.; Tsantikidi, K.; Metaxa-Mariatou, V.; Stamou, P.E.; Kladi-Skandali, A.; Kapeni, E.; Tsaousis, G.; Pentheroudakis, G.; Petrakis, D.; et al. Clinical feasibility of NGS liquid biopsy analysis in NSCLC patients. PLoS ONE 2019, 14, e0226853. [Google Scholar] [CrossRef] [Green Version]
- Sabari, J.K.; Offin, M.; Stephens, D.; Ni, A.; Lee, A.; Pavlakis, N.; Clarke, S.; Diakos, C.I.; Datta, S.; Tandon, N.; et al. A prospective study of circulating tumor DNA to guide matched targeted therapy in lung cancers. J. Natl. Cancer Inst. 2019, 111, 575–583. [Google Scholar] [CrossRef]
- Tailor, T.D.; Rao, X.; Campa, M.J.; Wang, J.; Gregory, S.G.; Patz, E.F., Jr. Whole exome sequencing of cell-free DNA for early lung cancer: A pilot study to differentiate benign from malignant CT-detected pulmonary lesions. Front. Oncol. 2019, 9, 317. [Google Scholar] [CrossRef]
- Tsui, D.W.Y.; Murtaza, M.; Wong, A.S.C.; Rueda, O.M.; Smith, C.G.; Chandrananda, D.; Soo, R.A.; Lim, H.L.; Goh, B.C.; Caldas, C.; et al. Dynamics of multiple resistance mechanisms in plasma DNA during EGFR-targeted therapies in non-small cell lung cancer. EMBO Mol. Med. 2018, 10, e7945. [Google Scholar] [CrossRef]
- Uchida, J.; Kato, K.; Kukita, Y.; Kumagai, T.; Nishino, K.; Daga, H.; Nagatomo, I.; Inoue, T.; Kimura, M.; Oba, S.; et al. Diagnostic accuracy of noninvasive genotyping of EGFR in lung cancer patients by deep sequencing of plasma cell-free DNA. Clin. Chem. 2015, 61, 1191–1196. [Google Scholar] [CrossRef] [Green Version]
- Weber, B.; Meldgaard, P.; Hager, H.; Wu, L.; Wei, W.; Tsai, J.; Khalil, A.; Nexo, E.; Sorensen, B.S. Detection of EGFR mutations in plasma and biopsies from non-small cell lung cancer patients by allele-specific PCR assays. BMC Cancer 2014, 14, 294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, M.; Topaloglu, U.; Petty, W.J.; Pagni, M.; Foley, K.L.; Grant, S.C.; Robinson, M.; Bitting, R.L.; Thomas, A.; Alistar, A.T.; et al. Circulating mutational portrait of cancer: Manifestation of aggressive clonal events in both early and late stages. J. Hematol. Oncol. 2017, 10, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Dong, A.; Li, S.; Ren, X.; Zhang, X. Consistency of genotyping data from simultaneously collected plasma circulating tumor DNA and tumor-DNA in lung cancer patients. J. Thorac. Dis. 2020, 12, 7290–7297. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Han, R.B.; Zhao, J.; Wang, J.; Yang, F.; Zhong, W.; Zhang, L.; Li, L.Y.; Wang, M.Z. Comparison of epidermal growth factor receptor mutation statuses in tissue and plasma in stage I-IV non-small cell lung cancer patients. Respiration 2013, 85, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Fenizia, F.; De Luca, A.; Pasquale, R.; Sacco, A.; Forgione, L.; Lambiase, M.; Iannaccone, A.; Chicchinelli, N.; Franco, R.; Rossi, A.; et al. EGFR mutations in lung cancer: From tissue testing to liquid biopsy. Future Oncol. 2015, 11, 1611–1623. [Google Scholar] [CrossRef]
- Aggarwal, C.; Thompson, J.C.; Black, T.A.; Katz, S.I.; Fan, R.; Yee, S.S.; Chien, A.L.; Evans, T.L.; Bauml, J.M.; Alley, E.W.; et al. Clinical implications of plasma-based genotyping with the delivery of personalized therapy in metastatic non-small cell lung cancer. JAMA Oncol. 2019, 5, 173–180. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, X.; Wang, F.; Zhang, M.; Sun, B.; Yin, W.; Deng, S.; Wan, Y.; Lu, W. The diagnostic accuracy of liquid biopsy in EGFR-mutated NSCLC: A systematic review and meta-analysis of 40 studies. SLAS Technol. 2020, 26, 42–54. [Google Scholar] [CrossRef]
- Mok, T.S.; Wu, Y.-L.; Ahn, M.-J.; Garassino, M.C.; Kim, H.R.; Ramalingam, S.S.; Shepherd, F.A.; He, Y.; Akamatsu, H.; Theelen, W.S.M.E.; et al. Osimertinib or platinum–pemetrexed in EGFR T790M–positive lung cancer. N. Engl. J. Med. 2016, 376, 629–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, T.; Nakashima, C.; Komiya, K.; Kitera, K.; Hirai, M.; Kimura, S.; Aragane, N. Mechanisms of acquired resistance to afatinib clarified with liquid biopsy. PLoS ONE 2018, 13, e0209384. [Google Scholar] [CrossRef] [PubMed]
- Hochmair, M.J.; Buder, A.; Schwab, S.; Burghuber, O.C.; Prosch, H.; Hilbe, W.; Cseh, A.; Fritz, R.; Filipits, M. Liquid-biopsy-based identification of EGFR T790M mutation-mediated resistance to Afatinib treatment in patients with advanced EGFR mutation-positive NSCLC, and subsequent response to Osimertinib. Target. Oncol. 2019, 14, 75–83. [Google Scholar] [CrossRef] [Green Version]
- Ito, K.; Suzuki, Y.; Saiki, H.; Sakaguchi, T.; Hayashi, K.; Nishii, Y.; Watanabe, F.; Hataji, O. Utility of liquid biopsy by improved PNA-LNA PCR clamp method for detecting EGFR mutation at initial diagnosis of non-small-cell lung cancer: Observational study of 190 consecutive cases in clinical practice. Clin Lung Cancer 2018, 19, 181–190. [Google Scholar] [CrossRef]
- Mezquita, L.; Swalduz, A.; Jovelet, C.; Ortiz-Cuaran, S.; Howarth, K.; Planchard, D.; Avrillon, V.; Recondo, G.; Marteau, S.; Benitez, J.C.; et al. Clinical relevance of an amplicon-based liquid biopsy for detecting ALK and ROS1 fusion and resistance mutations in patients with non-small-cell lung cancer. JCO Precis. Oncol. 2020, 4, 272–282. [Google Scholar] [CrossRef]
- Chen, Y.; Guo, W.; Fan, J.; Chen, Y.; Zhang, X.; Chen, X.; Luo, P. The applications of liquid biopsy in resistance surveillance of anaplastic lymphoma kinase inhibitor. Cancer Manag. Res. 2017, 9, 801–811. [Google Scholar] [CrossRef] [Green Version]
- Russo, M.; Misale, S.; Wei, G.; Siravegna, G.; Crisafulli, G.; Lazzari, L.; Corti, G.; Rospo, G.; Novara, L.; Mussolin, B.; et al. Acquired resistance to the TRK inhibitor entrectinib in colorectal cancer. Cancer Discov. 2016, 6, 36–44. [Google Scholar] [CrossRef] [Green Version]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phallen, J.; Sausen, M.; Adleff, V.; Leal, A.; Hruban, C.; White, J.; Anagnostou, V.; Fiksel, J.; Cristiano, S.; Papp, E.; et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl. Med. 2017, 9, eaan2415. [Google Scholar] [CrossRef] [Green Version]
- Cortiula, F.; Pasello, G.; Follador, A.; Nardo, G.; Polo, V.; Scquizzato, E.; Del Conte, A.; Miorin, M.; Giovanis, P.; D’Urso, A.; et al. A multi-center, real-life experience on liquid biopsy practice for EGFR testing in Non-Small Cell Lung Cancer (NSCLC) patients. Diagnostics 2020, 10, 765. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Hagiwara, K.; Han, B.; Tjulandin, S.; Grohe, C.; Yokoi, T.; Morabito, A.; Novello, S.; Arriola, E.; Molinier, O.; et al. ctDNA Determination of EGFR mutation status in European and Japanese patients with advanced NSCLC: The ASSESS study. J. Thorac. Oncol. 2016, 11, 1682–1689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietel, M.; Bubendorf, L.; Dingemans, A.M.; Dooms, C.; Elmberger, G.; Garcia, R.C.; Kerr, K.M.; Lim, E.; Lopez-Rios, F.; Thunnissen, E.; et al. Diagnostic procedures for non-small-cell lung cancer (NSCLC): Recommendations of the European Expert Group. Thorax 2016, 71, 177–184. [Google Scholar] [CrossRef] [Green Version]
- Xiang, C.; Huo, M.; Ma, S.; Guo, L.; Zhao, R.; Teng, H.; Zhang, J.; Han, Y. Molecular profiling for supernatants and matched cell pellets of pleural effusions in non-small-cell lung cancer. J. Mol. Diagn. 2020, 22, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Shao, D.; Deng, Q.; Tang, H.; Wang, J.; Liu, J.; Guo, F.; Lin, Y.; Peng, Z.; Mao, M.; et al. Next generation sequencing-based molecular profiling of lung adenocarcinoma using pleural effusion specimens. J. Thorac. Dis. 2018, 10, 2631–2637. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.R.; Mooney, K.L.; Libiran, P.; Jones, C.D.; Joshi, R.; Lau, H.D.; Stehr, H.; Berry, G.J.; Zehnder, J.L.; Long, S.R.; et al. Targeted deep sequencing of cell-free DNA in serous body cavity fluids with malignant, suspicious, and benign cytology. Cancer Cytopathol. 2020, 128, 43–56. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.; Hua, P.; Liu, N.; Li, Q.; Zhu, X.; Jiang, L.; Zheng, K.; Su, X. Targeted next-generation sequencing in cytology specimens for molecular profiling of lung adenocarcinoma. Int. J. Clin. Exp. Pathol. 2018, 11, 3647–3655. [Google Scholar]
- Wang, C.G.; Zeng, D.X.; Huang, J.A.; Jiang, J.H. Effective assessment of low times MET amplification in pleural effusion after epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs) acquired resistance: Cases report. Medicine 2018, 97, e9021. [Google Scholar] [CrossRef]
- Liao, Y.; Ma, Z.; Zhang, Y.; Li, D.; Lv, D.; Chen, Z.; Li, P.; Ai-Dherasi, A.; Zheng, F.; Tian, J.; et al. Targeted deep sequencing from multiple sources demonstrates increased NOTCH1 alterations in lung cancer patient plasma. Cancer Med. 2019, 8, 5673–5686. [Google Scholar] [CrossRef] [Green Version]
- Smalley, K.S.; Fedorenko, I.V.; Kenchappa, R.S.; Sahebjam, S.; Forsyth, P.A. Managing leptomeningeal melanoma metastases in the era of immune and targeted therapy. Int. J. Cancer. 2016, 139, 1195–1201. [Google Scholar] [CrossRef] [Green Version]
- Cheng, H.; Perez-Soler, R. Leptomeningeal metastases in non-small-cell lung cancer. Lancet Oncol. 2018, 19, e43–e55. [Google Scholar] [CrossRef]
- Bruno, M.K.; Raizer, J. Leptomeningeal metastases from solid tumors (meningeal carcinomatosis). Cancer Treat. Res. 2005, 125, 31–52. [Google Scholar] [PubMed]
- Kesari, S.; Batchelor, T.T. Leptomeningeal metastases. Neurol. Clin. 2003, 21, 25–66. [Google Scholar] [CrossRef]
- Ernani, V.; Stinchcombe, T.E. Management of brain metastases in non-small-cell lung cancer. J. Oncol. Pract. 2019, 15, 563–570. [Google Scholar] [CrossRef]
- Sperduto, P.W.; Yang, T.J.; Beal, K.; Pan, H.; Brown, P.D.; Bangdiwala, A.; Shanley, R.; Yeh, N.; Gaspar, L.E.; Braunstein, S.; et al. Estimating survival in patients with lung cancer and brain metastases: An update of the graded prognostic assessment for lung cancer using molecular markers (Lung-molGPA). JAMA Oncol. 2017, 3, 827–831. [Google Scholar] [CrossRef]
- Xiao, F.; Lv, S.; Zong, Z.; Wu, L.; Tang, X.; Kuang, W.; Zhang, P.; Li, X.; Fu, J.; Xiao, M.; et al. Cerebrospinal fluid biomarkers for brain tumor detection: Clinical roles and current progress. Am. J. Transl. Res. 2020, 12, 1379–1396. [Google Scholar]
- McEwen, A.E.; Leary, S.E.S.; Lockwood, C.M. Beyond the blood: CSF-derived cfDNA for diagnosis and characterization of CNS tumors. Front. Cell. Dev. Biol. 2020, 8, 45. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; He, J.Y.; Zou, Y.L.; Guo, X.S.; Cui, J.Z.; Guo, L.; Bu, H. Evaluating the cerebrospinal fluid ctDNA detection by next-generation sequencing in the diagnosis of meningeal Carcinomatosis. BMC Neurol. 2019, 19, 331. [Google Scholar] [CrossRef] [Green Version]
- Zheng, M.M.; Li, Y.S.; Jiang, B.Y.; Tu, H.Y.; Tang, W.F.; Yang, J.J.; Zhang, X.C.; Ye, J.Y.; Yan, H.H.; Su, J.; et al. Clinical utility of cerebrospinal fluid cell-free DNA as liquid biopsy for leptomeningeal metastases in ALK-rearranged NSCLC. J. Thorac. Oncol. 2019, 14, 924–932. [Google Scholar] [CrossRef]
- Li, N.; Liu, Y.; Duan, J.; Yang, B.; Bai, H.; Sun, R.; Yu, L.; Wang, J. Prognostic significance of molecular characteristics of cerebrospinal fluid for non-small cell lung cancer patients with leptomeningeal metastasis. Thorac. Cancer 2019, 10, 1673–1682. [Google Scholar] [CrossRef] [Green Version]
- Balasubramanian, S.K.; Sharma, M.; Venur, V.A.; Schmitt, P.; Kotecha, R.; Chao, S.T.; Suh, J.H.; Angelov, L.; Mohammadi, A.M.; Vogelbaum, M.A.; et al. Impact of EGFR mutation and ALK rearrangement on the outcomes of non-small cell lung cancer patients with brain metastasis. Neuro. Oncol. 2020, 22, 267–277. [Google Scholar] [CrossRef]
- Ballard, P.; Yates, J.W.; Yang, Z.; Kim, D.W.; Yang, J.C.; Cantarini, M.; Pickup, K.; Jordan, A.; Hickey, M.; Grist, M.; et al. Preclinical comparison of Osimertinib with other EGFR-TKIs in EGFR-mutant NSCLC brain metastases models, and early evidence of clinical brain metastases activity. Clin. Cancer Res. 2016, 22, 5130–5140. [Google Scholar] [CrossRef] [Green Version]
- Iuchi, T.; Shingyoji, M.; Sakaida, T.; Hatano, K.; Nagano, O.; Itakura, M.; Kageyama, H.; Yokoi, S.; Hasegawa, Y.; Kawasaki, K.; et al. Phase II trial of gefitinib alone without radiation therapy for Japanese patients with brain metastases from EGFR-mutant lung adenocarcinoma. Lung Cancer 2013, 82, 282–287. [Google Scholar] [CrossRef]
- Petrelli, F.; Lazzari, C.; Ardito, R.; Borgonovo, K.; Bulotta, A.; Conti, B.; Cabiddu, M.; Capitanio, J.F.; Brighenti, M.; Ghilardi, M.; et al. Efficacy of ALK inhibitors on NSCLC brain metastases: A systematic review and pooled analysis of 21 studies. PLoS ONE 2018, 13, e0201425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ying, S.; Ke, H.; Ding, Y.; Liu, Y.; Tang, X.; Yang, D.; Li, M.; Liu, J.; Yu, B.; Xiang, J.; et al. Unique genomic profiles obtained from cerebrospinal fluid cell-free DNA of non-small cell lung cancer patients with leptomeningeal metastases. Cancer Biol. Ther. 2019, 20, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Steensma, D.P.; Bejar, R.; Jaiswal, S.; Lindsley, R.C.; Sekeres, M.A.; Hasserjian, R.P.; Ebert, E.L. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015, 126, 9–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jan, M.; Ebert, B.L.; Jaiswal, S. Clonal hematopoiesis. Semin. Hematol. 2017, 54, 43–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, M.; Lu, C.; Wang, J.; McLellan, M.D.; Johnson, K.J.; Wendl, M.C.; McMichael, J.F.; Schmidt, H.K.; Yellapantula, V.; Miller, C.A.; et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 2014, 20, 1472–1478. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.G.; Lindsley, R.C.; Mermel, C.H.; Burtt, N.; Chavez, A.; et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014, 371, 2488–2498. [Google Scholar] [CrossRef] [Green Version]
- Chabon, J.J.; Hamilton, E.G.; Kurtz, D.M.; Esfahani, M.S.; Moding, E.J.; Stehr, H.; Schroers-Martin, J.; Nabet, B.Y.; Chen, B.; Chaudhuri, A.A.; et al. Integrating genomic features for non-invasive early lung cancer detection. Nature 2020, 580, 245–251. [Google Scholar] [CrossRef]
- Hu, Y.; Ulrich, B.C.; Supplee, J.; Kuang, Y.; Lizotte, P.H.; Feeney, N.B.; Guibert, N.M.; Awad, M.M.; Wong, K.K.; Janne, P.A.; et al. False-positive plasma genotyping due to clonal hematopoiesis. Clin. Cancer Res. 2018, 24, 4437–4443. [Google Scholar] [CrossRef] [Green Version]
- Mayrhofer, M.; De Laere, B.; Whitington, T.; Van Oyen, P.; Ghysel, C.; Ampe, J.; Ost, P.; Demey, W.; Hoekx, L.; Schrijvers, D.; et al. Cell-free DNA profiling of metastatic prostate cancer reveals microsatellite instability, structural rearrangements and clonal hematopoiesis. Genome Med. 2018, 10, 85. [Google Scholar] [CrossRef]
- Liu, J.; Chen, X.; Wang, J.; Zhou, S.; Wang, C.L.; Ye, M.Z.; Wang, X.Y.; Song, Y.; Wang, Y.Q.; Zhang, L.T.; et al. Biological background of the genomic variations of cf-DNA in healthy individuals. Ann. Oncol. 2019, 30, 464–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, H.T.; Nagayama, S.; Chin, Y.M.; Otaki, M.; Hayashi, R.; Kiyotani, K.; Fukunaga, Y.; Ueno, M.; Nakamura, Y.; Low, S.K. Clinical significance of clonal hematopoiesis in the interpretation of blood liquid biopsy. Mol. Oncol. 2020, 14, 1719–1730. [Google Scholar] [CrossRef] [PubMed]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar] [PubMed]
- Stroun, M.; Lyautey, J.; Lederrey, C.; Olson-Sand, A.; Anker, P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin. Chim. Acta 2001, 313, 139–142. [Google Scholar] [CrossRef]
- Lin, C.C.; Shih, J.Y.; Yu, C.J.; Ho, C.C.; Liao, W.Y.; Lee, J.H.; Tsai, T.H.; Su, K.Y.; Hsieh, M.S.; Chang, Y.L.; et al. Outcomes in patients with non-small-cell lung cancer and acquired Thr790Met mutation treated with osimertinib: A genomic study. Lancet Respir. Med. 2018, 6, 107–116. [Google Scholar] [CrossRef]
- Guo, N.; Lou, F.; Ma, Y.; Li, J.; Yang, B.; Chen, W.; Hua, Y.; Zhang, J.-B.; Zhao, M.-Y.; Wu, W.-J.; et al. Circulating tumor DNA detection in lung cancer patients before and after surgery. Sci. Rep. 2016, 6, 33519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Zhang, J.; Guan, T.; Yang, F.; Lou, F.; Chen, W.; Zhao, M.; Zhang, J.; Chen, S.; Wang, J. Comparison of plasma to tissue DNA mutations in surgical patients with non-small cell lung cancer. J. Thorac. Cardiovasc. Surg. 2017, 154, 1123–1131. [Google Scholar] [CrossRef] [Green Version]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451. [Google Scholar] [CrossRef]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2019, 20, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Luchini, C.; Lawlor, R.T.; Milella, M.; Scarpa, A. Molecular tumor boards in clinical practice. Trends Cancer 2020, 6, 738–744. [Google Scholar] [CrossRef] [PubMed]

| Author | Number of Patients | Isolation Method | Main Findings |
|---|---|---|---|
| Crosbie et al. [55] | 33 | CellSearch | CTC clusters associated with increased overall CTC counts. High CTC counts (>18/7.5 mL blood) associated with reduced DFS and OS. |
| Hofman et al. [26] | 208 | ISET | >50 circulating non-hematological cells were associated with shorter OS and DFS. |
| Janning et al. [56] | 127 | Parsotrix and CellSearch | Parsotix detected at least 1 CTC in 61% of samples compared to 32% for CellSearch. CTCs were PD-L1+ in 47% of cases, PD-L1+ and PD-L1− in 47% and PD-L1− in 7%. Increase in PD-L1+ CTCs seen in disease progression. |
| Jin et al. [52] | 45 | CanPatrol | CTC counts increased with disease progression. CTCs with mesenchymal features more abundant in tumors >2 cm. higher post-operative CTCs associated with tumor progression. |
| Krebs et al. [57] | 101 | CellSearch | Stage IV NSCLC had higher CTCs than stage III NSCLC. <5 CTCs pre-chemotherapy were associated with longer PFS and better OS. |
| Li et al. [53] | 174 | Negative enrichment | CTCs detected in 79.3% of patients. CTCs showed a higher diagnostic efficacy compared to serum tumor markers. |
| Lindsay et al. [58] | 550 | CellSearch | CTC counts of >2/7.5 mL blood was associated with reduced PFS, and >5/7.5 mL of blood with worse OS. |
| Manjunath et al. [59] | 60 | Microfiltration | CTC clusters noted in 41.2% of NSCLC patients. No CTC clusters identified in patients with radiographically benign lesions. |
| Murlidhar et al. [60] | 36 | OncoBean chip | CTC clusters associated with worse prognosis. Clusters displayed genotypic characteristics of therapeutic resistance. |
| Tamminga et al. [61] | 31 | CellSearch | CTCs were detected more frequently and in greater numbers from pulmonary vein compared to radial artery. Post-operative decrease in CTCs noted in blood from radial artery, but not the pulmonary vein. 1.2% of cells isolated showed aneuploidy, indicating majority of cells likely epithelial cells. |
| Wei et al. [50] | 73 | Nano-enrichment, direct visualization | Average CTC numbers were 5.7/7.5 mL of blood and decreased to 2.4/7.5 mL of blood after chemotherapy. <5 CTC/7.5 mL of blood showed better PFS. EGFR mutations were associated with greater number of CTCs. |
| Xu et al. [62] | 20 | Microfludic chip | 75% of patients had detectable CTCs. Successful WES on single isolated CTC, with detection of 6 new mutations, concordant with surgical specimen. |
| Author | Number of Patients | Platform | Main Findings |
|---|---|---|---|
| Chaudhuri et al. [109] | 94 | CAPP-seq (NGS) | Detectable ctDNA post-treatment preceded radiological evidence of progression in 72% of cases. Of the patients that relapsed, 94% had detectable ctDNA after treatment with curative intent. |
| Cho et al. [110] | 36 | PANAmutyper (PCR) | Factors associated with higher ctDNA in plasma included higher pathological tumor stage, nodal metastasis, solid adenocarcinoma subtype, tumor necrosis, greater tumor volume and frequent mitoses. |
| Li et al. [111] | 26 | WGS | Driver genes detected in all CSF ctDNA samples. 92.3% of patients had higher allele fractions in CSF than CSF precipitates or plasma. EGFR T790M was detected in CSF of 30.4% samples from patients who progressed on TKI. |
| Oxnard et al. [105] | 216 | BEAMing | Plasma detection of T790M was 70% sensitive. OOR and PFS were similar T790M positive tumors detected through plasma ctDNA or biopsy. |
| Papadopoulou et al. [112] | 171 | NGS | 49% of NSCLC patients had at least 1 mutation detected at diagnosis by NGS. 86.1% concordance in clinically relevant mutations between ctDNA and tissue biopsy. |
| Sabari et al. [113] | 210 | ResBio ctDx-Lung | ctDNA detection lower in patients on systemic treatment. High concordance of ctDNA detected oncogenic drivers with tissue detection (91.6%). |
| Tailor et al. [114] | 33 | SureSelect All Exon V5 + UTR | Patients with malignant nodules showed a significantly higher number of somatic mutations. 82% of malignant lesions identified through mutational analysis. |
| Tsui et al. [115] | 50 | Tam-Seq PCR, digital PCR | Low levels of EGFR mutations in TKI naïve patients resulted in better PFS and OS. Pre-treatment mutations in both EGFR and TP53 correlated with worse prognosis. Progression without T790M mutation resulted in worse survival. |
| Uchida et al. [116] | 288 | NGS | EGFR exon 19 deletion sensitivity was 50.9% and specificity was 98.0%. L858R mutation sensitivity was 51.9% and specificity was 94.1%. |
| Weber et al. [117] | 199 | Cobas EGFR test | 91% concordance of EGFR mutations between tissue and plasma ctDNA samples. Six EGFR mutations detected in ctDNA samples only. |
| Yang et al. [118] | 103 | Gardant360 | Poor survival if >3 mutations detected in ctDNA |
| Zhang et al. [119] | 27 | NGS | Overall ctDNA and tissue concordance for driver gene mutations was 85.2%, sensitivity and specificity was 87.0% and 75%, respectively. Concordance reached 100% in cases of boney metastasis and/or concurrent TP53 mutations. |
| Zhao et al. [120] | 111 | Mutant-enriched PCR | EGFR mutation concordance between paired plasma and tissue samples was 71.2%. Sensitivity was higher for poorly differentiated tumors (77.8%) compared to well differentiated (20%) and moderately differentiated (19%) tumors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Capua, D.; Bracken-Clarke, D.; Ronan, K.; Baird, A.-M.; Finn, S. The Liquid Biopsy for Lung Cancer: State of the Art, Limitations and Future Developments. Cancers 2021, 13, 3923. https://doi.org/10.3390/cancers13163923
Di Capua D, Bracken-Clarke D, Ronan K, Baird A-M, Finn S. The Liquid Biopsy for Lung Cancer: State of the Art, Limitations and Future Developments. Cancers. 2021; 13(16):3923. https://doi.org/10.3390/cancers13163923
Chicago/Turabian StyleDi Capua, Daniel, Dara Bracken-Clarke, Karine Ronan, Anne-Marie Baird, and Stephen Finn. 2021. "The Liquid Biopsy for Lung Cancer: State of the Art, Limitations and Future Developments" Cancers 13, no. 16: 3923. https://doi.org/10.3390/cancers13163923
APA StyleDi Capua, D., Bracken-Clarke, D., Ronan, K., Baird, A.-M., & Finn, S. (2021). The Liquid Biopsy for Lung Cancer: State of the Art, Limitations and Future Developments. Cancers, 13(16), 3923. https://doi.org/10.3390/cancers13163923






