Maturation State-Specific Alternative Splicing in FLT3-ITD and NPM1 Mutated AML
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Samples
2.2. Genetic Profile
2.3. RNA Sequencing
2.4. Differential Gene Expression and Splicing Analysis
3. Results
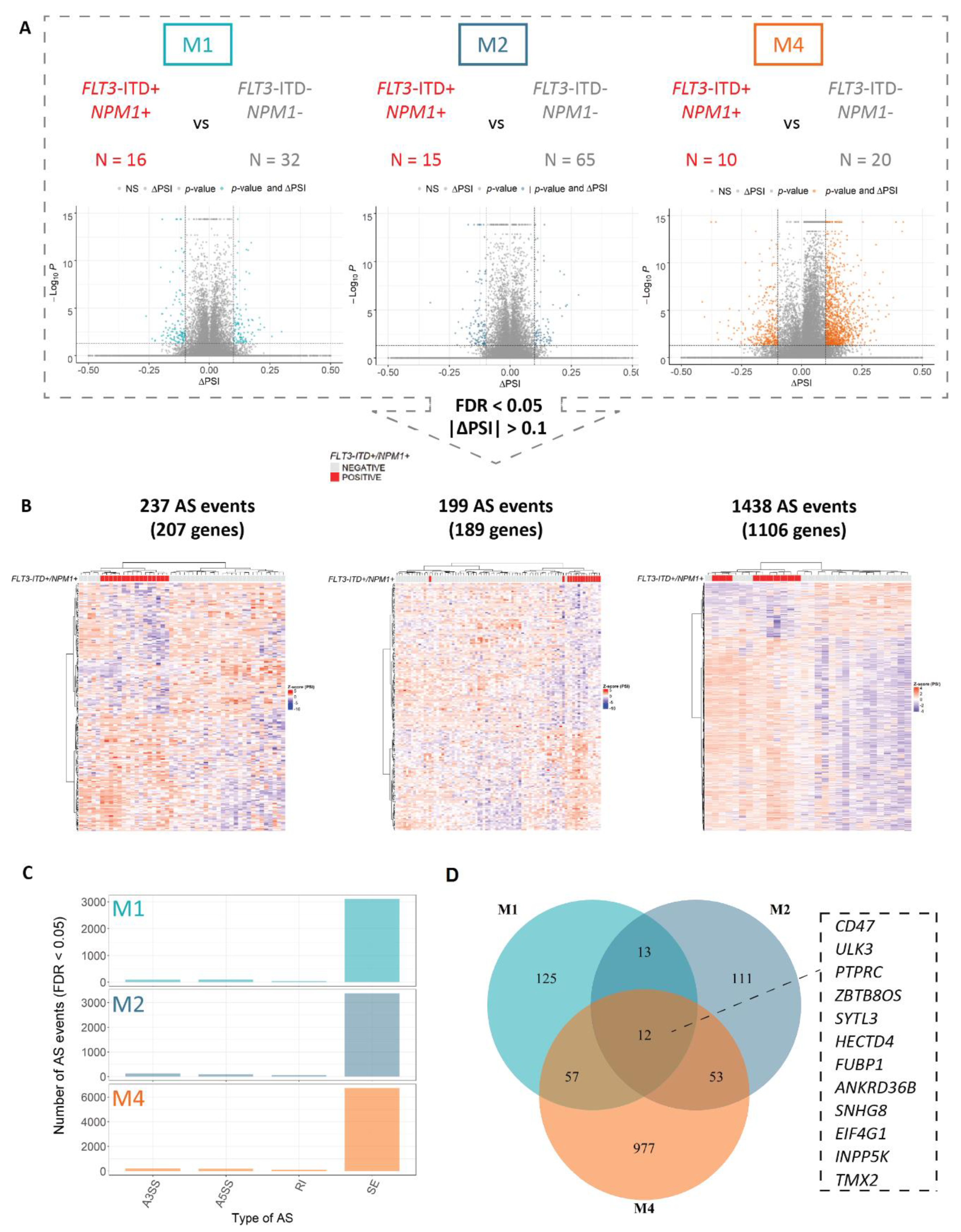
3.1. Alternative Splicing Profiles of FLT3-ITD and NPM1 Double Mutated Cells Show High FAB-Type Specificity
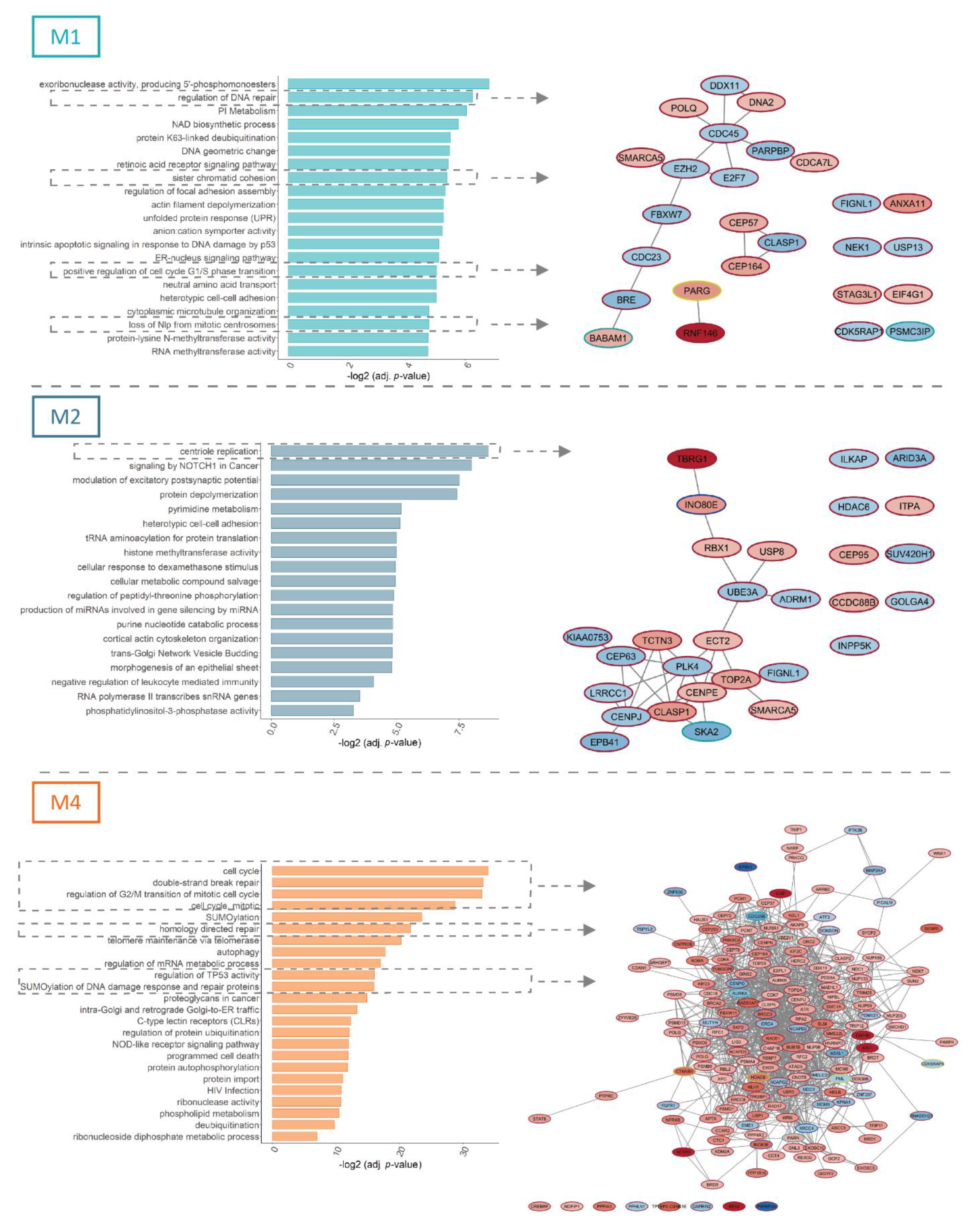
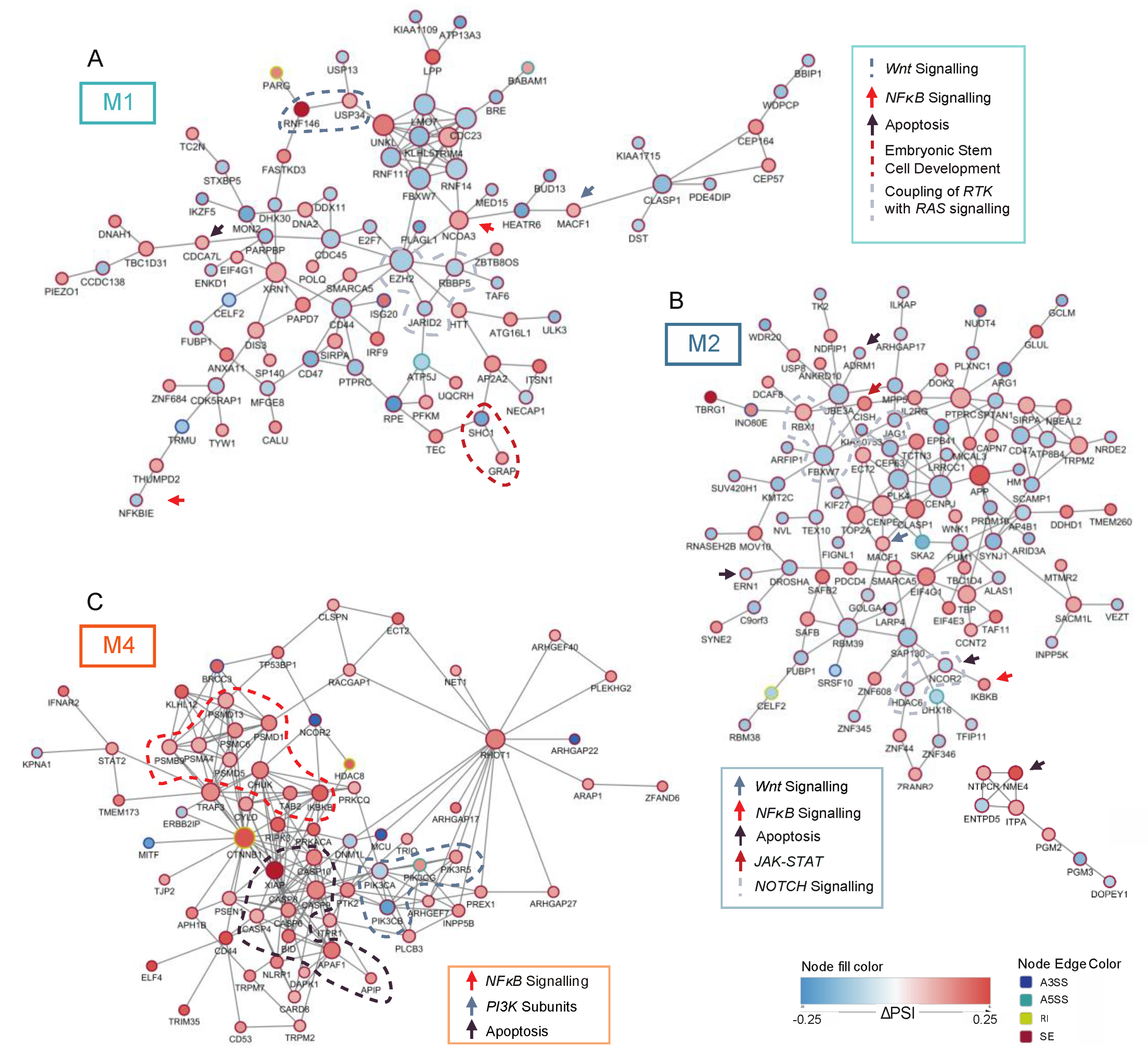
3.2. FLT3-ITD and NPM1 Double Mutated Cells Display Altered Splicing of Genes Involved in Cell Cycle Control, DNA Damage Response and Signaling Pathways
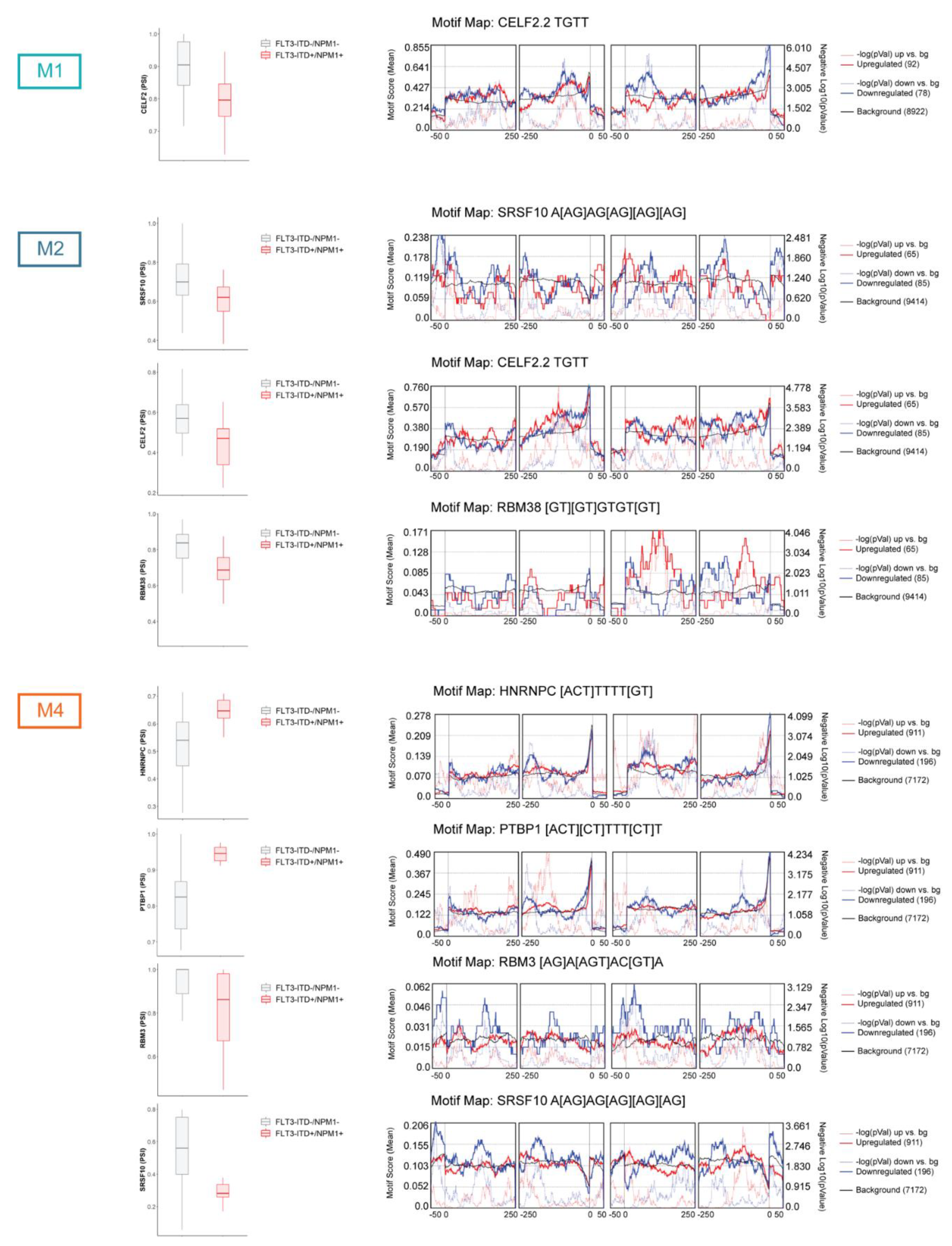
3.3. Factors Regulating Differential Splicing in the Context of FLT3-ITD and NPM1 Mutations
3.4. Evaluation of the Relevance of Differential Splicing Events
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Genomic and Epigenomic Landscapes of Adult De Novo Acute Myeloid Leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef]
- Corces-Zimmerman, M.R.; Majeti, R. Pre-leukemic evolution of hematopoietic stem cells: The importance of early mutations in leukemogenesis. Leukemia 2014, 28, 2276–2282. [Google Scholar] [CrossRef] [PubMed]
- Corces-Zimmerman, M.R.; Hong, W.J.; Weissman, I.L.; Medeiros, B.C.; Majeti, R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc. Natl. Acad. Sci. USA 2014, 111, 2548–2553. [Google Scholar] [CrossRef] [PubMed]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef]
- Ajith, S.; Gazzara, M.R.; Cole, B.S.; Shankarling, G.; Martinez, N.M.; Mallory, M.J.; Lynch, K.W. Position-dependent activity of CELF2 in the regulation of splicing and implications for signal-responsive regulation in T cells. RNA Biol. 2016, 13, 569–581. [Google Scholar] [CrossRef]
- Zhang, T.; Lin, Y.; Liu, J.; Zhang, Z.G.; Fu, W.; Guo, L.Y.; Pan, L.; Kong, X.; Zhang, M.K.; Lu, Y.H.; et al. Rbm24 Regulates Alternative Splicing Switch in Embryonic Stem Cell Cardiac Lineage Differentiation. Stem Cells 2016, 34, 1776–1789. [Google Scholar] [CrossRef]
- Wojtuszkiewicz, A.; Assaraf, Y.G.; Maas, M.J.P.; Kaspers, G.J.L.; Jansen, G.; Cloos, J. Pre-mRNA splicing in cancer: The relevance in oncogenesis, treatment and drug resistance. Expert Opin. Drug Metab. Toxicol. 2015, 11, 673–689. [Google Scholar] [CrossRef] [PubMed]
- Pederiva, C.; Böhm, S.; Julner, A.; Farnebo, M. Splicing controls the ubiquitin response during DNA double-strand break repair. Cell Death Differ. 2016, 23, 1648–1657. [Google Scholar] [CrossRef]
- Blencowe, B.J. Splicing regulation: The cell cycle connection. Curr. Biol. 2003, 13, R149–R151. [Google Scholar] [CrossRef][Green Version]
- Moore, M.J.; Wang, Q.; Kennedy, C.J.; Silver, P.A. An alternative splicing network links cell-cycle control to apoptosis. Cell 2010, 142, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Cambindo Botto, A.E.; Muñoz, J.C.; Giono, L.E.; Nieto-Moreno, N.; Cuenca, C.; Kornblihtt, A.R.; Muñoz, M.J. Reciprocal regulation between alternative splicing and the DNA damage response. Genet. Mol. Biol. 2020, 43. [Google Scholar] [CrossRef]
- Kahles, A.; Van Lehmann, K.; Toussaint, N.C.; Hüser, M.; Stark, S.G.; Sachsenberg, T.; Stegle, O.; Kohlbacher, O.; Sander, C.; Caesar-Johnson, S.J.; et al. Comprehensive Analysis of Alternative Splicing Across Tumors from 8705 Patients. Cancer Cell 2018, 34, 211–224. [Google Scholar] [CrossRef]
- Climente-González, H.; Porta-Pardo, E.; Godzik, A.; Eyras, E. The Functional Impact of Alternative Splicing in Cancer. Cell Rep. 2017, 20, 2215–2226. [Google Scholar] [CrossRef] [PubMed]
- Adamia, S.; Haibe-Kains, B.; Pilarski, P.M.; Bar-Natan, M.; Pevzner, S.; Avet-Loiseau, H.; Lode, L.; Verselis, S.; Fox, E.A.; Burke, J.; et al. AGenome-wide aberrantRNASplicing in patients with acute myeloid leukemia identifies novel potential disease markers and therapeutic targets. Clin. Cancer Res. 2014, 20, 1135–1145. [Google Scholar] [CrossRef]
- Darman, R.B.; Seiler, M.; Agrawal, A.A.; Lim, K.H.; Peng, S.; Aird, D.; Bailey, S.L.; Bhavsar, E.B.; Chan, B.; Colla, S.; et al. Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3’ Splice Site Selection through Use of a Different Branch Point. Cell Rep. 2015, 13, 1033–1045. [Google Scholar] [CrossRef]
- Ilagan, J.O.; Ramakrishnan, A.; Hayes, B.; Murphy, M.E.; Zebari, A.S.; Bradley, P.; Bradley, R.K. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 2015, 25, 14–26. [Google Scholar] [CrossRef]
- Kim, E.; Ilagan, J.O.; Liang, Y.; Daubner, G.M.; Lee, S.C.W.; Ramakrishnan, A.; Li, Y.; Chung, Y.R.; Micol, J.B.; Murphy, M.E.; et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell 2015, 27, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lieu, Y.K.; Ali, A.M.; Penson, A.; Reggio, K.S.; Rabadan, R.; Raza, A.; Mukherjee, S.; Manley, J.L. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc. Natl. Acad. Sci. USA 2015, 112, E4726–E4734. [Google Scholar] [CrossRef]
- Metzeler, K.H.; Herold, T.; Rothenberg-Thurley, M.; Amler, S.; Sauerland, M.C.; Görlich, D.; Schneider, S.; Konstandin, N.P.; Dufour, A.; Bräundl, K.; et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood 2016, 128, 686–698. [Google Scholar] [CrossRef] [PubMed]
- Masson, K.; Rönnstrand, L. Oncogenic signaling from the hematopoietic growth factor receptors c-Kit and FLT3. Cell. Signal. 2009, 21, 1717–1726. [Google Scholar] [CrossRef] [PubMed]
- Zarka, J.; Short, N.J.; Kanagal-Shamanna, R.; Issa, G.C. Nucleophosmin 1 mutations in acute myeloid leukemia. Genes (Basel) 2020, 11, 649. [Google Scholar] [CrossRef] [PubMed]
- Dovey, O.M.; Cooper, J.L.; Mupo, A.; Grove, C.S.; Lynn, C.; Conte, N.; Andrews, R.M.; Pacharne, S.; Tzelepis, K.; Vijayabaskar, M.S.; et al. Molecular synergy underlies the co-occurrence patterns and phenotype of NPM1-mutant acute myeloid leukemia. Blood 2017, 130, 1911–1922. [Google Scholar] [CrossRef] [PubMed]
- Rau, R.; Magoon, D.; Greenblatt, S.; Li, L.; Annesley, C.; Duffield, A.S.; Huso, D.; McIntyre, E.; Clohessy, J.G.; Reschke, M.; et al. NPMc+ cooperates with FLT3/ITD mutations to cause acute leukemia recapitulating human disease. Exp. Hematol. 2014, 42, 101–113. [Google Scholar] [CrossRef]
- Valk, P.J.M.; Verhaak, R.G.W.; Beijen, M.A.; Erpelinck, C.A.J.; van Doorn-Khosrovani, S.B.V.W.; Boer, J.M.; Beverloo, H.B.; Moorhouse, M.J.; van der Spek, P.J.; Löwenberg, B.; et al. Prognostically Useful Gene-Expression Profiles in Acute Myeloid Leukemia. N. Engl. J. Med. 2004, 350, 1617–1628. [Google Scholar] [CrossRef]
- Cauchy, P.; James, S.R.; Zacarias-Cabeza, J.; Ptasinska, A.; Imperato, M.R.; Assi, S.A.; Piper, J.; Canestraro, M.; Hoogenkamp, M.; Raghavan, M.; et al. Chronic FLT3-ITD Signaling in Acute Myeloid Leukemia Is Connected to a Specific Chromatin Signature. Cell Rep. 2015, 12, 821–836. [Google Scholar] [CrossRef]
- Luco, R.F.; Allo, M.; Schor, I.E.; Kornblihtt, A.R.; Misteli, T. Epigenetics in alternative pre-mRNA splicing. Cell 2011, 144, 16–26. [Google Scholar] [CrossRef]
- Luco, R.F.; Pan, Q.; Tominaga, K.; Blencowe, B.J.; Pereira-Smith, O.M.; Misteli, T. Regulation of alternative splicing by histone modifications. Science 2010, 327, 996–1000. [Google Scholar] [CrossRef]
- Lev Maor, G.; Yearim, A.; Ast, G. The alternative role of DNA methylation in splicing regulation. Trends Genet. 2015, 31, 274–280. [Google Scholar] [CrossRef]
- Sciarrillo, R.; Wojtuszkiewicz, A.; Kooi, I.E.; Leon, L.G.; Sonneveld, E.; Kuiper, R.P.; Jansen, G.; Giovannetti, E.; Kaspers, G.J.L.; Cloos, J. Glucocorticoid resistant pediatric acute lymphoblastic leukemia samples display altered splicing profile and vulnerability to spliceosome modulation. Cancers 2020, 12, 723. [Google Scholar] [CrossRef]
- Wiggers, C.R.M.; Baak, M.L.; Sonneveld, E.; Nieuwenhuis, E.E.S.; Bartels, M.; Creyghton, M.P. AML Subtype Is a Major Determinant of the Association between Prognostic Gene Expression Signatures and Their Clinical Significance. Cell Rep. 2019, 28, 2866–2877. [Google Scholar] [CrossRef]
- Höllein, A.; Jeromin, S.; Meggendorfer, M.; Fasan, A.; Nadarajah, N.; Kern, W.; Haferlach, C.; Haferlach, T. Minimal residual disease (MRD) monitoring and mutational landscape in AML with RUNX1-RUNX1T1: A study on 134 patients. Leukemia 2018, 32, 2270–2274. [Google Scholar] [CrossRef]
- Cappelli, L.V.; Meggendorfer, M.; Dicker, F.; Jeromin, S.; Hutter, S.; Kern, W.; Haferlach, T.; Haferlach, C.; Höllein, A. DNMT3A mutations are over-represented in young adults with NPM1 mutated AML and prompt a distinct co-mutational pattern. Leukemia 2019, 33, 2741–2746. [Google Scholar] [CrossRef]
- Stengel, A.; Kern, W.; Haferlach, T.; Meggendorfer, M.; Fasan, A.; Haferlach, C. The impact of TP53 mutations and TP53 deletions on survival varies between AML, ALL, MDS and CLL: An analysis of 3307 cases. Leukemia 2017, 31, 705–711. [Google Scholar] [CrossRef]
- Baer, C.; Pohlkamp, C.; Haferlach, C.; Kern, W.; Haferlach, T. Molecular patterns in cytopenia patients with or without evidence of myeloid neoplasm—A comparison of 756 cases. Leukemia 2018, 32, 2295–2298. [Google Scholar] [CrossRef]
- Pabst, T.; Vellenga, E.; Van Putten, W.; Schouten, H.C.; Graux, C.; Vekemans, M.C.; Biemond, B.; Sonneveld, P.; Passweg, J.; Verdonck, L.; et al. Favorable effect of priming with granulocyte colony-stimulating factor in remission induction of acute myeloid leukemia restricted to dose escalation of cytarabine. Blood 2012, 119, 5367–5373. [Google Scholar] [CrossRef] [PubMed]
- Löwenberg, B.; Pabst, T.; Maertens, J.; Van Norden, Y.; Biemond, B.J.; Schouten, H.C.; Spertini, O.; Vellenga, E.; Graux, C.; Havelange, V.; et al. Therapeutic value of clofarabine in younger and middle-aged (18–65 years) adults with newly diagnosed AML. Blood 2017, 129, 1636–1645. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Park, J.W.; Lu, Z.X.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2 Elegant Graphics for Data Analysis; Use R! Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-24275-0. [Google Scholar]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Bioconductor—Maser. Available online: https://bioconductor.org/packages/release/bioc/html/maser.html (accessed on 24 June 2021).
- Bioconductor—AnnotationHub. Available online: https://bioconductor.org/packages/release/bioc/html/AnnotationHub.html (accessed on 24 June 2021).
- Lawrence, M.; Huber, W.; Pagès, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.T.; Carey, V.J. Software for Computing and Annotating Genomic Ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.; Gentleman, R.; Carey, V. rtracklayer: An R package for interfacing with genome browsers. Bioinformatics 2009, 25, 1841–1842. [Google Scholar] [CrossRef]
- Park, J.W.; Jung, S.; Rouchka, E.C.; Tseng, Y.T.; Xing, Y. rMAPS: RNA map analysis and plotting server for alternative exon regulation. Nucleic Acids Res. 2016, 44, W333–W338. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.Y.; Jung, S.; Kook, T.L.; Rouchka, E.C.; Bok, J.; Park, J.W. rMAPS2: An update of the RNA map analysis and plotting server for alternative splicing regulation. Nucleic Acids Res. 2021, 48, W300–W306. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Verhaak, R.G.W.; Goudswaard, C.S.; Van Putten, W.; Bijl, M.A.; Sanders, M.A.; Hugens, W.; Uitterlinden, A.G.; Erpelinck, C.A.J.; Delwel, R.; Löwenberg, B.; et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): Association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood 2005, 106, 3747–3754. [Google Scholar] [CrossRef] [PubMed]
- Yun, H.; Vohra, S.; Mupo, A.; Giotopoulos, G.; Sasca, D.; Horton, S.J.; Agrawal-Singh, S.; Meduri, E.; Basheer, F.; Marando, L.; et al. Mutational Synergy Coordinately Remodels Chromatin Accessibility, Enhancer Landscape and 3-Dimensional DNA Topology to Alter Gene Expression during Leukemia Induction. Blood 2019, 134, 278. [Google Scholar] [CrossRef]
- Gao, Y.; Vasic, R.; Halene, S. Role of alternative splicing in hematopoietic stem cells during development. Stem Cell Investig. 2018, 5, 26. [Google Scholar] [CrossRef] [PubMed]
- Lemischka, I.R.; Pritsker, M. Alternative splicing increases complexity of stem cell transcriptome. Cell Cycle 2006, 5, 347–351. [Google Scholar]
- Ullrich, S.; Guigó, R. Dynamic changes in intron retention are tightly associated with regulation of splicing factors and proliferative activity during B-cell development. Nucleic Acids Res. 2021, 48, 1327–1340. [Google Scholar] [CrossRef] [PubMed]
- Reimer, K.A.; Neugebauer, K.M. Blood relatives: Splicing mechanisms underlying erythropoiesis in health and disease [version 1; referees: 3 approved]. F1000Research 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Zhou, B.; Thol, F.; Zhou, Y.; Chen, L.; Shao, C.; Deboever, C.; Hou, J.; Li, H.; Chaturvedi, A.; et al. Distinct splicing signatures affect converged pathways in myelodysplastic syndrome patients carrying mutations in different splicing regulators. RNA 2016, 22, 1535–1549. [Google Scholar] [CrossRef]
- Yoshimi, A.; Lin, K.T.; Wiseman, D.H.; Rahman, M.A.; Pastore, A.; Wang, B.; Lee, S.C.W.; Micol, J.B.; Zhang, X.J.; de Botton, S.; et al. Coordinated alterations in RNA splicing and epigenetic regulation drive leukaemogenesis. Nature 2019, 574, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-J.; Yan, M.; Chen, J.-Y.; Chen, L.; Kim, E.; Shima, T.; Davis, A.G.; Miyauchi, S.; Stoner, S.A.; Fu, X.-D.; et al. RUNX1 Deficiency Cooperates with SRSF2 Mutation to Further Disrupt RNA Splicing and Exacerbate Myelodysplastic Syndromes in Mouse Models. Blood 2018, 132, 1796. [Google Scholar] [CrossRef]
- Xu, Y.; Zhao, W.; Olson, S.D.; Prabhakara, K.S.; Zhou, X. Alternative splicing links histone modifications to stem cell fate decision. Genome Biol. 2018, 19, 133. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wojtuszkiewicz, A.; van der Werf, I.; Hutter, S.; Walter, W.; Baer, C.; Kern, W.; Janssen, J.J.W.M.; Ossenkoppele, G.J.; Haferlach, C.; Cloos, J.; et al. Maturation State-Specific Alternative Splicing in FLT3-ITD and NPM1 Mutated AML. Cancers 2021, 13, 3929. https://doi.org/10.3390/cancers13163929
Wojtuszkiewicz A, van der Werf I, Hutter S, Walter W, Baer C, Kern W, Janssen JJWM, Ossenkoppele GJ, Haferlach C, Cloos J, et al. Maturation State-Specific Alternative Splicing in FLT3-ITD and NPM1 Mutated AML. Cancers. 2021; 13(16):3929. https://doi.org/10.3390/cancers13163929
Chicago/Turabian StyleWojtuszkiewicz, Anna, Inge van der Werf, Stephan Hutter, Wencke Walter, Constance Baer, Wolfgang Kern, Jeroen J. W. M. Janssen, Gert J. Ossenkoppele, Claudia Haferlach, Jacqueline Cloos, and et al. 2021. "Maturation State-Specific Alternative Splicing in FLT3-ITD and NPM1 Mutated AML" Cancers 13, no. 16: 3929. https://doi.org/10.3390/cancers13163929
APA StyleWojtuszkiewicz, A., van der Werf, I., Hutter, S., Walter, W., Baer, C., Kern, W., Janssen, J. J. W. M., Ossenkoppele, G. J., Haferlach, C., Cloos, J., & Haferlach, T. (2021). Maturation State-Specific Alternative Splicing in FLT3-ITD and NPM1 Mutated AML. Cancers, 13(16), 3929. https://doi.org/10.3390/cancers13163929





