Abstract
The reuse of stabilized (under thermophilic conditions) sewage sludge and manure on agricultural soils is a common practice. The aim of this study was to evaluate the risks associated with their repeated applications on the spread of pathogenic bacteria and antibiotic resistance genes (ARGs) that encode resistance to tetracycline (tetA and tetW), sulphonamide (sul1 and sul2), erythromycin (ermB), vancomycin (vanA) and integron genetic element (intI1). The trial fields has been regularly fertilized every 3rd year since 1996 with manure (MF; 330 kg N/ha) and sewage sludge (SF; 330 kg N/ha and SF3; 990 kg N/ha). Unfertilized soil (CF) served as a control. Samples were collected at different time points: (i) right before fertilization (which was also 3 years after the last fertilization), (ii) 5 months after fertilization, and (iii) 11 months after fertilization. The relative abundance of amplicon sequence variants (ASVs) assigned to potentially pathogenic bacteria was low (0.3% and 0.25% in sludge and manure, respectively), and no association with the application of these fertilizers was found. On the other hand, our data indicate that an increased relative abundance of the ARGs sul1 and tetW was significantly associated with these fertilizer applications, and sul1 was increased in all treatments regardless of the time. It is suggested that sul1 should be monitored in organically fertilized soils to prevent its spread and possible further accumulation in crops.
1. Introduction
A common practice to maximize crop yields is the application of organic fertilizers on arable soil. Among the well-known benefits of their application is the improvement of soil fertility by enriching it with nutrients (especially phosphorus and nitrogen), pH alteration [1,2], and the replenishment of soil organic matter, the depletion of which is a serious problem [3]. Stabilised sewage sludge and animal manure are traditionally used as organic fertilizers which, in some European countries, are the primary means of municipal and industrial organic waste reuse [4]. For example, in the Czech Republic, 13 million tons of manure and 61,000 tons of sludge were applied to agronomic soils in 2018 [5,6].
The reuse of organic waste in agriculture has been motivated not only by its beneficial impact on soil fertility, but also by the recent approval of Directive 2018/851/EC, amending Directive 2008/98/EC on waste. This directive guides waste utilisation management and strategies by promoting the minimisation of waste production and supporting the principles of the circular economy [7]. Therefore, the reuse of organic waste, such as stabilised sewage sludge (biosolids) and animal manure, in agriculture seems to be a feasible and cheap way to meet the goals of “Closing the loop”, an EU action plan for the circular economy [8,9].
Although such fertilizers are beneficial in many aspects, they can pose a risk due to their contamination with pollutants or pathogenic microorganisms that can be transferred into the arable soil. The reuse and recycling of organic waste, therefore, has to be implemented with consideration for the environment in order to avoid causing a potential contamination burden. Directive 86/278/EEC sets the limits for heavy metals and several organic pollutants, e.g., polychlorinated biphenyls and polyaromatic hydrocarbons [10,11,12]. However, the presence of many other contaminants is not regulated.
Emerging pollutants that are currently the focus of worldwide attention include antibiotic resistance genes (ARGs) and antibiotic residuals [13,14,15,16]. Although the occurrence of ARGs in soil bacterial communities is natural, it is thoroughly established that the application of animal manure and sewage sludge increases the diversity and the number of ARGs in the soil environment [17,18,19]. The elevated presence of antibiotics in sludge and manure exists because only 10 to 40% of antibiotics are metabolised, and the rest are released into the environment either in urine or faeces [19,20,21]. In several studies, sewage sludge and manure were identified as hotspots for the dissemination of ARGs when applied to soil [2,13,16,22]. Furthermore, it is known that ARGs can persist in soil and be further spread even when the levels of antibiotics are at sub-inhibitory concentrations [23].
Despite the implemented restrictions on antibiotic (ATBs) usage in veterinary and healthcare clinics (the World Health Organization (WHO) developed a methodology to monitor antibiotic consumption [24]), worldwide production of antibiotics is still rising. The global animal consumption of ATBs was estimated at 63,200 tons in 2010, representing a 30% increase in antibiotic consumption over the previous 10 years [25]. With no policy changes, global antibiotic consumption is predicted to rise by 200% between 2015 and 2030 [26]. Due to population growth and the need for intensification of crop and animal production [25], higher production of sewage sludge and manure together with their continuous reuse in agronomy is expected. Therefore, it is necessary to monitor ARG levels in regularly fertilized arable soils over time and determine indicator ARGs that signal an increased health risk.
Besides ATBs and ARGs, sludge and manure amendments can also pose a potential risk due to the presence of pathogenic microorganisms originating from human or animal faeces [27,28]. Directive 86/278/EEC does not specify the limits for pathogens; however, most countries have their own regulations that include limits for several indicator microorganisms (e.g., Escherichia coli, Salmonella spp., Enterococci species, or Clostridium perfringens) [7]. The indicator microorganisms, though, do not reflect the actual contamination by a diverse spectrum of pathogens. To minimise the risk of any pathogen transmission from organic fertilizers to fresh produce, the time between manure application and crop harvest is limited to 120 days with and 90 days without direct contact of the edible parts of the crop with the fertilized soil [29,30]. Unfortunately, these limits do not seem to provide sufficient control over possible pathogen transmission in general. The survival duration of Escherichia coli O157:H7 was reported to be significantly influenced by spatiotemporal factors (e.g., site, year, season) supporting its survival beyond 90 days in fertilized soils [31]. Furthermore, pathogen transfer despite the 90- and 120-day limitations has also been demonstrated by a number of reported outbreaks, for example: vegetable contamination with E. coli from raw bovine manure [32]; recent E. coli O157:H7 outbreaks associated with romaine lettuce in 2018 and 2019 from waterborne contamination [33,34]; or Salmonella outbreaks related to organic tomato production from irrigation water and soil amendments [35]. Although it is known that animal manure and sewage sludge can transfer a spectrum of potential human pathogens into the soil, there is still limited information about the survival of such pathogens in long-term, regularly fertilized soil.
2. Materials and Methods
2.1. Samples Collection
The soil samples were collected from a field located in the Czech Republic (50°7′40″ N, 14°22′33″ E), that was covered with chernozem with a pH 7.8 ± 0.4 and contents of clay 2.18%, silt 71.8% and sand 26.03%. The field had been organically fertilized since 1996 with: (i) sewage sludge (330 kg N/ha/3 years; SF), (ii) sewage sludge (990 kg N/ha/3 years; SF3), and (iii) cattle manure (330 kg N/ha/3 years; MF). The unfertilized soil was used as a control group (CF). Potatoes (Solanum tuberosum L.), winter wheat (Triticum aestivum L.), and spring barley (Hordeum vulgare L.) were grown in the field using 3-year rotation systems that also included fallow periods. The fertilizers were only applied to soil before the potato sowing, i.e., every 3 years.
Soil sampling was performed 3 times per year: right before fertilization (September 2015), 5 months after fertilization (March 2016), and 11 months after fertilization (September 2016). Given the repeated fertilizer application, soil samples collected in September 2015 also represented soil 3 years after fertilization performed in September 2012.
For the microbial community analyses, 4 replicates of soil samples were collected from the soil of each fertilized variant. Each sample was a homogenized mixture of 6 soil sub-samples, sampled with a sampler probe. In total, 16 composed soil samples were collected each time and stored at −20 °C until further analyses. The sewage sludge applied to the soils in October 2012 and 2015 was obtained from a wastewater treatment plant in Prague. The University Farm Estate Lány (https://lany.czu.cz/en, accessed on 6 June 2021) provided the cow manure. The samples were stored at −20 °C until further analyses. More information about the experimental field, the fertilizer application, and the sample collection procedure can be found in our previous publication [36]. For the ARG abundance mapping, 2 out of the 4 collected replicates were analysed, each of which was run in 3 technical replicates.
2.2. DNA Isolation and 16S rRNA Amplicons Generation
Metagenomic DNA from samples of soil, manure, and sewage sludge was isolated using a FastDNA Spin Kit for Soil (MP Biomedicals, Solon, OH, USA) and purified with a Genomic DNA Clean and Concentrator kit (ZYMO Research, Irvine, CA, USA) according to the manufacturer’s instructions. The DNA concentration was normalised to 10 ng/µL before the amplicon generation process was initiated.
Amplicons were generated with two polymerase chain reactions (PCRs), the procedure of which was adopted from a previous study [37]. Briefly, the first PCR master mix contained: 0.02 U/µL KAPA HiFi HotStart ReadyMix (Kapa Biosystems, USA), 0.3 µM of both 515 forward (5′-GTGYCAGCMGCNGCGG-3′) and 926 reverse (5′-CCGYCAATTYMTTTRAGTTT-3′) primers, and purified metagenomic DNA (~10 ng/µL). Water for molecular biology (Sigma-Aldrich, USA) was used to fill the master mix volume up to 15 µL. For the second PCR, the primers were modified with internal barcode and adaptor sequences [38]. The second PCR master mix (25 µL) contained 0.02 U/µL KAPA HiFi HotStart ReadyMix, 0.5 µL of the product of the previous PCR, 1 µM of each modified primer (Sigma-Aldrich, St. Louis, MI, USA), and water for molecular biology. The temperature programs used for the first and second PCR, respectively, were as follows: (i) 5 min at 95 °C, 20 s at 98 °C, 15 s at 56 °C (28–30 cycles), 15 s at 72 °C, and a final extension of 5 min at 72 °C; and (ii) 5 min at 95 °C, 20 s at 98 °C, 15 s at 50 °C (8–10 cycles), 15 s at 72 °C, and a final extension of 5 min at 72 °C. The generated amplicons were eventually purified with SPRIselect magnetic beads (Beckman Coulter, Brea, CA, USA), and sent on ice packs to the Core Facility for Nucleic Acid Analysis at the University of Alaska Fairbanks for Illumina Miseq sequencing. The DNA concentration was normalized to 1–2 ng/µL (SequalPrep Kit, Thermo Fisher Scientific, Waltham, MA, USA) prior to the sequencing process.
2.3. Data Processing and Analysis
Raw sequences obtained after sequencing were processed using the DADA2 package and procedure following the DADA2 pipeline version 1.16 [39] in the program R [40]. During this step, the sequences were trimmed and filtered (truncLen function) according to the parameters that were calculated automatically based on the sequences’ quality. Sequences were dereplicated with the following removal of sequencing errors: The forward and reverse reads were merged, and the amplicon sequence variant (ASV) table was constructed. Eventually, the chimeric sequences were identified and removed. The taxonomy was assigned to the ASVs using the rdp_train_set_16 [41] database for 16S rRNA gene sequences (assignTaxonomy function, minimal bootstrap value 50). As a result, the taxonomic assignment of individual ASVs was performed, and individual genera in each sample were determined. All obtained MiSeq reads were deposited in the NCBI Short Read Archive under BioProject accession number PRJNA681506 (organic fertilizers, samples collected in September 2015 and 2016) and PRJNA587449 (samples collected in March 2016).
The potential human pathogens were identified by comparing the ASVs to a database of 122 potential pathogenic bacteria adopted from previous studies [27,36]. The comparison was performed using a local BLAST+ search, while the sequence identity was set to 99%, and the expected value cutoff was set to 1.0 × 10−1 [36]. The alpha-diversity of the bacterial community in soil samples, described by the Shannon diversity index [42], was assessed using the phyloseq package [43] and vegan package in R [44], and the significant difference of the index among fertilization treatments, time, or their interaction was tested using the Kruskal–Wallis rank sum test. The association of fertilization regime and prokaryotic community structure was analysed using an ANOVA-like permutation test. The tests were performed on data transformed to relative abundances, and each time point was analysed separately.
2.4. qPCR Assays
The antibiotic resistance genes (ARGs) subjected to quantitative real-time PCR (qPCR) analysis included sul1, sul2, tetA, tetW, intl1, ermB, and vanA. The qPCR reactions were conducted in 96-well plates and analysed with a CFX96 Touch qPCR System (Bio-Rad, USA) according to an adapted protocol described by Xu et al. [45]. Primers targeting the ARGs of interest together with 16S rRNA gene are listed in Supplementary Table S1. The qPCRs assays were carried out with the following cycle conditions (35 cycles): 3 min at 98 °C, 3 s at 95 °C, 20 s under annealing temperature (Supplementary Table S1), 15 s at 72 °C, and a final extension of 5 min at 72 °C. Master mix (20 μL) for one reaction was prepared as follows: 10 μL KAPA SYBR FAST qPCR Kit (Kapa Biosystems, Wilmington, MA, USA), 0.6 μL of ROX High Reference dye (Kapa Biosystems, USA), 4.74 μL of water for molecular biology, 0.33 μL of each primer (100 mM) targeted to the ARGs (Supplementary Table S1), and 4 μL of DNA (3 ng/μL). The qPCR amplification was performed using SYBR Green and ROX as fluorophores. Each sample was analysed in 3 technical replicates.
Ten-fold serial diluted samples of reference genes were tested on the same qPCR plate. The purpose of such samples was to construct standard calibration curves for determining the absolute abundance of ARGs or 16S rRNA gene in real samples. The preparation procedure of the standard calibration curves (both ARGs and 16S rRNA) for qPCR assays is described in detail in the supplementary material.
2.5. Statistical Analysis of ARGs Abundance
The abundances of ARGs were normalized to 16S rRNA in order to compare relative abundance in the different samples. The influence of the fertilization regime on the relative abundance of the ARGs was tested using the non-parametric Kruskal–Wallis test and pairwise Wilcoxon signed-rank test because the data did not meet the conditions of normality (Shapiro–Wilk test). The FDR correction was applied to obtained p-values [46]. The boxplots showing the relative abundance of ARGs among fertilization regimes through time was conducted using ggplot2 package version 3.2.1 [47] in R. Concentrations of ARGs in soils and organic fertilizers were compared using one-way analysis of variance (ANOVA), and a significant difference was assumed at the level p ≤ 0.05.
3. Results and Discussion
3.1. Microbiological Safety of the Organic Fertilizers
Microorganisms occur in manure and sewage sludge naturally due to their involvement in anaerobic digestion processes leading to the breakdown of complex organic polymers [48]. However, because animal manure and sewage sludge are of urine and faecal origin, some of the microorganisms may be pathogenic to humans or animals [49]. In this study, manure harboured 501 unique 16S rRNA gene ASVs belonging to 150 bacterial genera, and sewage sludge harboured 161 unique ASVs assigned to 71 bacterial genera. While the manure microbial population mainly consisted of three phyla (45% of Bacteroidetes, 20.5% of Proteobacteria, and 20% of Firmicutes), sewage sludge was predominantly inhabited by Firmicutes (88%).
The Kruskal–Wallis test revealed that bacterial diversity had significantly changed through time (p < 0.001), but was not significantly influenced by the fertilization regime (Figure 1). The Shannon diversity index showed that the bacterial diversity in all samples was highest in soils sampled in September 2016 (Figure 1), which may have been caused by differences in meteorological conditions (Supplementary Table S2), e.g., air temperature and total precipitation. The microbial community structure, though, was associated with both fertilization and time (p ≤ 0.01).
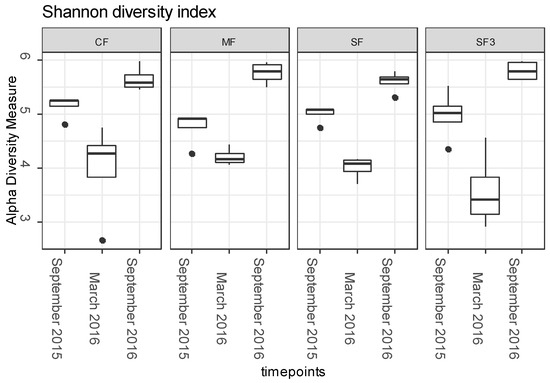
Figure 1.
Shannon diversity index calculated from sequence data obtained from soils sampled at different time points. Fertilization treatments: control soil (CF), manure (MF; 330 kg N/ha), sewage sludge (SF; 330 kg N/ha), sewage sludge (SF3; 990 kg N/ha).
Application of manure and sludge to soil did not influence the alpha-diversity, but it did significantly shape the structure of the microbial communities living in both the soil and the endosphere of the cultivated crop. Similar trends were found in our previous research [36]. Opitutus, Chitinophaga, Solirubrobacter, Romboutsia, Rhodanobacter, and Pseudoxanthomonas or Nitrosospira were all found to be significantly enriched in organically treated soils (MF, SF, and/or SF3) [37]. Whereas such changes were largely attributed to changes in soil nutrient profile, also reflected by alterations in enzymatic activity [37], some studies point to the possible introduction of Allochthonous microorganisms from organic fertilizers into the soil environment [50,51]. To avoid potential pathogen transmission, the digestion process of fertilizers can be placed under mesophilic (35–37 °C) or thermophilic (55–57 °C) conditions [52]. The thermophilic conditions are known to have a better hygienization effect than the mesophilic, and therefore the occurrence of pathogens in materials processed under these conditions is generally lower [27,53]. Furthermore, the thermophilic stabilization is also sufficient to reduce indicator microorganisms such as E. coli, Salmonella spp., or Enterococcus sp. [53,54,55]. In this study, both manure and sewage sludge were stabilized under thermophilic conditions before their soil application [36]; hence, the relative abundance of ASVs assigned to potentially pathogenic bacteria was low (0.3% and 0.25% in sludge and manure, respectively). These sequences were only assigned to Mycobacterium phlei (both fertilizers) and Alcaligenes faecalis (identified in sewage sludge). Mycobacterium phlei, which is a nontuberculous mycobacterium [56], was previously detected in sludge and cattle faeces [27,57], and the opportunistic pathogen Alcaligenes faecalis [58] was isolated from various environments, such as water and soil.
Potentially pathogenic microorganisms were monitored in soils before fertilization (September 2015), 5 months afterwards (March 2016), and 11 months afterwards (September 2016). In contrast to other studies that reported an increased prevalence of pathogens after organic fertilizer application [59,60,61], no potential human or animal pathogens were detected in fertilized soils in September 2015 and March 2016. However in September 2016, Mycobacterium phlei, Mycobacterium fortuitum, and Bacillus anthracis/Bacillus cereus were detected in soil samples across all fertilized variants (MF, SF, SF3). Given the time interval between the fertilizer application and the first detection of potential pathogens in soils, we do not think that their occurrence was associated directly with the fertilizer application. Besides, Mycobacterium fortuitum was also detected in CF. The detection of the potential pathogens can be also attributed to bacterial diversity (regardless of the fertilization treatment), which was highest in September 2016, suggesting that a broader spectrum of microorganisms, including pathogens, was present.
Such findings correspond with our previous study [36], in which the transfer of potential pathogens from fertilized soils into the tubers of Solanum tuberosum L. was not observed. The thermophilic anaerobic stabilization of organic waste therefore seemed to be sufficient to prevent soil contamination by pathogens [62]. However, as the meta-analysis shows, when raw (not stabilized) manures are used as amendments, the soils are contaminated with pathogens (e.g., E. coli and Salmonella spp.) and their survival is mainly affected by the daily average temperatures. Their log reduction was significantly higher at temperatures above 20 °C [63].
3.2. Abundance of ARGs in the Organic Fertilizers
The ARGs whose abundance was mapped in the organic fertilizers and soils included genes encoding resistance to tetracycline (tetA and tetW), sulphonamide (sul1 and sul2), erythromycin (ermB), vancomycin (vanA), and integron genetic element (intI1). Their relative abundance in the organic fertilizers applied to soil in September 2012 and 2015 is listed in Table 1. Their abundance differed not only between the manure and sewage sludge, but also depending on the fertilization year (2012 vs. 2015). It is the variation between the years that reflects the fact that even though the organic fertilizers are produced in order to meet nutrient and selected pollutants content regulations (e.g., Nitrate Regulation 91/676/EEC, Directive 86/278/EEC), ATB and ARG contamination is poorly controlled.

Table 1.
The relative abundances of ARGs in fertilizers applied in the years 2012 and 2015. Grey highlights mean that the relative abundances of ARGs in manure or sewage sludge did not statistically differ (one-way ANOVA, p ≥ 0.05) between the years 2012 and 2015.
Generally, the highest relative abundances were found for intI1, tetA, tetW, and sul1. The relative abundance of intl1, ranging from 2.96 × 10−1 to 5.30 × 10−2, is not surprising for several reasons. Integrons play a major role in bacterial adaptation and are associated with the horizontal gene transfer of antibiotic or metal resistance [64,65]. Intl1, which is a class 1 integron, encodes integrase protein, primarily widespread among Gram-negative bacteria [66]. Similar levels of intl1 in sludge were also documented by An et al. [67] and are usually predominant over intI2 and intI3 [67,68].
The higher levels of sul1, sul2, tetA, and tetW in organic fertilizers are often detected because sulphonamides and tetracyclines are among the most extensively used antibiotics [69,70]. They are used not only in healthcare clinics, but also in veterinary clinics, intensive animal farming, and agriculture for crop protection [71,72]. In sludge and manure, the abundance of tetA, tetW, and sul1 was higher than other ARGs (Table 1). The lower prevalence of sul2 compared to sul1 (12- to 145-fold difference) might be explained by the structure of the microbial community or the mechanisms of gene mobility. Different species were found to prefer one resistance mechanism over the other; e.g., while Salmonella spp. use sul1 as a dominant resistance mechanism against sulphonamides, sul2 was found more frequently in E. coli [73]. Therefore, the sul1 and sul2 distribution in the environment may be attributed to the microbial composition [73,74]. In contrast to sul2, sul1 is typically associated with integrons (class 1), which facilitate ARG transfer [73,75,76]. However, both genes can be co-located with other resistance genes; thus, the selective pressure of other antibiotics could also induce their spreading [75].
ErmB encodes the resistance to macrolides traditionally used in human and veterinary medicine against both Gram-positive and Gram-negative bacteria [77]. Although the macrolides are often used against bovine mastitis, the abundance of ermB in cattle manure ranged only from 4.67 × 10−4 to 8.30 × 10−6. These concentrations were more than 28 and 264 times lower than in the sewage sludge used in 2015 and 2012, respectively. The relative amount of vanA, encoding the resistance against vancomycin that is used against Gram-positive bacteria [78], was detected at similar levels in both fertilizers (Table 1).
3.3. ARGs Fate in Fertilized Soils
In this study, the two organic fertilizers had been applied to soil regularly every 3rd year since 1996. Therefore, the samples collected in September 2015 represent not only the soil right before fertilization in 2015, but also the soil 3 years after fertilization in 2012. In these samples, sul1 was the only ARG with a significantly higher relative abundance (pfdr < 0.05, pairwise Wilcoxon test) in all the fertilization treatments (MF, SF, SF3) compared to CF. The increased abundance of sul1 was previously associated with the application of sewage sludge or manure [79,80,81], but it is the 3-year persistence of sul1 in soil that was alarming. The possible reasons for this are as follows: (i) sul1 is frequently transferred between different bacterial taxa, allowing the gene to be ubiquitous [82]; (ii) the sul1 gene is often associated with class 1 integrons, which facilitate its spread [73], and; (iii) sulfonamides bound to soil particles [83] and organic matter in fertilizers significantly increase the sorption tendency [84], which might lead to the natural selection of resistant bacteria in soil. The significantly higher abundance of sul1 (pfdr < 0.05, pairwise Wilcoxon test) in SF-, SF3-, and MF-treated soils compared to CF was also observed in March 2016 and September 2016 (Figure 2). The highest abundance, detected in March 2016 (5 months after fertilization) implies that sul1 dissipates over time. However, that dissipation might take more than 5 years [85]. That is probably also the reason why the sul1 abundance was still significantly increased in September 2015. Particular attention should therefore be paid to the crop that follows the fertilization treatment, as the transfer of ARGs into the plant from the soil was also documented [86,87]. Compared to sul1, sul2 was not detected in any of the treatments, suggesting that sul2 dissemination does not pose a health threat.
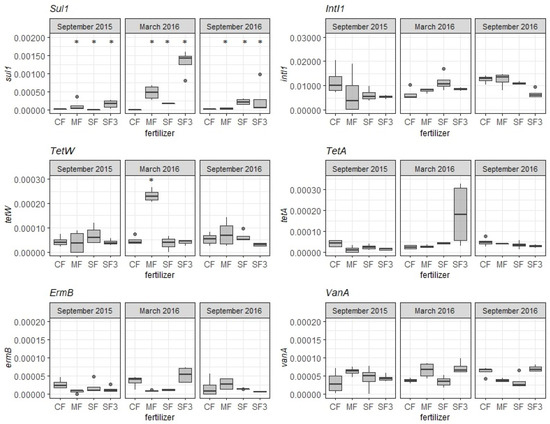
Figure 2.
The relative abundance of ARGs and integron Int1 in soils collected over 1 year. When there is a significant difference (pfdr < 0.05) in relative abundance of ARGs in amended soil versus control soil, the treatment is marked with asterisk. Fertilization treatments: control soil (CF), manure (MF; 330 kg N/ha), sewage sludge (SF; 330 kg N/ha), sewage sludge (SF3; 990 kg N/ha).
Tetracyclines were often detected in soil at higher concentrations than sulphonamides, as summarized in a review of [69], which could be due to their longer half-life [20]. Thus, the resistance genes for tetracyclines were prevalent in various environments [88,89]. A significant enrichment of tetW in MF-treated soil was only detected in March 2016, 5 months after fertilization (Figure 2). This enrichment was probably caused by the 1000-fold higher relative abundance of tetW in manure than in the control (CF). In contrast, fertilization was not significantly associated with the increased relative abundance of tetA in fertilized soils. Despite the connection between anthropogenic activities and the increased abundance of ARGs, pristine environments were also confirmed to harbour an indigenous soil resistome [90]. Furthermore, Tang et al. [91] found low ARG variation in bulk and rhizosphere soils over 69 years without anthropogenic activities. Thus, the background abundances of ARGs in CF soil (Figure 2) might be connected with indigenous soil microbiota, previous contamination, or other environmental conditions.
In contrast to other studies [92,93] that showed the abundance of intl1 positively correlate with several ARGs, our study showed no significant increase in intl1 in any of the fertilized soils (Figure 2) at any time (pfdr > 0.05, pairwise Wilcoxon test). This may be due to the low relative abundance of intl1 in the organic fertilizers, which was only 25 and 4 times higher (manure and sewage sludge in 2015, respectively) than its abundance in CF (1.19 × 10−2) in September 2015. That is a remarkable difference in ratios compared, for example, to sul1, the abundance of which was approximately 2000 times higher in both fertilizers compared to CF. The presence of intl1 in CF can be explained by the fact that only 14.6% of function proteins encoded on integron cassettes are related to antibiotic resistance, while the rest of the cassettes encode proteins involved in processes such as biosynthesis, regulation, transport, repair mechanisms, and resistance to heavy metals [67]. Therefore, the occurrence of intl1 may not necessarily be only associated with ARGs, as they also support other cell processes.
Also, the abundance of vanA and ermB in amended soils (MF, SF, SF3) was not significantly higher than that in CF (Figure 2) (pfdr > 0.05, pairwise Wilcoxon test). With vanA, such results can be explained by its low abundance in manure and sludge applied in 2015, ranging from 10−5 to 10−6, which is not significantly different from the vanA abundance in CF. In contrast to that, the relative abundance of ermB in sewage sludge applied in 2015 was almost 480 times higher than in CF, and therefore, its increase in fertilized soils would be expected. However no significant difference was found in any of the fertilization treatments (SF, SF3, and MF) compared to CF (pfdr > 0.05, pairwise Wilcoxon test). Such findings might be due to the gene being predominantly located on a chromosome [94,95], making the gene transfer less frequent than genes located on plasmids or associated with mobile genetic cassettes [79]. Based on the obtained results, it seems that two factors appeared to play a crucial role in the enrichment of ARGs in fertilized soils—the difference in ARG abundance in organic fertilizers versus CF and the gene mobility mechanism.
4. Conclusions
We proved that the regular application of animal manure and sewage sludge (treated under thermophilic conditions) did not introduce any potential human or animal pathogens to agronomical soil, and therefore did not pose a direct health threat to final consumers. Furthermore, repeated fertilization did not significantly increase the relative abundance of tetA, sul2, intl1, ermB, or vanA in soil. However, both manure and sewage sludge were significantly associated with an increased relative abundance of sul1 and tetW. While the abundance of tetW was significantly increased in MF-treated soil only 5 months after fertilization, sul1 was significantly higher in MF, SF, and SF3 treatments regardless of the time. Therefore, sul1 should be monitored in organically fertilized soils to prevent its spread and possible further accumulation in crops.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/agronomy11071423/s1. Materials and methods: Preparation of samples for standard calibration curves (ARGs and 16s RNA gene); Supplementary Table S1: List of primer sets (Sigma Aldrich, Sigma-Aldrich, USA) targeted to 16S rRNA and antibiotic resistance genes (ARGs) used in qPCR assays; Supplementary Table S2: Comparison of meteorological data at different sampling points.
Author Contributions
Conceptualization, H.S., J.C., and M.K.; methodology, H.S. and M.K.; formal analysis, M.K. and H.S.; investigation, M.K., T.V., and M.B.; data curation, M.K.; writing—original draft preparation, H.S. and M.K.; writing—review and editing, K.D.; visualization, M.K.; supervision, H.S. and K.D.; project administration, H.S. and J.B.; funding acquisition, K.D. and J.B. All authors have read and agreed to the published version of the manuscript.
Funding
We wish to thank the Czech Science Foundation (project no. 19-02836S) for funding this research, within which the methodology applied for data analysis was developed.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors wish to also thank Benjamin John Watson-Jones MEng for English language correction and the Institute of Arctic Biology’s Genomics Core Lab at the University of Alaska Fairbanks for the Illumina-sequencing of generated amplicons (INBRE 2P20GM103395).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhang, Y.T.; Shen, H.; He, X.H.; Thomas, B.; Lupwayi, N.Z.; Hao, X.Y.; Thomas, M.C.; Shi, X.J. Fertilization Shapes Bacterial Community Structure by Alteration of Soil pH. Front. Microbiol. 2017, 8, 1325. [Google Scholar] [CrossRef] [Green Version]
- Sorinolu, A.J.; Tyagi, N.; Kumar, A.; Munir, M. Antibiotic resistance development and human health risks during wastewater reuse and biosolids application in agriculture. Chemosphere 2021, 265, 129032. [Google Scholar] [CrossRef]
- Gómez-Sagasti, M.T.; Hernández, A.; Artetxe, U.; Garbisu, C.; Becerril, J.M. How Valuable Are Organic Amendments as Tools for the Phytomanagement of Degraded Soils? The Knowns, Known Unknowns, and Unknowns. Front. Sustain. Food Syst. 2018, 2, 68. [Google Scholar] [CrossRef] [Green Version]
- Eurostat. Sewage Sludge Production and Disposal from Urban Wastewater. 2017. Available online: https://ec.europa.eu/eurostat/statistics-explained/index.php?title=File:Sewage_sludge_disposal_from_urban_wastewater_treatment,_by_treatment_method,_2017_(%25_of_total_dry_mass).png (accessed on 5 May 2021).
- Czech Statistical Office, C.S. Consumption of Fertilisers in the Crop Year. 2018. Available online: https://vdb.czso.cz/vdbvo2/faces/index.jsf?page=vystup-objekt&pvo=ZEM11#w= (accessed on 5 May 2021).
- Czech Statistical Office, C.S. Production and Disposal of Wastewater Digested Sludge. 2018. Available online: https://www.czso.cz/csu/czso/3-environment-v440y0ba3f (accessed on 5 May 2021).
- Collivignarelli, M.C.; Abba, A.; Frattarola, A.; Miino, M.C.; Padovani, S.; Katsoyiannis, I.; Torretta, V. Legislation for the Reuse of Biosolids on Agricultural Land in Europe: Overview. Sustainability 2019, 11, 6015. [Google Scholar] [CrossRef] [Green Version]
- EUR-Lex. Closing the Loop—An EU Action Plan for the Circular Economy. 2015. Available online: https://eur-lex.europa.eu/resource.html?uri=cellar:8a8ef5e8-99a0-11e5-b3b7-01aa75ed71a1.0012.02/DOC_1&format=PDF (accessed on 5 May 2021).
- Urra, J.; Alkorta, I.; Garbisu, C. Potential Benefits and Risks for Soil Health Derived From the Use of Organic Amendments in Agriculture. Agronomy 2019, 9, 542. [Google Scholar] [CrossRef] [Green Version]
- Stiborova, H.; Kolar, M.; Vrkoslavova, J.; Pulkrabova, J.; Hajslova, J.; Demnerova, K.; Uhlik, O. Linking toxicity profiles to pollutants in sludge and sediments. J. Hazard. Mater. 2017, 321, 672–680. [Google Scholar] [CrossRef] [PubMed]
- Clarke, R.M.; Cummins, E. Evaluation of “Classic” and Emerging Contaminants Resulting from the Application of Biosolids to Agricultural Lands: A Review. Hum. Ecol. Risk Assess. Int. J. 2015, 21, 492–513. [Google Scholar] [CrossRef]
- Lee, W.; Park, S.H.; Kim, J.; Jung, J.Y. Occurrence and removal of hazardous chemicals and toxic metals in 27 industrial wastewater treatment plants in Korea. Desalin. Water Treat. 2015, 54, 1141–1149. [Google Scholar] [CrossRef]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stiborova, H.; Bacakova, A.; Musilova, L.; Demnerova, K. From Natural Phenomenon of Antibiotic Resistance in the Environment to Emergence of Multiresistant Strains. Chem. Listy 2018, 112, 833–839. [Google Scholar]
- Checcucci, A.; Trevisi, P.; Luise, D.; Modesto, M.; Blasioli, S.; Braschi, I.; Mattarelli, P. Exploring the Animal Waste Resistome: The Spread of Antimicrobial Resistance Genes Through the Use of Livestock Manure. Front. Microbiol. 2020, 11, 9. [Google Scholar] [CrossRef]
- Pazda, M.; Kumirska, J.; Stepnowski, P.; Mulkiewicz, E. Antibiotic resistance genes identified in wastewater treatment plant systems—A review. Sci. Total Environ. 2019, 697, 134023. [Google Scholar] [CrossRef]
- Chen, C.Q.; Pankow, C.A.; Oh, M.; Heath, L.S.; Zhang, L.Q.; Du, P.; Xia, K.; Pruden, A. Effect of antibiotic use and composting on antibiotic resistance gene abundance and resistome risks of soils receiving manure-derived amendments. Environ. Int. 2019, 128, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Y.; Zhang, W.; Yang, L.X.; Stedtfeld, R.D.; Peng, A.P.; Gu, C.; Boyd, S.A.; Li, H. Antibiotic resistance genes and bacterial communities in cornfield and pasture soils receiving swine and dairy manures. Environ. Pollut. 2019, 248, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Han, X.-M.; Hu, H.-W.; Chen, Q.-L.; Yang, L.-Y.; Li, H.-L.; Zhu, Y.-G.; Li, X.-Z.; Ma, Y.-B. Antibiotic resistance genes and associated bacterial communities in agricultural soils amended with different sources of animal manures. Soil Biol. Biochem. 2018, 126, 91–102. [Google Scholar] [CrossRef]
- Cycoń, M.; Mrozik, A.; Piotrowska-Seget, Z. Antibiotics in the Soil Environment—Degradation and Their Impact on Microbial Activity and Diversity. Front. Microbiol. 2019, 10, 338. [Google Scholar] [CrossRef]
- Pei, M.K.; Zhang, B.; He, Y.L.; Su, J.Q.; Gin, K.; Lev, O.; Shen, G.X.; Hu, S.Q. State of the art of tertiary treatment technologies for controlling antibiotic resistance in wastewater treatment plants. Environ. Int. 2019, 131, 105026. [Google Scholar] [CrossRef]
- Schluter, A.; Szczepanowski, R.; Puhler, A.; Top, E.M. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. Fems Microbiol. Rev. 2007, 31, 449–477. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microl. 2014, 12, 465–478. [Google Scholar] [CrossRef]
- WHO. WHO Report on Surveillance of Antibiotic Consumption 2016–2018. 2018. Available online: https://www.who.int/medicines/areas/rational_use/oms-amr-amc-report-2016-2018/en/ (accessed on 5 May 2021).
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [Green Version]
- Klein, E.Y.; Van Boeckel, T.P.; Martinez, E.M.; Pant, S.; Gandra, S.; Levin, S.A.; Goossens, H.; Laxminarayan, R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, E3463–E3470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stiborova, H.; Wolfram, J.; Demnerova, K.; Macek, T.; Uhlik, O. Bacterial community structure in treated sewage sludge with mesophilic and thermophilic anaerobic digestion. Folia Microbiol. 2015, 60, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Goberna, M.; Simón, P.; Hernández, M.T.; García, C. Prokaryotic communities and potential pathogens in sewage sludge: Response to wastewaster origin, loading rate and treatment technology. Sci. Total Environ. 2018, 615, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Ramos, T.M.; Jay-Russell, M.T.; Millner, P.D.; Shade, J.; Misiewicz, T.; Sorge, U.S.; Hutchinson, M.; Lilley, J.; Pires, A.F.A. Assessment of Biological Soil Amendments of Animal Origin Use, Research Needs, and Extension Opportunities in Organic Production. Front. Sustain. Food Syst. 2019, 3. [Google Scholar] [CrossRef] [Green Version]
- Clements, D.P.; Bihn, E.A. Chapter 16—The Impact of Food Safety Training on the Adoption of Good Agricultural Practices on Farms. In Safety and Practice for Organic Food; Biswas, D., Micallef, S.A., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 321–344. [Google Scholar]
- Sharma, M.; Millner, P.D.; Hashem, F.; Vinyard, B.T.; East, C.L.; Handy, E.T.; White, K.; Stonebraker, R.; Cotton, C.P. Survival of Escherichia coli in Manure-Amended Soils Is Affected by Spatiotemporal, Agricultural, and Weather Factors in the Mid-Atlantic United States. Appl. Environ. Microbiol. 2019, 85, e02392-18. [Google Scholar] [CrossRef] [Green Version]
- Ingham, S.C.; Losinski, J.A.; Andrews, M.P.; Breuer, J.E.; Breuer, J.R.; Wood, T.M.; Wright, T.H. Escherichia coli contamination of vegetables grown in soils fertilized with noncomposted bovine manure: Garden-scale studies. Appl. Environ. Microbiol. 2004, 70, 6420–6427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FDA. Multistate Outbreak of E. coli O157:H7 Infections Linked to Romaine Lettuce (Final Update). 2018. Available online: https://www.cdc.gov/ecoli/2018/o157h7-04-18/index.html (accessed on 26 November 2020).
- FDA. Outbreak Investigation of E. coli: Romaine from Salinas, California (November 2019). Available online: https://www.fda.gov/food/outbreaks-foodborne-illness/outbreak-investigation-e-coli-romaine-salinas-california-november-2019 (accessed on 26 November 2020).
- Gu, G.; Strawn, L.K.; Oryang, D.O.; Zheng, J.; Reed, E.A.; Ottesen, A.R.; Bell, R.L.; Chen, Y.; Duret, S.; Ingram, D.T.; et al. Agricultural Practices Influence Salmonella Contamination and Survival in Pre-harvest Tomato Production. Front. Microbiol. 2018, 9, 2451. [Google Scholar] [CrossRef] [Green Version]
- Kracmarova, M.; Karpiskova, J.; Uhlik, O.; Strejcek, M.; Szakova, J.; Balik, J.; Demnerova, K.; Stiborova, H. Microbial Communities in Soils and Endosphere of Solanum tuberosum L. and their Response to Long-Term Fertilization. Microorganisms 2020, 8, 1377. [Google Scholar] [CrossRef]
- Kracmarova, M.; Kratochvilova, H.; Uhlik, O.; Strejcek, M.; Szakova, J.; Cerny, J.; Tlustos, P.; Balik, J.; Demnerova, K.; Stiborova, H. Response of soil microbes and soil enzymatic activity to 20 years of fertilization. Agronomy 2020, 10, 1542. [Google Scholar] [CrossRef]
- Fraraccio, S.; Strejcek, M.; Dolinova, I.; Macek, T.; Uhlik, O. Secondary compound hypothesis revisited: Selected plant secondary metabolites promote bacterial degradation of cis-1,2-dichloroethylene (cDCE). Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. A language and environment for statistical computing in R Foundation for Statistical Computing. In Vienna, Austria. Available online: https://_wwwR-projectorg/ (accessed on 26 November 2020).
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.L.; McGarrell, D.M.; Sun, Y.N.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callahan, B.J.; Sankaran, K.; Fukuyama, J.A.; McMurdie, P.J.; Holmes, S.P. Bioconductor Workflow for Microbiome Data Analysis: From raw reads to community analyses. F1000Research 2016, 5, 1492. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oksanen, J.B.F.G.; Kindt, R.; Legendre, P.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.5-6. 2019. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 9 May 2021).
- Xu, J.; Xu, Y.; Wang, H.M.; Guo, C.S.; Qiu, H.Y.; He, Y.; Zhang, Y.; Li, X.C.; Meng, W. Occurrence of antibiotics and antibiotic resistance genes in a sewage treatment plant and its effluent-receiving river. Chemosphere 2015, 119, 1379–1385. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot 2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Schommer, V.A.; Wenzel, B.M.; Daroit, D.J. Anaerobic co-digestion of swine manure and chicken feathers: Effects of manure maturation and microbial pretreatment of feathers on methane production. Renew. Energy 2020, 152, 1284–1291. [Google Scholar] [CrossRef]
- Heinonen-Tanski, H.; Mohaibes, M.; Karinen, P.; Koivunen, J. Methods to reduce pathogen microorganisms in manure. Livest. Sci. 2006, 102, 248–255. [Google Scholar] [CrossRef]
- Chen, Z.; Jiang, X. Microbiological Safety of Chicken Litter or Chicken Litter-Based Organic Fertilizers: A Review. Agriculture 2014, 4, 1–29. [Google Scholar] [CrossRef] [Green Version]
- Usman, K.; Khan, S.; Ghulam, S.; Khan, M.U.; Khan, N.; Khan, M.A.; Khalil, S.K. Sewage Sludge: An Important Biological Resource for Sustainable Agriculture and Its Environmental Implications. Am. J. Plant Sci. 2012, 3, 14. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.L.; Ortiz, R.; Steele, T.W.J.; Stuckey, D.C. Toxicants inhibiting anaerobic digestion: A review. Biotechnol. Adv. 2014, 32, 1523–1534. [Google Scholar] [CrossRef]
- López, A.; Rodríguez-Chueca, J.; Mosteo, R.; Gómez, J.; Ormad, M.P. Microbiological quality of sewage sludge after digestion treatment: A pilot scale case of study. J. Clean. Prod. 2020, 254, 120101. [Google Scholar] [CrossRef]
- Piterina, A.V.; Bartlett, J.; Pembroke, T.J. Evaluation of the removal of indicator bacteria from domestic sludge processed by Autothermal Thermophilic Aerobic Digestion (ATAD). Int. J. Environ. Res. Public Health 2010, 7, 3422–3441. [Google Scholar] [CrossRef] [Green Version]
- Lloret, E.; Pastor, L.; Martínez-Medina, A.; Blaya, J.; Pascual, J.A. Evaluation of the removal of pathogens included in the Proposal for a European Directive on spreading of sludge on land during autothermal thermophilic aerobic digestion (ATAD). Chem. Eng. J. 2012, 198, 171–179. [Google Scholar] [CrossRef]
- Abdallah, A.M.; Rashid, M.; Adroub, S.A.; Arnoux, M.; Ali, S.; van Soolingen, D.; Bitter, W.; Pain, A. Complete Genome Sequence of Mycobacterium phlei Type Strain RIVM601174. J. Bacteriol. 2012, 194, 3284–3285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghielmetti, G.; Friedel, U.; Scherrer, S.; Sarno, E.; Landolt, P.; Dietz, O.; Hilbe, M.; Zweifel, C.; Stephan, R. Non-tuberculous Mycobacteria isolated from lymph nodes and faecal samples of healthy slaughtered cattle and the abattoir environment. Transbound. Emerg. Dis. 2018, 65, 711–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jahansepas, A.; Aghazadeh, M.; Rezaee, M.A.; Hasani, A.; Sharifi, Y.; Aghazadeh, T.; Mardaneh, J. Occurrence of Enterococcus faecalis and Enterococcus faecium in Various Clinical Infections: Detection of Their Drug Resistance and Virulence Determinants. Microb. Drug Resist. 2018, 24, 76–82. [Google Scholar] [CrossRef]
- Gu, G.Y.; Strawn, L.K.; Zheng, J.; Reed, E.A.; Rideout, S.L. Diversity and Dynamics of Salmonella enterica in Water Sources, Poultry Litters, and Field Soils Amended With Poultry Litter in a Major Agricultural Area of Virginia. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Alegbeleye, O.O.; Singleton, I.; Sant’Ana, A.S. Sources and contamination routes of microbial pathogens to fresh produce during field cultivation: A review. Food Microbiol. 2018, 73, 177–208. [Google Scholar] [CrossRef]
- Maffei, D.F.; Batalha, E.Y.; Landgraf, M.; Schaffner, D.W.; Franco, B.D.G.M. Microbiology of organic and conventionally grown fresh produce. Braz. J. Microbiol. 2016, 47, 99–105. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.M.; Zhang, S.W.; Yuan, Z.W. Adoption of solid organic waste composting products: A critical review. J. Clean. Prod. 2020, 272, 122712. [Google Scholar] [CrossRef]
- Tran, D.T.Q.; Bradbury, M.I.; Van Ogtrop, F.F.; Bozkurt, H.; Jones, B.J.; McConchie, R. Environmental Drivers for Persistence of Escherichia coli and Salmonella in Manure-Amended Soils: A Meta-Analysis. J. Food Prot. 2020, 83, 1268–1277. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, A.Z.; Cen, T.Y.; Li, X.Y.; He, M.; Li, D.; Chen, J.M. Sub-inhibitory concentrations of heavy metals facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes in water environment. Environ. Pollut. 2018, 237, 74–82. [Google Scholar] [CrossRef]
- Li, J.; Phulpoto, I.A.; Zhang, G.; Yu, Z. Acceleration of emergence of E. coli antibiotic resistance in a simulated sublethal concentration of copper and tetracycline co-contaminated environment. AMB Express 2021, 11, 14. [Google Scholar] [CrossRef]
- Stalder, T.; Barraud, O.; Casellas, M.; Dagot, C.; Ploy, M.-C. Integron Involvement in Environmental Spread of Antibiotic Resistance. Front. Microbiol. 2012, 3, 119. [Google Scholar] [CrossRef] [Green Version]
- An, X.-L.; Chen, Q.-L.; Zhu, D.; Zhu, Y.-G.; Gillings, M.R.; Su, J.-Q. Impact of Wastewater Treatment on the Prevalence of Integrons and the Genetic Diversity of Integron Gene Cassettes. Appl. Environ. Microbiol. 2018, 84, e02766-17. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Wang, P.L.; Yang, Q.X. Occurrence and diversity of antibiotic resistance in untreated hospital wastewater. Sci. Total Environ. 2018, 621, 990–999. [Google Scholar] [CrossRef]
- Conde-Cid, M.; Nunez-Delgado, A.; Fernandez-Sanjurjo, M.J.; Alvarez-Rodriguez, E.; Fernandez-Calvino, D.; Arias-Estevez, M. Tetracycline and Sulfonamide Antibiotics in Soils: Presence, Fate and Environmental Risks. Processes 2020, 8, 1479. [Google Scholar] [CrossRef]
- Granados-Chinchilla, F.; Rodríguez, C. Tetracyclines in Food and Feedingstuffs: From Regulation to Analytical Methods, Bacterial Resistance, and Environmental and Health Implications. J. Anal. Methods Chem. 2017, 2017, 1315497. [Google Scholar] [CrossRef]
- Ibrahim, M.; Ahmad, F.; Yaqub, B.; Ramzan, A.; Imran, A.; Afzaal, M.; Mirza, S.A.; Mazhar, I.; Younus, M.; Akram, Q.; et al. Current trends of antimicrobials used in food animals and aquaculture. In Antibiotics and Antimicrobial Resistance Genes in the Environment; Elsevier: Amsterdam, The Netherlands, 2020; pp. 39–69. [Google Scholar] [CrossRef]
- Taylor, P.; Reeder, R. Antibiotic use on crops in low and middle-income countries based on recommendations made by agricultural advisors. CABI Agric. Biosci. 2020, 1, 1–14. [Google Scholar] [CrossRef]
- Antunes, P.; Machado, J.; Sousa, J.C.; Peixe, L. Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica strains and relation with integrons. Antimicrob. Agents Chemother. 2005, 49, 836–839. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Shin, S.G.; Jang, H.M.; Kim, Y.B.; Lee, J.; Kim, Y.M. Characterization of antibiotic resistance genes in representative organic solid wastes: Food waste-recycling wastewater, manure, and sewage sludge. Sci. Total Environ. 2017, 579, 1692–1698. [Google Scholar] [CrossRef]
- Jiang, H.; Cheng, H.; Liang, Y.; Yu, S.; Yu, T.; Fang, J.; Zhu, C. Diverse Mobile Genetic Elements and Conjugal Transferability of Sulfonamide Resistance Genes (sul1, sul2, and sul3) in Escherichia coli Isolates From Penaeus vannamei and Pork From Large Markets in Zhejiang, China. Front. Microbiol. 2019, 10, 1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domínguez, M.; Miranda, C.D.; Fuentes, O.; de la Fuente, M.; Godoy, F.A.; Bello-Toledo, H.; González-Rocha, G. Occurrence of Transferable Integrons and sul and dfr Genes Among Sulfonamide-and/or Trimethoprim-Resistant Bacteria Isolated From Chilean Salmonid Farms. Front. Microbiol. 2019, 10, 748. [Google Scholar] [CrossRef]
- Fyfe, C.; Grossman, T.H.; Kerstein, K.; Sutcliffe, J. Resistance to macrolide antibiotics in public health pathogens. Cold Spring Harb. Perspect. Med. 2016, 6, a025395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wijesekara, P.N.K.; Kumbukgolla, W.W.; Jayaweera, J.A.A.S.; Rawat, D. Review on Usage of Vancomycin in Livestock and Humans: Maintaining Its Efficacy, Prevention of Resistance and Alternative Therapy. Vet. Sci. 2017, 4, 6. [Google Scholar] [CrossRef] [Green Version]
- Murray, R.; Tien, Y.-C.; Scott, A.; Topp, E. The impact of municipal sewage sludge stabilization processes on the abundance, field persistence, and transmission of antibiotic resistant bacteria and antibiotic resistance genes to vegetables at harvest. Sci. Total Environ. 2019, 651, 1680–1687. [Google Scholar] [CrossRef]
- Heuer, H.; Smalla, K. Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ. Microbiol. 2007, 9, 657–666. [Google Scholar] [CrossRef]
- Heuer, H.; Schmitt, H.; Smalla, K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 2011, 14, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Feng, K.; Li, S.; Zhang, Y.; Chen, H.; Yin, H.; Xu, M.; Deng, Y. Exploring abundance, diversity and variation of a widespread antibiotic resistance gene in wastewater treatment plants. Environ. Int. 2018, 117, 186–195. [Google Scholar] [CrossRef]
- Schwarz, J.; Thiele-Bruhn, S.; Eckhardt, K.U.; Schulten, H.R. Sorption of Sulfonamide Antibiotics to Soil Organic Sorbents: Batch Experiments with Model Compounds and Computational Chemistry. ISRN Soil Sci. 2012, 2012, 159189. [Google Scholar] [CrossRef] [Green Version]
- Sukul, P.; Lamshöft, M.; Zühlke, S.; Spiteller, M. Sorption and desorption of sulfadiazine in soil and soil-manure systems. Chemosphere 2008, 73, 1344–1350. [Google Scholar] [CrossRef]
- Tien, Y.-C.; Drury, C.; Reynolds, D.; Topp, E. Enrichment of antibiotic resistance genes in soil receiving composts derived from swine manure, yard wastes, or food wastes, and evidence for multi-year persistence of swine Clostridium spp. Can. J. Microbiol. 2017, 64, 201–208. [Google Scholar] [CrossRef]
- Cerqueira, F.; Matamoros, V.; Bayona, J.M.; Berendonk, T.U.; Elsinga, G.; Hornstra, L.M.; Pina, B. Antibiotic resistance gene distribution in agricultural fields and crops. A soil-to-food analysis. Environ. Res. 2019, 177, 108608. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Hu, H.W.; Chen, Q.L.; Singh, B.K.; Yan, H.; Chen, D.L.; He, J.Z. Transfer of antibiotic resistance from manure-amended soils to vegetable microbiomes. Environ. Intern. 2019, 130, 104912. [Google Scholar] [CrossRef]
- Zhu, Y.G.; Johnson, T.A.; Su, J.Q.; Qiao, M.; Guo, G.X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.Y.; Li, G.X.; Chang, J.L.; Kong, Y.L.; Jiang, T.; Wang, J.N.; Yuan, J. Enrichment of antibiotic resistance genes after sheep manure aerobic heap composting. Bioresour. Technol. 2021, 323, 124620. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Chen, Z.; Zhang, F.; Liu, Y.; Yan, T. Abundance, diversity and mobility potential of antibiotic resistance genes in pristine Tibetan Plateau soil as revealed by soil metagenomics. FEMS Microbiol. Ecol. 2020, 96, fiaa172. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Shen, M.; Zhang, Y.; Zhu, D.; Wang, H.; Zhao, Y.; Kang, Y. The changes in antibiotic resistance genes during 86 years of the soil ripening process without anthropogenic activities. Chemosphere 2021, 266, 128985. [Google Scholar] [CrossRef] [PubMed]
- Szekeres, E.; Chiriac, C.M.; Baricz, A.; Szőke-Nagy, T.; Lung, I.; Soran, M.L.; Rudi, K.; Dragos, N.; Coman, C. Investigating antibiotics, antibiotic resistance genes, and microbial contaminants in groundwater in relation to the proximity of urban areas. Environ. Pollut. 2018, 236, 734–744. [Google Scholar] [CrossRef]
- Sabri, N.A.; Schmitt, H.; Van der Zaan, B.; Gerritsen, H.W.; Zuidema, T.; Rijnaarts, H.H.M.; Langenhoff, A.A.M. Prevalence of antibiotics and antibiotic resistance genes in a wastewater effluent-receiving river in the Netherlands. J. Environ. Chem. Eng. 2020, 8, 102245. [Google Scholar] [CrossRef]
- Jackson, C.R.; Fedorka-Cray, P.J.; Barrett, J.B.; Ladely, S.R. Effects of Tylosin Use on Erythromycin Resistance in Enterococci Isolated from Swine. Appl. Environ. Microbiol. 2004, 70, 4205–4210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, A.; Nawaz, M.; Khan, S.; Steele, R. Detection and characterization of erythromycin-resistant methylase genes in Gram-positive bacteria isolated from poultry litter. Appl. Microbiol. Biotechnol. 2002, 59, 377–381. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).