Exogenous GbHMGS1 Overexpression Improves the Contents of Three Terpenoids in Transgenic Populus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Extraction of DNA and RNA
2.3. Expression Analyses
2.4. Subcellular Localization of HMGS1
2.5. Plant Transformation
2.6. Metabolome Analysis
3. Results
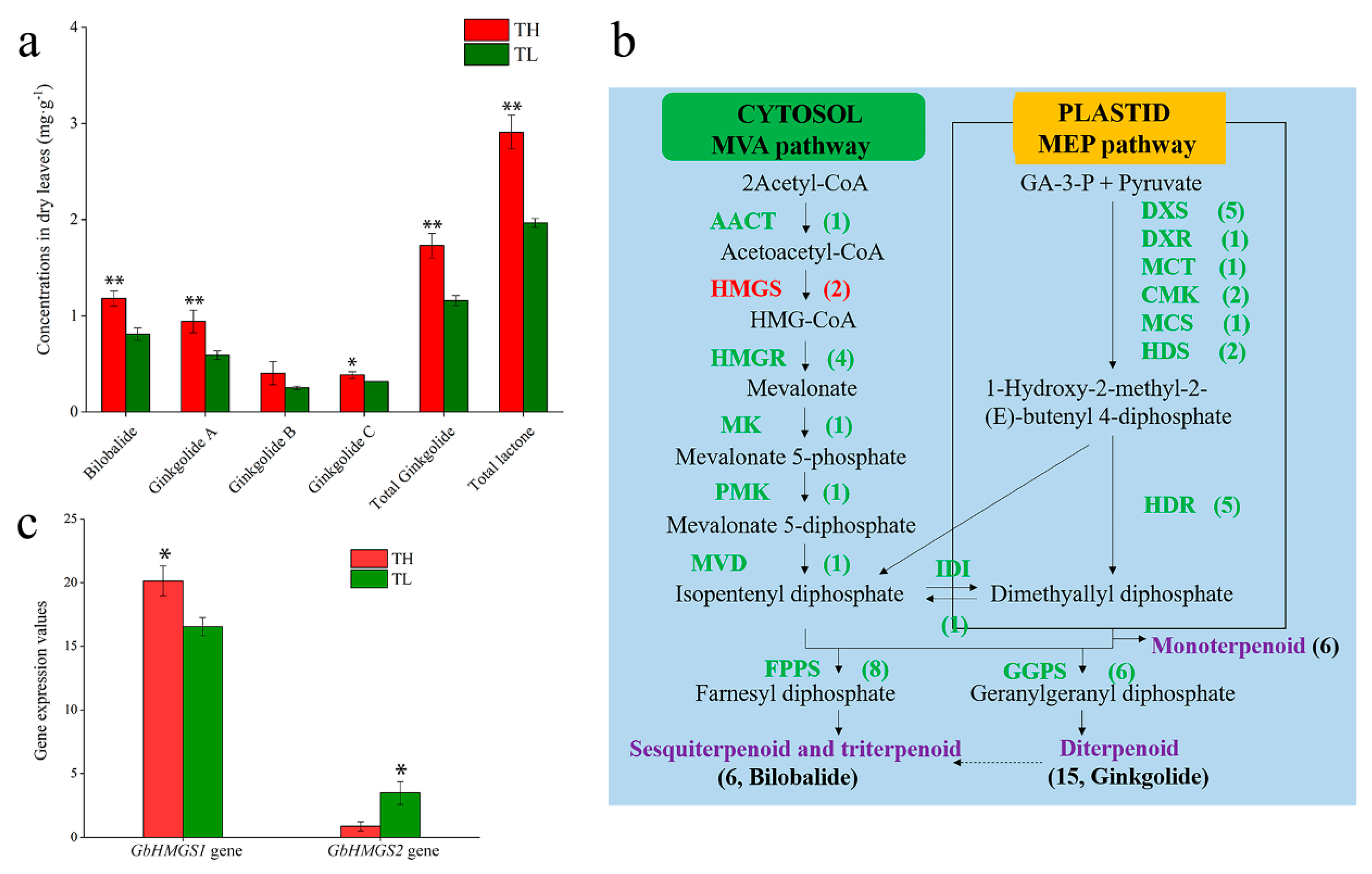
3.1. Determination of Total Lactone Content in Ginkgo
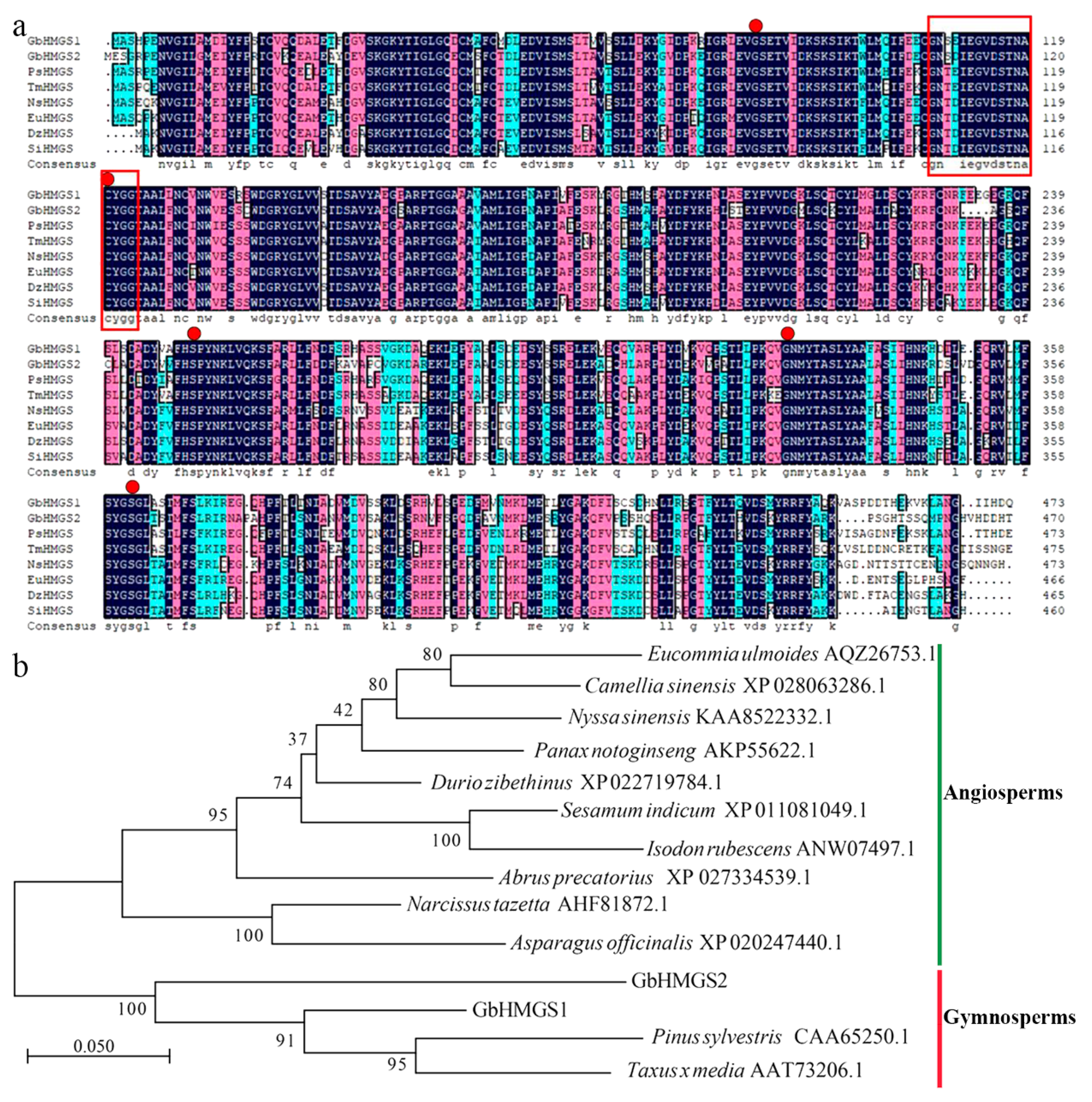
3.2. Identification and Characterization of Candidate Terpenoid Biosynthesis Genes
3.3. Expression Patterns of GbHMGS1 in Ginkgo
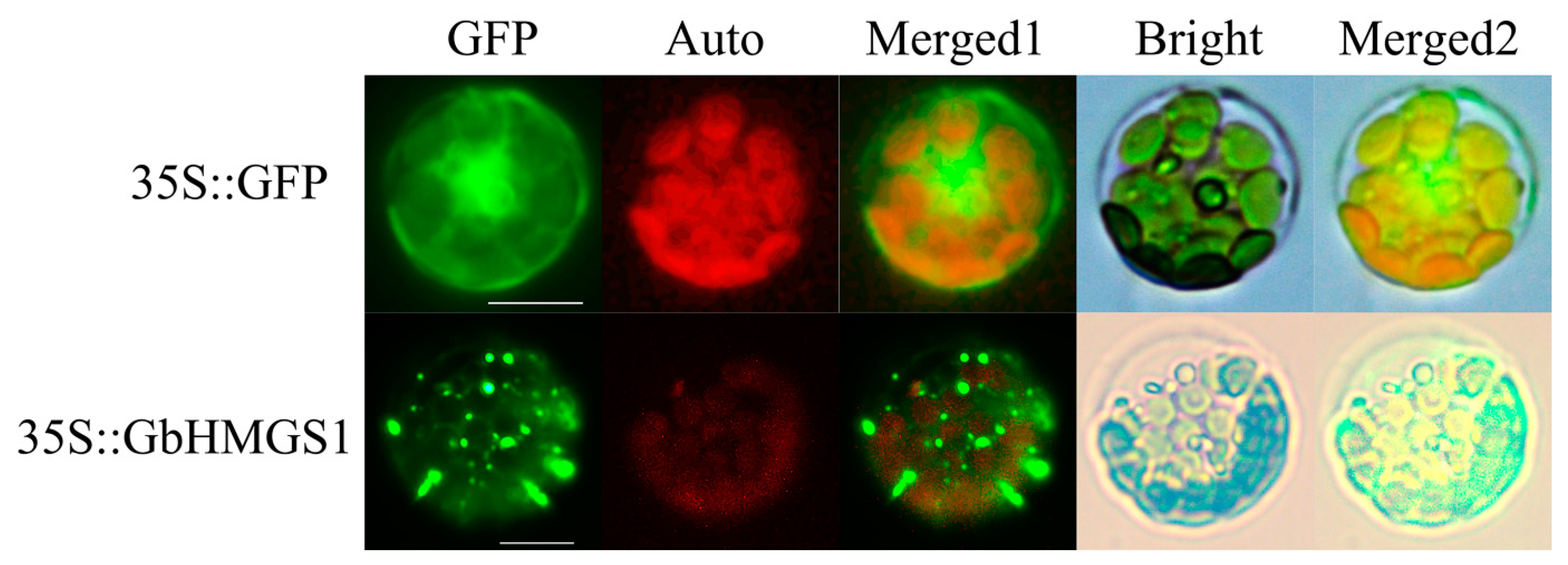
3.4. Subcellular Localization of the GbHMGS1 Protein
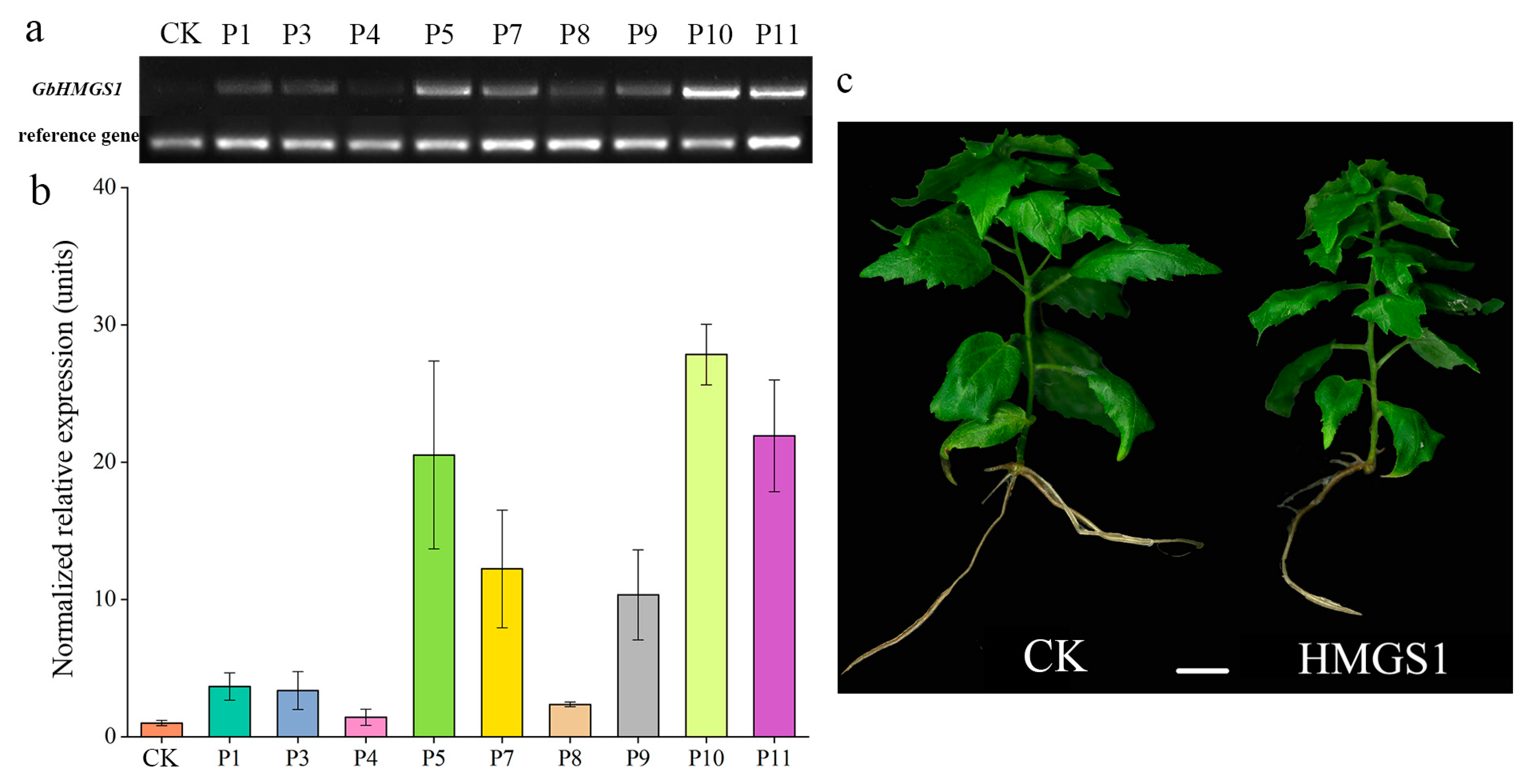
3.5. Selection of Targeted Transgenic Lines of HMGS1 in Populus
3.6. Metabolic Differences between the Transgenic and CK Poplar Plants
4. Discussion
4.1. Terpenoid Backbone Biosynthesis
4.2. Expression Patterns
4.3. Gene Functions
4.4. Differences in Metabolites
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Role of the Funding Source
Conflicts of Interest
References
- Šmarda, P.; Horová, L.; Knápek, O.; Dieck, H.; Dieck, M.; Ražná, K.; Hrubík, P.; Orlóci, L.; Papp, L.; Veselá, K.; et al. Multiple haploids, triploids, and tetraploids found in modern-day “living fossil” Ginkgo biloba. Hortic. Res. 2018, 5, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Cao, F.; Yin, T.; Chen, Y. A highly dense genetic map for Ginkgo biloba constructed using sequence-based markers. Front. Plant Sci. 2017, 8, 1041. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Guo, J.; Zhou, Q.; Xin, Y.; Wang, G.; Xu, L.-A. De novo transcriptome analysis revealed genes involved in flavonoid biosynthesis, transport and regulation in Ginkgo biloba. Ind. Crops Prod. 2018, 124, 226–235. [Google Scholar] [CrossRef]
- DeFeudis, F.V.; Papadopoulos, V.; Drieu, K. Ginkgo biloba extracts and cancer: A research area in its infancy. Fundam. Clin. Pharmacol. 2003, 17, 405–417. [Google Scholar] [CrossRef]
- Rodriguez, M.; Ringstad, L.; Schafer, P.; Just, S.; Hofer, H.W.; Malmsten, M.; Siegel, G. Reduction of atherosclerotic nanoplaque formation and size by Ginkgo biloba (EGb 761) in cardiovascular high-risk patients. Atherosclerosis 2007, 192, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Song, H.; Sun, M.; Zhu, Z.; Xing, G.; Xu, X.; Gao, M.; Hou, L.; Li, M. Digital gene expression analysis during floral transition in pak choi (Brassica rapa subsp. chinensis). Biotechnol. Biotechnol. Equip. 2017, 31, 670–678. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Jin, X.; Chen, Y.; Wei, M.; Liao, W.; Zhao, S.; Fu, C.; Yu, L. TcMYC2a, a basic helix-loop-helix transcription factor, transduces JA-signals and regulates taxol biosynthesis in Taxus chinensis. Front. Plant. Sci. 2018, 9, 863. [Google Scholar] [CrossRef]
- Miziorko, H.M. Enzymes of the mevalonate pathway of isoprenoid biosynthesis. Arch. Biochem. Biophys. 2011, 505, 131–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pulido, P.; Perello, C.; Rodriguez-Concepcion, M. New insights into plant isoprenoid metabolism. Mol. Plant 2012, 5, 964–967. [Google Scholar] [CrossRef] [Green Version]
- Shin, B.L.; Kim, M.; Han, J. Exceptionally high percentage of IPP synthesis by Ginkgo biloba IspH is mainly due to Phe residue in the active site. Phytochemistry 2017, 136, 9–14. [Google Scholar] [CrossRef]
- Bongers, M.; Perez-Gil, J.; Hodsom, M.P.; Schrübbers, L.; Wulff, T.; Sommer, M.O.; Nielsen, L.K.; Vickers, C.E. Adaptation of hydroxymethylbutenyl diphosphate reductase enables volatile isoprenoid production. ELIFE 2020, e48685. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Xu, F.; Huang, X.; Zhang, W.; Cheng, H.; Wang, X.; Cheng, S.; Shen, Y. Characterization and transcriptional profiling of Ginkgo biloba mevalonate diphosphate decarboxylase gene (GbMVD) promoter towards light and exogenous hormone treatments. Plant Mol. Biol. Rep. 2016, 34, 566–581. [Google Scholar] [CrossRef]
- Meng, X.; Xu, F.; Song, Q.; Ye, J.; Liao, Y.; Zhang, W. Isolation, characterization and functional analysis of a novel 3-hydroxy-3-methylglutaryl-coenzyme A synthase gene (GbHMGS2) from Ginkgo biloba. Acta Physiol. Plant. 2018, 40, 72. [Google Scholar] [CrossRef]
- Montamat, F.; Guilloton, M.; Karst, F.; Delrot, S. Isolation and characterization of a cDNA encoding Arabidopsis thaliana 3-hydroxy-3-methylglutaryl-coenzyme A synthase. Gene 1995, 167, 197–201. [Google Scholar] [CrossRef]
- Alex, D.; Bach, T.J.; Chye, M.L. Expression of Brassica juncea 3-hydroxy-3-methylglutaryl CoA synthase is developmentally regulated and stress-responsive. Plant J. 2000, 22, 415–426. [Google Scholar] [CrossRef]
- Kai, G.Y.; Li, S.S.; Wang, W.; Lu, Y.; Wang, J.; Liao, P.; Cui, L.J. Molecular cloning and expression analysis of a gene encoding 3-hydroxy-3-methylglutaryl-CoA synthase from Camptotheca acuminata. Russ. J. Plant Physiol. 2013, 60, 131–138. [Google Scholar] [CrossRef]
- Ren, A.; Ouyang, X.; Shi, L.; Jiang, A.L.; Mu, D.S.; Li, M.J.; Han, Q.; Zhao, M.W. Molecular characterization and expression analysis of GlHMGS, a gene encoding hydroxymethylglutaryl-CoA synthase from Ganoderma lucidum (Ling-zhi) in ganoderic acid biosynthesis pathway. World J. Microbiol. Biotechnol. 2013, 29, 523–531. [Google Scholar] [CrossRef]
- Wang, H.; Nagegowda, D.A.; Rawat, R.; Bouvier-Nave, P.; Guo, D.; Bach, T.J.; Chye, M.L. Overexpression of Brassica juncea wild-type and mutant HMG-CoA synthase 1 in Arabidopsis up-regulates genes in sterol biosynthesis and enhances sterol production and stress tolerance. Plant. Biotechnol. J. 2012, 10, 31–42. [Google Scholar] [CrossRef]
- Meng, X.; Song, Q.; Ye, J.; Wang, L.; Xu, F. Characterization, function, and transcriptional profiling analysis of 3-Hydroxy-3-methylglutaryl-CoA synthase gene (GbHMGS1) towards stresses and exogenous hormone treatments in Ginkgo biloba. Molecules 2017, 22, 1706. [Google Scholar] [CrossRef] [Green Version]
- Ayadi, R.; Trémouillaux-Guiller, J. Root formation from transgenic calli of Ginkgo biloba. Tree Physiol. 2003, 23, 713–718. [Google Scholar] [CrossRef] [Green Version]
- Dupré, P.; Lacoux, J.; Neutelings, G.; Mattar-Laurain, D.; Fliniaux, M.-A.; David, A.; Jacquin-Dubreuil, A. Genetic transformation of Ginkgo biloba by Agrobacterium tumefaciens. Physiol. Plant. 2000, 108, 413–419. [Google Scholar] [CrossRef]
- Wu, Y.; Guo, J.; Zhou, Q.; Xu, M.; Wang, G.; Xu, L.-A. Variation analysis of flavonoids and related active substances content in ginkgo leaves from different regions. J. Nanjing For. Univ. (Nat. Sci. Ed.) 2019, 43, 183–188. [Google Scholar] [CrossRef]
- Zhou, Q.; Mu, K.; Xu, M.; Ma, X.; Ni, Z.; Wang, J.; Xu, L.-A. Variation in the concentrations of major secondary metabolites in Ginkgo leaves from different geographical populations. Forests 2017, 8, 266. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Chen, C.; Cai, H.; Wu, L. Overexpression of PeHKT1;1 improves salt tolerance in Populus. Genes 2018, 9, 475. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Ma, S.; Kong, X.; Takano, T.; Liu, S. Efficient agrobacterium-mediated transformation of hybrid poplar Populus davidiana dode X Populus bollena lauche. Int. J. Mol. Sci. 2013, 14, 2515–2528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Wang, T.; Xin, Y.; Wang, G.; Xu, L.-A. Overexpression of the GbF3′H1 gene enhanced the epigallocatechin, gallocatechin, and catechin contents in transgenic Populus. J. Agr. Food Chem. 2020, 68, 998–1006. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, B.; Su, X.; Zhang, S.; Huang, M. Reference gene selection for quantitative real-time polymerase chain reaction in Populus. Anal. Biochem. 2011, 408, 337–339. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Xu, M.; Chen, Y.; Huang, M. Transient expression for functional gene analysis using Populus protoplasts. Plant Cell Tissue Organ Cult. 2013, 114, 11–18. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, W.; Wang, M.; Cheng, Q. Cloning and characterization of the PtVIP1 gene in Populus. J. For. Res. 2018, 30, 2259–2266. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, T.; Xin, Y.; Wang, G.; Xu, L.-A. Overexpression of GbF3′5′H1 provides a potential to improve the content of epicatechin and gallocatechin. Molecules 2020, 25, 4836. [Google Scholar] [CrossRef]
- Chen, T.; Zhao, Q.; Wang, L.; Xu, Y.; Wei, W. Comparative metabolomic analysis of the green microalga Chlorella sorokiniana cultivated in the single culture and a consortium with bacteria for wastewater remediation. Appl. Biochem. Biotechnol. 2017, 183, 1062–1075. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Lu, L.; Jin, C.; Wang, S.; Zhou, J.; Ni, Y.; Fu, Z.; Jin, Y. Effects of short term lead exposure on gut microbiota and hepatic metabolism in adult Zebrafish. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2018, 209, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ning, M.; Wei, P.; Shen, H.; Wan, X.; Jin, M.; Li, X.; Shi, H.; Qiao, Y.; Jiang, G.; Gu, W.; et al. Proteomic and metabolomic responses in hepatopancreas of whiteleg shrimp Litopenaeus vannamei infected by microsporidian Enterocytozoon hepatopenaei. Fish Shellfish Immunol. 2019, 87, 534–545. [Google Scholar] [CrossRef]
- Xiong, Q.; Chen, X.; Shen, T.; Zhong, L.; Zhu, C.; Peng, X.; He, X.; Fu, J.; Ouyang, L.; Bian, J.; et al. Metabolomics and proteomics analyses of grain yield reduction in rice under abrupt drought-flood alternation. BioRxiv 2018, 271940. [Google Scholar] [CrossRef]
- Sabater-Jara, A.B.; Souliman-Youssef, S.; Novo-Uzal, E.; Almagro, L.; Belchí-Navarro, S.; Pedreño, M.A. Biotechnological approaches to enhance the biosynthesis of ginkgolides and bilobalide in Ginkgo biloba. Phytochem. Rev. 2013, 12, 191–205. [Google Scholar] [CrossRef]
- Chen, C.; Zheng, Y.; Zhong, Y.; Wu, Y.; Li, Z.; Xu, L.A.; Xu, M. Transcriptome analysis and identification of genes related to terpenoid biosynthesis in Cinnamomum camphora. BMC Genom. 2018, 19, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Yan, X.; Wang, J.; Li, S.; Liao, P.; Kai, G. Molecular cloning and expression analysis of a new putative gene encoding 3-hydroxy-3-methylglutaryl-CoA synthase from Salvia miltiorrhiza. Acta Physiol. Plant 2011, 33, 953–961. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, K.I.; Chang, Y.J.; Kim, S.U. Developmental pattern of Ginkgo biloba levopimaradiene synthase (GbLPS) as probed by promoter analysis in Arabidopsis thaliana. Plant Cell Rep. 2012, 31, 1119–1127. [Google Scholar] [CrossRef]
- Liu, Y.J.; Zhao, Y.J.; Zhang, M.; Su, P.; Wang, X.J.; Zhang, X.N.; Gao, W.; Huang, L.Q. Cloning and characterisation of the gene encoding 3-hydroxy-3-methylglutaryl-CoA synthase in Tripterygium wilfordii. Molecules 2014, 19, 19696–19707. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Yu, W.; Wang, G.; Cao, F.; Cai, J.; Wang, H. Comparative proteomic and physiological analysis reveals the variation mechanisms of leaf coloration and carbon fixation in a xantha mutant of Ginkgo biloba L. Int. J. Mol. Sci. 2016, 17, 1794. [Google Scholar] [CrossRef] [Green Version]
- Nagegowda, D.A.; Ramalingam, S.; Hemmerlin, A.; Bach, T.J.; Chye, M.L. Brassica juncea HMG-CoA synthase: Localization of mRNA and protein. Planta 2005, 221, 844–856. [Google Scholar] [CrossRef] [PubMed]
- Crisp, M.; Liu, Q.; Roux, K.; Rattner, J.B.; Shanahan, C.; Burke, B.; Stahl, P.D.; Hodzic, D. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J. Cell Biol. 2006, 172, 41–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, P.; Chen, X.; Wang, M.; Bach, T.J.; Chye, M.L. Improved fruit α-tocopherol, carotenoid, squalene and phytosterol contents through manipulation of Brassica juncea 3-HYDROXY-3-METHYLGLUTARYL-COA SYNTHASE1 in transgenic tomato. Plant Biotechnol. J. 2018, 16, 784–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aharoni, A.; Galili, G. Metabolic engineering of the plant primary-secondary metabolism interface. Curr. Opin. Biotechnol. 2011, 22, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.J.; Sun, M.H.; Lu, J.; Liu, Y.J.; Hu, D.G.; Hao, Y.J. Transcription factor AREB2 is involved in soluble sugar accumulation by activating sugar transporter and amylase genes. Plant Physiol. 2017, 174, 2348–2362. [Google Scholar] [CrossRef]

| Primer_ID | Forward PCR Primer (5′-3′) | Reverse PCR Primer (5′-3′) | Product Length (bp) |
|---|---|---|---|
| GbHMGS1_ORF | ATGGCGTCTCATCCAGAAAATG | CAATTGATCATGGATGATACCA | 1428 |
| GbHMGS1_qRT-PCR | CTTATGGGAGTGGTTTGGCTT | AAATCTTCAGGGGACAACACA | 142 |
| Ginkgo reference_qRT-PCR | GGTGCCAAAAAGGTGGTCAT | CAACAACGAACATGGGAGCAT | - |
| Populus reference_qRT-PCR | GGCAAGGAGAAGGTACACAT | CAATCACACGCTTGTCAATA | - |
| GbHMGS1_5′OUTER | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | GGAGGCTCAAGAATGCTATGTTAC | 1731 |
| GbHMGS1_5′INNER | CTAATACGACTCACTATAGGGC | CCTCCAGGGTATCATGCTTGTTA | 1236 |
| GbHMGS1_3′OUTER | ACACAATTGGCCTTGGACAAGAC | ACTCTGCGTTGATACCACTGCTTGCCCTATAGTGAGTCGTATTAG | 1974 |
| GbHMGS1_3′INNER | GCCAGCGAGTACCCAGTTGTGGA | GCCCTATAGTGAGTCGTATTAG | 1463 |
| GbHMGS2_5′OUTER | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | GGCAGAGACATCCATTACATTTG | 1347 |
| GbHMGS2_5′INNER | CTAATACGACTCACTATAGGGC | GGATAATGTGAAAGGATGGG | 1286 |
| GbHMGS2_3′OUTER | ATTGGGCGACTGGAGGTGGGAA | ACTCTGCGTTGATACCACTGCTTGCCCTATAGTGAGTCGTATTAG | 1374 |
| GbHMGS2_3′INNER | TTCACAGTTTTGTCTTGCGG | GCCCTATAGTGAGTCGTATTAG | 887 |
| Metabolite Name | Class | Sub Class | CK | GbHMGS1 |
|---|---|---|---|---|
| Lanosterol | Prenyl lipid | Triterpenoid | 78.89 ± 2.33 | 126.37 ± 14.34 * |
| Dehydroabietic acid | Prenyl lipid | Diterpenoid | 0.07 ± 0.03 | 0.17 ± 0.02 * |
| Phytol | Prenyl lipid | Diterpenoid | 0.00 ± 0.00 | 0.05 ± 0.02 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.-Q.; Wang, T.-L.; Xin, Y.; Huang, S.-J.; Wang, G.-B.; Xu, L.-A. Exogenous GbHMGS1 Overexpression Improves the Contents of Three Terpenoids in Transgenic Populus. Forests 2021, 12, 595. https://doi.org/10.3390/f12050595
Wu Y-Q, Wang T-L, Xin Y, Huang S-J, Wang G-B, Xu L-A. Exogenous GbHMGS1 Overexpression Improves the Contents of Three Terpenoids in Transgenic Populus. Forests. 2021; 12(5):595. https://doi.org/10.3390/f12050595
Chicago/Turabian StyleWu, Ya-Qiong, Tong-Li Wang, Yue Xin, Shu-Jing Huang, Gui-Bin Wang, and Li-An Xu. 2021. "Exogenous GbHMGS1 Overexpression Improves the Contents of Three Terpenoids in Transgenic Populus" Forests 12, no. 5: 595. https://doi.org/10.3390/f12050595
APA StyleWu, Y.-Q., Wang, T.-L., Xin, Y., Huang, S.-J., Wang, G.-B., & Xu, L.-A. (2021). Exogenous GbHMGS1 Overexpression Improves the Contents of Three Terpenoids in Transgenic Populus. Forests, 12(5), 595. https://doi.org/10.3390/f12050595









