First Experimental Measurements of Biophotons from Astrocytes and Glioblastoma Cell Cultures
Abstract
1. Introduction
2. Material and Methods
2.1. The Experimental Apparatuses
2.2. Cell Lines and Sample Preparation
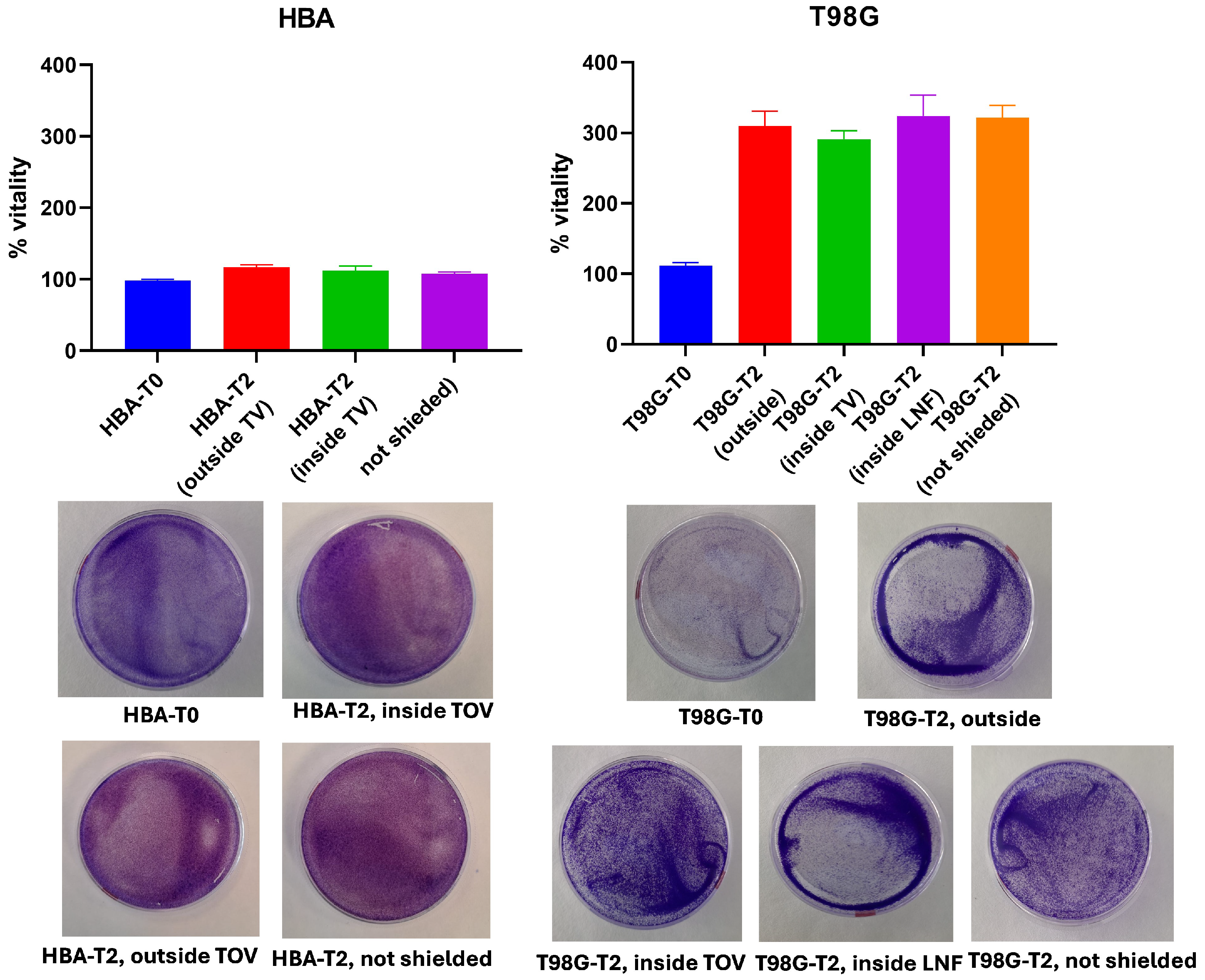
2.3. Crystal Violet Staining
2.4. The Data Taking Conditions
3. Collected Data and Preliminary Observations
3.1. The Data Taking Campaign
- (a)
- About two days of measurements in complete darkness, with no samples present in either setup, aimed at assessing residual luminescence decay and background levels.
- (b)
- Two days of measurements with astrocyte cell cultures placed in both setups.
- (c)
- Two days of measurements with glioblastoma cell cultures placed in both setups.
3.2. Statistical Analyses of the Experimental Data
4. Data Analysis and Results
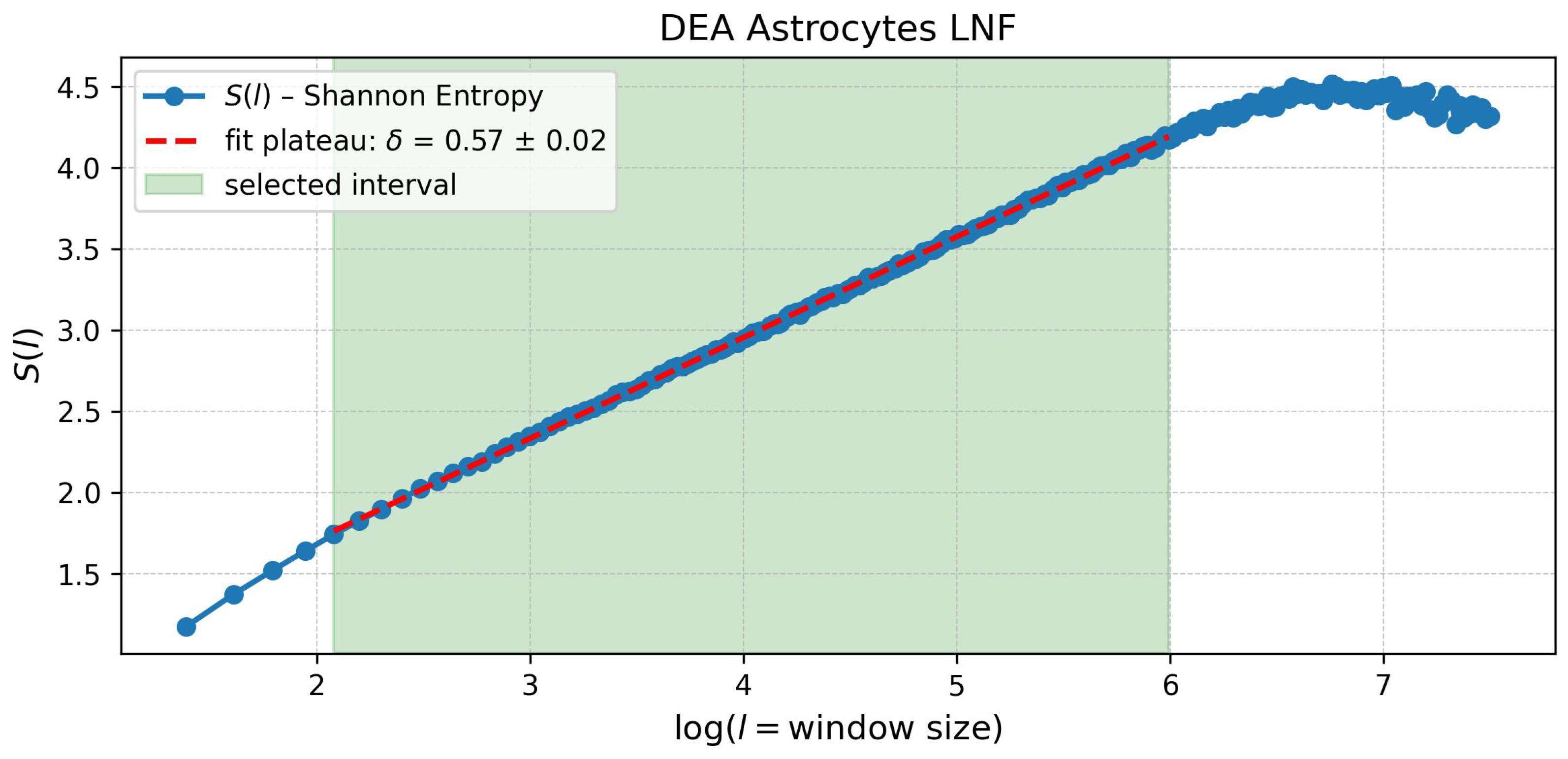
4.1. The Diffusion Entropy Analysis (DEA)
Mathematical Formulation of DEA
- : corresponds to a strongly non-stationary regime, where the waiting-time distribution has a divergent mean and variance. In this case, the processes lack a characteristic time scale, and long waiting times dominate the dynamics.
- : defines a weakly non-stationary regime in which the mean waiting time is finite while the variance diverges. The system exhibits subdiffusive behaviour with persistent temporal correlations and non-ergodic properties. This regime is of particular interest in the context of biophotonic signals, as it may indicate the presence of underlying temporal organization or self-structured dynamics rather than purely random activity.
- : indicates a stationary regime, with both finite mean and variance of the waiting-time distribution. In this case, the process becomes ergodic and effectively Markovian in the long-time limit, leading to standard (Gaussian) diffusion. This pattern is typically associated with purely random dynamics, consistent with unstructured noise that lacks long-range temporal correlations.
- Select a global interval along the horizontal axis.
- Define a moving window of width w, containing n evenly spaced data points.
- Perform a local linear fit within each window, estimating and its uncertainty .
| Setup | Measurement | DEA Without Stripes | DEA with Stripes |
|---|---|---|---|
| LNF | Background (Dark) | = 2.76 ± 0.04 = 0.62 ± 0.02 | = 3.04 ± 0.04 = 0.49 ± 0.01 |
| Astrocytes | = 2.42 ± 0.02 = 0.79 ± 0.01 | = 2.75 ± 0.06 = 0.57 ± 0.02 | |
| Glioblastoma | = 2.60 ± 0.04 = 0.70 ± 0.02 | = 2.85 ± 0.07 = 0.54 ± 0.02 | |
| TOV | Background (Dark) | = 2.90 ± 0.04 = 0.55 ± 0.02 | = 2.89 ± 0.07 = 0.53 ± 0.02 |
| Astrocytes | = 2.54 ± 0.04 = 0.73 ± 0.02 | = 2.64 ± 0.03 = 0.61 ± 0.01 | |
| Glioblastoma | = 2.70 ± 0.02 = 0.65 ± 0.01 | = 2.79 ± 0.06 = 0.56 ± 0.02 |
4.2. Results
4.2.1. GBM Cell Viability
4.2.2. DEA Results
5. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gurwitsch, A.G. Die Natur des Spezifischen Erregers der Zellteilung. Archiv für Mikroskopische Anatomie und Entwicklungsmechanik 1923, 100, 11–40. [Google Scholar] [CrossRef]
- Gurwitsch, A.G. Die Mitogenetische Strahlung. Am. J. Med. Sci. 1960, 239, 387. [Google Scholar] [CrossRef]
- Reiter, T.; Gabor, D. Zellteilung und Strahlung; Springer: Berlin/Heidelberg, Germany, 1993. [Google Scholar] [CrossRef]
- Rajewsky, B. Über Einen Empfindlichen Lichtzähler. Phys. Z. 1931, 32, 121. [Google Scholar]
- Siebert, W.W.; Seffert, H. Physikalisches Nachweis der Gurwitsch-Strahlung mit Hilfe Eines Differenzverfahrens. Naturwissenschaften 1933, 21, 193–194. [Google Scholar] [CrossRef]
- Audubert, R. Die Emission von Strahlung bei Chemische Reaktionen. Angew. Chem. 1938, 51, 153–163. [Google Scholar] [CrossRef]
- Colli, L.; Facchini, U. Light Emission by Germinating Plant. Il Nuovo Cimento 1954, 12, 150–153. [Google Scholar] [CrossRef]
- Colli, L.; Facchini, U.; Guidotti, G.; Dugnani Lonati, R.; Orsenigo, M.; Sommariva, O. Further Measurements on the Bioluminescence of the Seedlings. Experientia 1955, 11, 479–481. [Google Scholar] [CrossRef]
- Popp, F.A.; Gu, Q.; Li, K.H. Biophoton Emission: Experimental Background and Theoretical Approaches. Mod. Phys. Lett. B 1994, 8, 1269–1296. [Google Scholar] [CrossRef]
- Quickenden, T.I.; Tilbury, R.N. Luminescence Spectra of Exponential and Stationary Phase Cultures of Respiratory Deficient Saccharomyces cerevisiae. J. Photochem. Photobiol. B Biol. 1991, 8, 169–174. [Google Scholar] [CrossRef]
- Van Wijk, R. Light in Shaping Life: Biophotons in Biology and Medicine; Boekenservice: Almere, The Netherlands, 2014. [Google Scholar]
- Mayburov, S. Biophoton Production and Communications. In Proceedings of the International Conference on Nanotechnology and Nanomaterials; MGOU Publishing: Moscow, Russia, 2014; pp. 351–358. [Google Scholar]
- Slawinski, J. Necrotic Photon Emission in Stress and Lethal Interactions. Curr. Top. Biophys. 1990, 19, 8–27. [Google Scholar]
- Slawinski, J. Biophotons from Stressed and Dying Organisms: Toxicological Aspects. Indian J. Exp. Biol. 2003, 41, 483–493. [Google Scholar]
- Gallep, C.M.; Dos Santos, S.R. Photon-Count During Germination of Wheat (Triticum aestivum) Wastewater Sediment Solut. Correl. Seedl. Growth. Seed Sci. Technol. 2007, 35, 607–614. [Google Scholar] [CrossRef]
- Tessaro, L.W.E.; Dotta, B.T.; Persinger, M.A. Bacterial Biophotons as Non-Local Information Carriers: Species-Specific Spectral Characteristics of a Stress Response. Microbiol. Open 2019, 8, 761. [Google Scholar] [CrossRef]
- Popp, F.A. Cancer Growth and Its Inhibition in Terms of Coherence. Electromagn. Biol. Med. 2009, 28, 53–60. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. Classification of Tumors of the Central Nervous System: A Summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy Plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Cramer, S.W.; Chen, C.C. Photodynamic Therapy for the Treatment of Glioblastoma. Front. Surg. 2020, 6, 81. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.J.; Fakurnejad, S.; Ma, Q.; Hashizume, R. Bioluminescence Imaging of an Immunocompetent Animal Model for Glioblastoma. J. Vis. Exp. 2016, 107, 53287. [Google Scholar] [CrossRef]
- Luwor, R.B.; Stylli, S.S.; Kaye, A.H. Using Bioluminescence Imaging in Glioma Research. J. Clin. Neurosci. 2015, 22, 779–784. [Google Scholar] [CrossRef]
- Kobayashi, M.; Takeda, M.; Sato, T.; Yamazaki, Y.; Kaneko, K.; Ito, K.; Kato, H.; Inaba, H. In Vivo Imaging of Spontaneous Ultraweak Photon Emission from a Rat’s Brain Correlated with Cerebral Energy Metabolism and Oxidative Stress. Neurosci. Res. 1999, 34, 103–113. [Google Scholar] [CrossRef]
- Isojima, Y.; Isoshima, T.; Nagai, K.; Kikuchi, K.; Nakagawa, H. Ultraweak Biochemiluminescence Detected from Rat Hippocampal Slices. NeuroReport 1995, 6, 658–660. [Google Scholar] [CrossRef] [PubMed]
- De Paolis, L.; Francini, R.; Davoli, I.; De Matteis, F.; Scordo, A.; Clozza, A.; Grandi, M.; Pace, E.; Curceanu, C.; Grigolini, P.; et al. Biophotons: A Hard Problem. Appl. Sci. 2024, 14, 5496. [Google Scholar] [CrossRef]
- Benfatto, M.; Pace, E.; Curceanu, C.; Scordo, A.; Clozza, A.; Davoli, I.; Lucci, M.; Francini, R.; De Matteis, F.; Grandi, M.; et al. Biophotons and Emergence of Quantum Coherence—A Diffusion Entropy Analysis. Entropy 2021, 23, 554. [Google Scholar] [CrossRef]
- Benfatto, M.; Pace, E.; Davoli, I.; Francini, R.; De Matteis, F.; Scordo, A.; Clozza, A.; De Paolis, L.; Curceanu, C.; Grigolini, P. Biophotons: New Experimental Data and Analysis. Entropy 2023, 25, 1431. [Google Scholar] [CrossRef]
- Hamamatsu Photonics. Photon Counting Head H12386-210 Report. 2021. Available online: https://www.hamamatsu.com/eu/en/product/type/H12386-210/index.html (accessed on 7 November 2025).
- Hamamatsu Photonics. Test Sheet for the Phototube H12386-210, Serial Number 30050260. 2021. Available online: https://www.hamamatsu.com/resources/pdf/etd/H12386_TPMO1073E.pdf (accessed on 7 November 2025).
- Good, P. Permutation, Parametric, and Bootstrap Tests of Hypotheses; Springer: New York, NY, USA, 1993. [Google Scholar] [CrossRef]
- Benfatto, M.; De Paolis, L.; Tonello, L.; Grigolini, P. Advanced Data Analysis of Spontaneous Biophoton Emission: A Multi-Method Approach. arXiv 2025, arXiv:2511.11080. [Google Scholar] [CrossRef]
- Scafetta, N.; Grigolini, P. Scaling Detection in Time Series: Diffusion Entropy Analysis. Phys. Rev. E 2002, 66, 036130. [Google Scholar] [CrossRef]
- Cakir, R.; Grigolini, P.; Krokhin, A.A. Dynamical Origin of Memory and Renewal. Phys. Rev. E 2006, 74, 021108. [Google Scholar] [CrossRef]
- Grigolini, P.; Palatella, L.; Raffaelli, G. Anomalous Diffusion: An Efficient Way to Detect Memory in Time Series. Fractals 2001, 9, 439–449. [Google Scholar] [CrossRef]
- Shah, Y.H.; Palatella, L.; Mahmoodi, K.; Santonocito, O.S.; Morelli, M.; Ferri, G.; Mazzanti, C.M.; Grigolini, P.; West, B.J. Cell Motility in Cancer, Crucial Events, Criticality, and Lévy Walks. Chaos Solitons Fractals 2024, 183, 114899. [Google Scholar] [CrossRef]
- Tang, R.; Dai, J. Biophoton Signal Transmission and Processing in the Brain. J. Photochem. Photobiol. B 2014, 139, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Boone, K.; Tuszyński, J.; Barclay, P.; Simon, C. Possible Existence of Optical Communication Channels in the Brain. Sci. Rep. 2016, 6, 36508. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, T.; Wan, Q.; Wang, Q.; Chen, Z.; Gao, Y.; Ye, Y.; Lin, J.; Zhao, B.; Wang, H.; et al. TRAF4 Maintains Deubiquitination of Caveolin-1 to Drive Glioblastoma Progression. Cell Rep. 2022, 82, 3573–3587. [Google Scholar] [CrossRef]
- Murugan, N.J.; Persinger, M.A.; Karbowski, L.M.; Dotta, B.T. Ultraweak Photon Emissions as a Non-Invasive, Early-Malignancy Detection Tool: An In Vitro and In Vivo Study. Cancers 2020, 12, 1001. [Google Scholar] [CrossRef]
- Murugan, N.J.; Rouleau, N.; Karbowski, L.M.; Persinger, M.A. Biophotonic Markers of Malignancy: Discriminating Cancers Using Wavelength Specific Biophotons. Biotechnol. Rep. 2017, 13, 7–11. [Google Scholar] [CrossRef]
- Tong, J. Biophoton Signaling in Mediation of Cell-to-Cell Communication and Radiation-Induced Bystander Effects. Radiat. Med. Protect. 2024, 5, 145–160. [Google Scholar] [CrossRef]




| Setup | Data | Skewness | SNR | ||
|---|---|---|---|---|---|
| LNF | Background (Dark) | 13.76 | 4.19 | 0.41 | |
| Astrocytes | 22.82 | 5.51 | 0.33 | 1.68 | |
| Glioblastoma | 20.27 | 5.06 | 0.32 | 1.09 | |
| TOV | Background (Dark) | 16.27 | 4.38 | 0.34 | |
| Astrocytes | 20.34 | 4.81 | 0.31 | 0.54 | |
| Glioblastoma | 19.54 | 4.73 | 0.35 | 0.43 |
| Astr | 0.13 | 32.29 | 0.13 | 27.86 |
| Dark | 0.10 | 21.59 | 0.14 | 23.66 |
| Glio | 0.17 | 28.18 | 0.18 | 26.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Paolis, L.D.; Pace, E.; Mazzanti, C.M.; Morelli, M.; Lorenzo, F.D.; Tonello, L.; Curceanu, C.; Clozza, A.; Grandi, M.; Davoli, I.; et al. First Experimental Measurements of Biophotons from Astrocytes and Glioblastoma Cell Cultures. Entropy 2026, 28, 112. https://doi.org/10.3390/e28010112
Paolis LD, Pace E, Mazzanti CM, Morelli M, Lorenzo FD, Tonello L, Curceanu C, Clozza A, Grandi M, Davoli I, et al. First Experimental Measurements of Biophotons from Astrocytes and Glioblastoma Cell Cultures. Entropy. 2026; 28(1):112. https://doi.org/10.3390/e28010112
Chicago/Turabian StylePaolis, Luca De, Elisabetta Pace, Chiara Maria Mazzanti, Mariangela Morelli, Francesca Di Lorenzo, Lucio Tonello, Catalina Curceanu, Alberto Clozza, Maurizio Grandi, Ivan Davoli, and et al. 2026. "First Experimental Measurements of Biophotons from Astrocytes and Glioblastoma Cell Cultures" Entropy 28, no. 1: 112. https://doi.org/10.3390/e28010112
APA StylePaolis, L. D., Pace, E., Mazzanti, C. M., Morelli, M., Lorenzo, F. D., Tonello, L., Curceanu, C., Clozza, A., Grandi, M., Davoli, I., Gemignani, A., Grigolini, P., & Benfatto, M. (2026). First Experimental Measurements of Biophotons from Astrocytes and Glioblastoma Cell Cultures. Entropy, 28(1), 112. https://doi.org/10.3390/e28010112








