Review of Recent (2015–2024) Popular Entropy Definitions Applied to Physiological Signals
Abstract
1. Introduction
2. Recent Entropy Definitions
2.1. Distance-Based Entropy Definitions
2.1.1. Range Entropy
2.1.2. Cosine Similarity Entropy
2.1.3. Diversity Entropy
2.1.4. Distribution Entropy
2.2. Symbolic and Ordinal Pattern-Based Entropy Definitions
2.2.1. Increment Entropy
2.2.2. Dispersion Entropy
2.2.3. Fluctuation-Based Dispersion Entropy
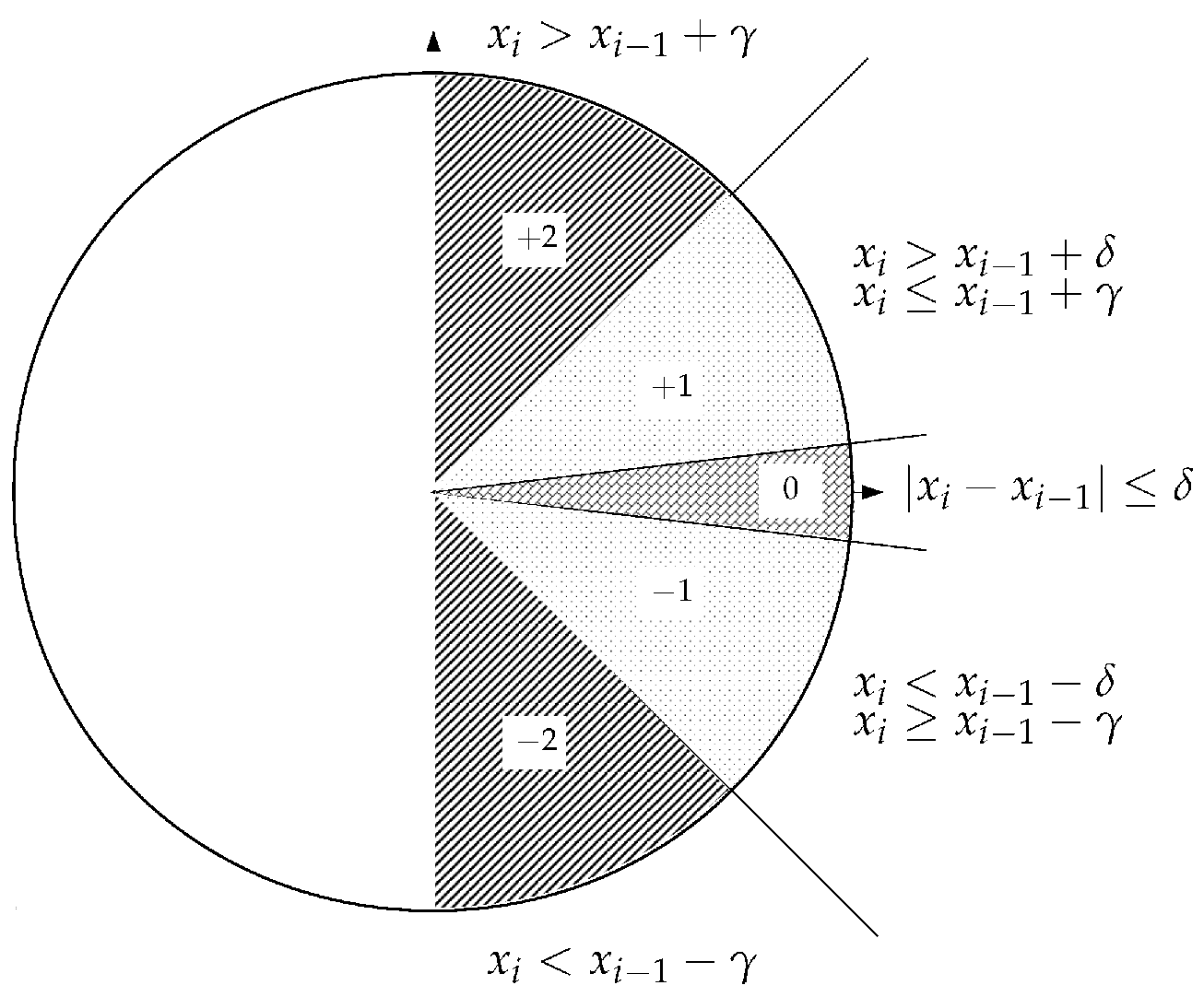
2.2.4. Slope Entropy
2.2.5. Symbolic Dynamic Entropy
2.3. Complexity Estimation Based on Sorting Effort
Bubble Entropy
2.4. Multiscale and Hierarchical Definitions
Entropy of Entropy
2.5. Geometric or Phase-Space Definitions
2.5.1. Phase Entropy
2.5.2. Gridded Distribution Entropy
2.6. Pattern-Detection Definitions
Attention Entropy
3. Methodology
- Does it propose a new entropy definition?
- Has it been published during the last decade (2015–2024)?
- Has the definition been used to analyze physiological signals (ECG/HRV, CTG, EEG, PPG, EHG, EMG)?
- Refer to any entropy definition investigating or including the name of the entropy definition in the title, abstract, or paper keywords.
- Are related at any point with either EEG, ECG/HRV, CTG, EMG, EHG, or PPG, or the word “Biomedical”.
- Belong to the “Computer Science and Engineering” field, as the most relevant available superset of Biomedical Engineering.
- (TITLE-ABS-KEY ("entropy definition")) AND
- (ALL ("EEG" OR "ECG" OR "HRV" OR "PPG" OR "Biomedical"
- OR "CTG" OR "EHG" OR "EMG"))
- AND PUBYEAR > 2014 AND PUBYEAR < 2025 AND
- (LIMIT-TO (SUBJAREA,"ENGI") OR LIMIT-TO (SUBJAREA,"COMP"))
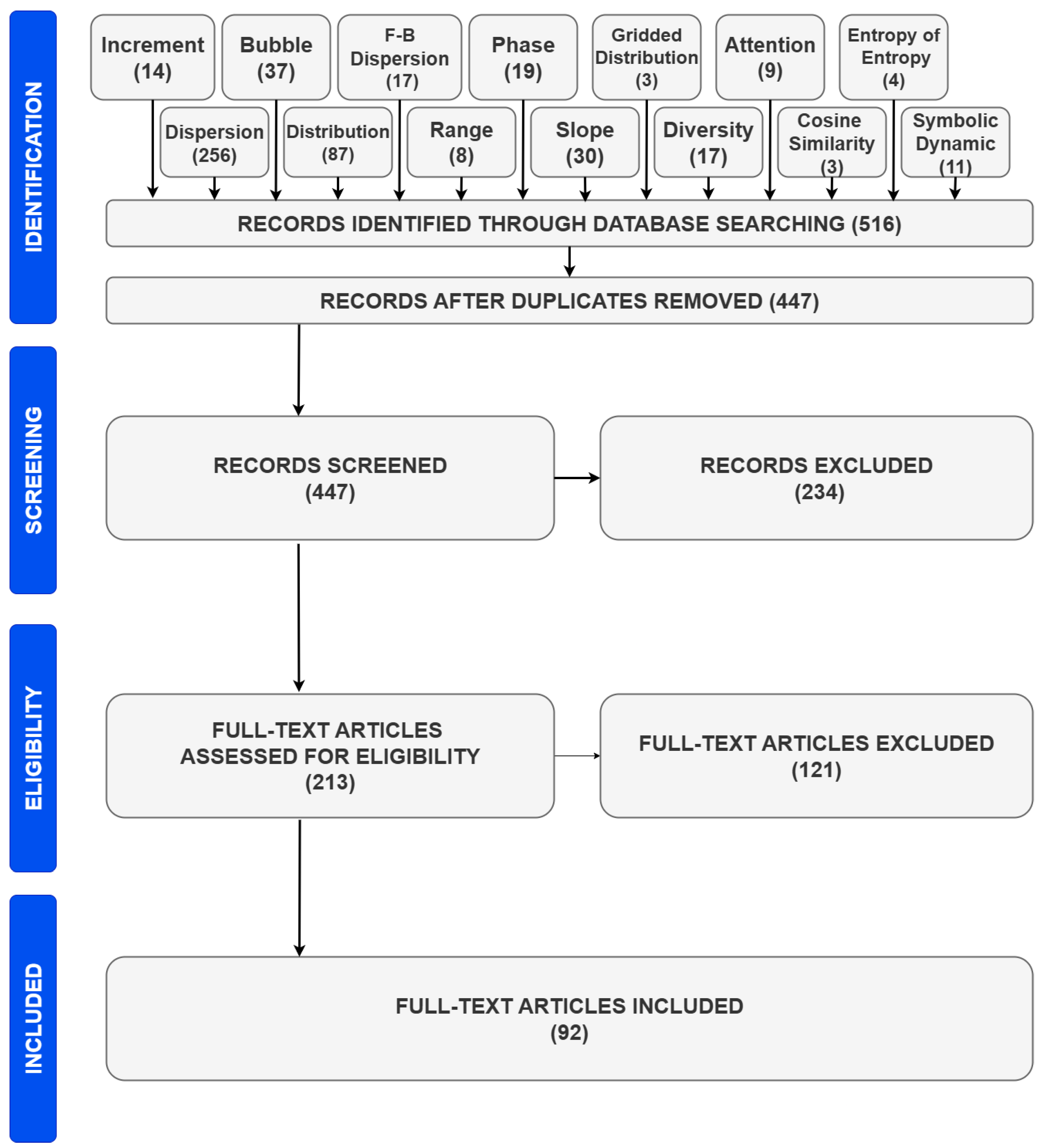
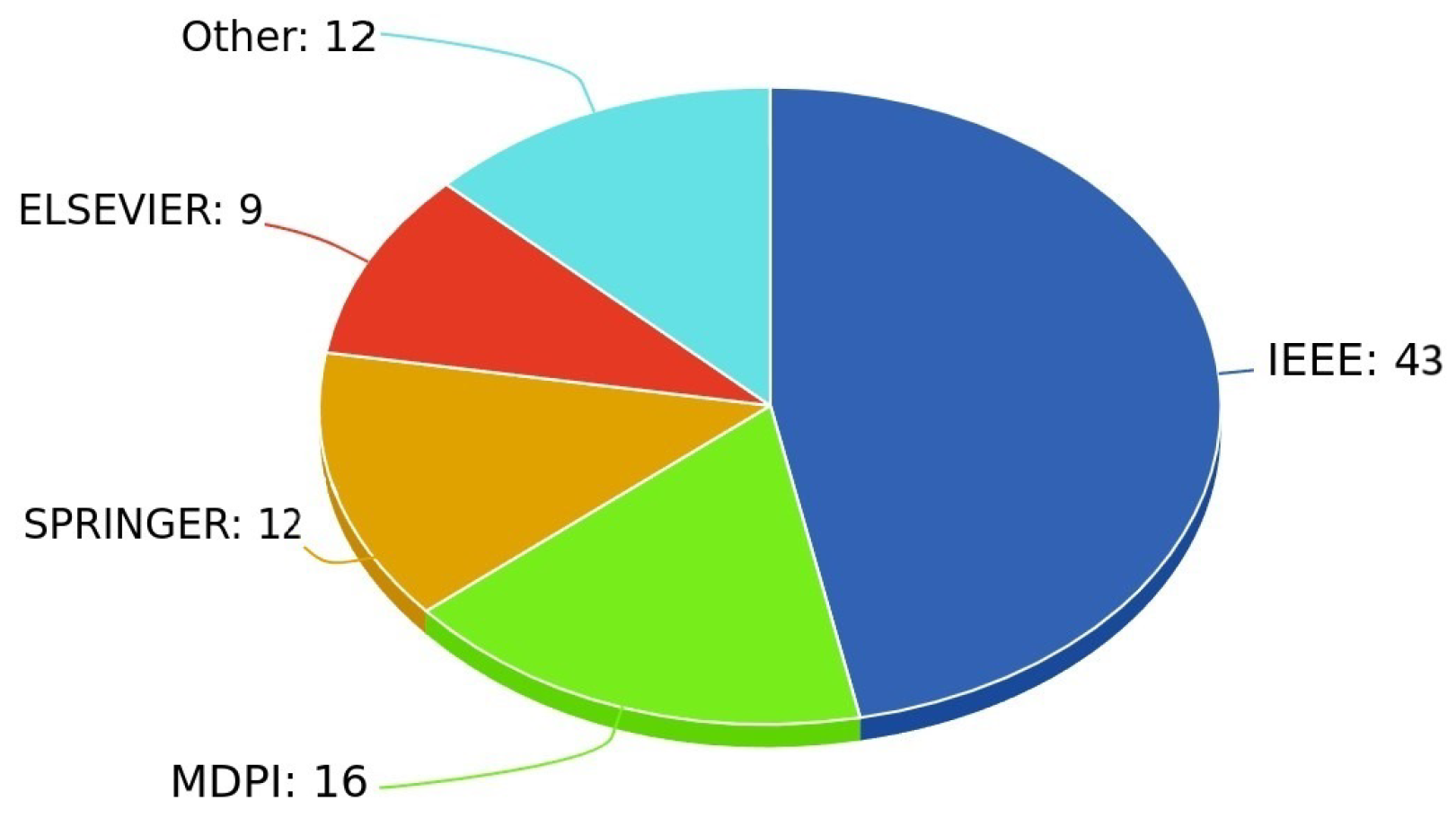
4. Search, Selection, and Descriptive Statistics
5. The Literature Review After the Systematic Article Selection
5.1. Articles on Electroencephalogram
5.2. Articles on Heart Signals
5.3. Articles on Cardiotocogram
5.4. Articles on Photoplethysmogram
5.5. Articles on Electrohysterography
5.6. Articles on Electromyography
6. Discussion
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Clausius, R. Ueber verschiedene für die Anwendung bequeme Formen der Hauptgleichungen der mechanischen Wärmetheorie. Ann. Der Phys. Und Chem. 1865, 125, 353–400. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 623–656. [Google Scholar] [CrossRef]
- Pincus, S.M. Approximate entropy as a measure of system complexity. Proc. Natl. Acad. Sci. USA 1991, 88, 2297–2301. [Google Scholar] [CrossRef]
- Richman, J.S.; Moorman, J.R. Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol.-Heart Circ. Physiol. 2000, 278, H2039–H2049. [Google Scholar] [CrossRef]
- Lake, D.E.; Richman, J.S.; Griffin, M.P.; Moorman, J.R. Sample entropy analysis of neonatal heart rate variability. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002, 283, R789–R797. [Google Scholar] [CrossRef] [PubMed]
- Bandt, C.; Pompe, B. Permutation Entropy: A Natural Complexity Measure for Time Series. Phys. Rev. Lett. 2002, 88, 174102. [Google Scholar] [CrossRef]
- Azami, H.; Faes, L.; Escudero, J.; Humeau-Heurtier, A.; Silva, L. Entropy Analysis of Univariate Biomedical Signals:Review and Comparison of Methods. In Frontiers in Entropy Across the Disciplines; Contemporary Mathematics and Its Applications: Monographs, Expositions and Lecture Notes; World Scientific: Singapore, 2022; Volume 4, pp. 233–286. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Omidvarnia, A.; Mesbah, M.; Pedersen, M.; Jackson, G. Range Entropy: A bridge between signal complexity and self-similarity. Entropy 2018, 20, 962. [Google Scholar] [CrossRef] [PubMed]
- Chanwimalueang, T.; Mandic, D.P. Cosine similarity entropy: Self-correlation-based complexity analysis of dynamical systems. Entropy 2017, 19, 652. [Google Scholar] [CrossRef]
- Wang, X.; Si, S.; Li, Y. Multiscale Diversity Entropy: A novel dynamical measure for fault diagnosis of rotating machinery. IEEE Trans. Ind. Inform. 2021, 17, 5419–5429. [Google Scholar] [CrossRef]
- Chen, W.; Wang, Z.; Xie, H.; Yu, W. Characterization of surface EMG signal based on fuzzy entropy. IEEE Trans. Neural Syst. Rehabil. Eng. 2007, 15, 266–272. [Google Scholar] [CrossRef]
- Li, P.; Liu, C.; Li, K.; Zheng, D.; Liu, C.; Hou, Y. Assessing the complexity of short-term heartbeat interval series by distribution entropy. Med. Biol. Eng. Comput. 2015, 53, 77–87. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, A.; Xu, N.; Xue, J. Increment entropy as a measure of complexity for time series. Entropy 2016, 18, 22. [Google Scholar] [CrossRef]
- Liu, X.; Wang, X.; Zhou, X.; Jiang, A. Appropriate use of the increment entropy for electrophysiological time series. Comput. Biol. Med. 2018, 95, 13–23. [Google Scholar] [CrossRef]
- Rostaghi, M.; Azami, H. Dispersion Entropy: A measure for time-series analysis. IEEE Signal Process. Lett. 2016, 23, 610–614. [Google Scholar] [CrossRef]
- Azami, H.; Escudero, J. Amplitude-and fluctuation-based dispersion entropy. Entropy 2018, 20, 210. [Google Scholar] [CrossRef] [PubMed]
- Cuesta-Frau, D. Slope Entropy: A new time series complexity estimator based on both symbolic patterns and amplitude information. Entropy 2019, 21, 1167. [Google Scholar] [CrossRef]
- Kouka, M.; Cuesta-Frau, D. Slope entropy characterisation: The role of the δ parameter. Entropy 2022, 24, 1456. [Google Scholar] [CrossRef] [PubMed]
- Kouka, M.; Cuesta-Frau, D.; Moltó-Gallego, V. Slope Entropy characterisation: An asymmetric approach to threshold parameters role analysis. Entropy 2024, 26, 82. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Y.; Li, G.; Xu, M.; Huang, W. A fault diagnosis scheme for planetary gearboxes using modified multi-scale symbolic dynamic entropy and mRMR feature selection. Mech. Syst. Signal Process. 2017, 91, 295–312. [Google Scholar] [CrossRef]
- Wang, J.; Li, T.; Xie, R.; Wang, X.M.; Cao, Y.Y. Fault feature extraction for multiple electrical faults of aviation electro-mechanical actuator based on symbolic dynamics entropy. In Proceedings of the 2015 IEEE International Conference on Signal Processing, Communications and Computing (ICSPCC), Ningbo, China, 19 September 2015; IEEE: Shanghai, China, 2015; pp. 1–6. [Google Scholar] [CrossRef]
- Manis, G.; Aktaruzzaman, M.; Sassi, R. Bubble entropy: An entropy almost free of parameters. IEEE Trans. Biomed. Eng. 2017, 64, 2711–2718. [Google Scholar] [CrossRef]
- Manis, G.; Bodini, M.; Rivolta, M.W.; Sassi, R. A two-steps-ahead estimator for Bubble entropy. Entropy 2021, 23, 761. [Google Scholar] [CrossRef] [PubMed]
- Manis, G.; Platakis, D.; Sassi, R. Exploration on Bubble entropy. IEEE J. Biomed. Health Inform. 2025, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.F.; Wei, S.Y.; Huang, H.P.; Hsu, L.; Chi, S.; Peng, C.K. Entropy of entropy: Measurement of dynamical complexity for biological systems. Entropy 2017, 19, 550. [Google Scholar] [CrossRef]
- Costa, M.; Goldberg, A.L.; Peng, C.-K. Multiscale entropy analysis of complex physiologic time series. Phys. Rev. Lett. 2002, 89, 068102. [Google Scholar] [CrossRef]
- Rohila, A.; Sharma, A. Phase entropy: A new complexity measure for heart rate variability. Physiol. Meas. 2019, 40, 105006. [Google Scholar] [CrossRef]
- Yan, C.; Li, P.; Liu, C.; Wang, X.; Yin, C.; Yao, L. Novel gridded descriptors of poincaré plot for analyzing heartbeat interval time-series. Comput. Biol. Med. 2019, 109, 280–289. [Google Scholar] [CrossRef]
- Yang, J.; Choudhary, G.I.; Rahardja, S.; Fränti, P. Classification of interbeat interval time-series using Attention entropy. IEEE Trans. Affect. Comput. 2023, 14, 321–330. [Google Scholar] [CrossRef]
- Arı, A. Analysis of EEG signal for seizure detection based on WPT. Electron. Lett. 2020, 56, 1381–1383. [Google Scholar] [CrossRef]
- Sukriti; Chakraborty, M.; Mitra, D. Dispersion entropy for the automated detection of epileptic seizures. In Proceedings of the 2020 IEEE 15th International Conference on Industrial and Information Systems (ICIIS), Rupnagar, India, 26–28 November 2020; IEEE: Dhanbad, India, 2020; pp. 204–207. [Google Scholar] [CrossRef]
- Huang, Y.; Zhao, Y.; Capstick, A.; Palermo, F.; Haddadi, H.; Barnaghi, P. Analyzing entropy features in time-series data for pattern recognition in neurological conditions. Artif. Intell. Med. 2024, 150, 102821. [Google Scholar] [CrossRef]
- Nabila, Y.; Zakaria, H. Epileptic seizure prediction from EEG signal recording using energy and dispersion entropy with SVM classifier. In Proceedings of the 2024 International Conference on Information Technology Research and Innovation (ICITRI), Jakarta, Indonesia, 5–6 September 2024; IEEE: Bandung, Indonesia, 2024; pp. 1–6. [Google Scholar] [CrossRef]
- Zaylaa, A.J.; Harb, A.; Khatib, F.I.; Nahas, Z.; Karameh, F.N. Entropy complexity analysis of electroencephalographic signals during pre-ictal, seizure and post-ictal brain events. In Proceedings of the 2015 International Conference on Advances in Biomedical Engineering (ICABME), Beirut, Lebanon, 16–18 September 2015; IEEE: Beirut, Lebanon, 2015; pp. 134–137. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, A.; Xu, N. Automated epileptic seizure detection in EEGs using increment entropy. In Proceedings of the 2017 IEEE 30th Canadian Conference on Electrical and Computer Engineering (CCECE), Windsor, ON, Canada, 30 April–3 May 2017; IEEE: Changzhou, China, 2017; pp. 1–4. [Google Scholar] [CrossRef]
- Chen, Z.; Ma, X.; Fu, J.; Li, Y. Ensemble improved permutation entropy: A new approach for time series analysis. Entropy 2023, 25, 1175. [Google Scholar] [CrossRef]
- Li, P.; Yan, C.; Karmakar, C.; Liu, C. Distribution entropy analysis of epileptic EEG signals. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; IEEE: Jinan, China, 2015; pp. 4170–4173. [Google Scholar] [CrossRef]
- Ali, E.; Udhayakumar, R.K.; Angelova, M.; Karmakar, C. Performance analysis of entropy methods in detecting epileptic seizure from surface Electroencephalograms. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Guadalajara, Mexico, 1–5 November 2021; IEEE: Dinajpur, Bangladesh, 2021; pp. 1082–1085. [Google Scholar] [CrossRef]
- Parui, S.; Samanta, D.; Chakravorty, N.; Mansoor, W.; Ghosh, U. A study on seizure detection performance in an automated process by extracting entropy features. In Proceedings of the 2022 5th International Conference on Signal Processing and Information Security (ICSPIS), Dubai, United Arab Emirates, 7–8 December 2022; IEEE: Kharagpur, India, 2022; pp. 86–91. [Google Scholar] [CrossRef]
- Khan, Y.A.; Tahreem, M.; Farooq, O. Single channel EEG based binary sleep and wake classification using entropy based features. In Proceedings of the 2023 International Conference on Recent Advances in Electrical, Electronics and Digital Healthcare Technologies (REEDCON), New Delhi, India, 1–3 May 2023; IEEE: Aligarh, India, 2023; pp. 100–105. [Google Scholar] [CrossRef]
- Manis, G.; Dudysova, D.; Gerla, V.; Lhotska, L. Detecting sleep spindles using entropy. In Proceedings of the European Medical and Biological Engineering Conference, Portorož, Slovenia, 29 November–3 December 2020; Springer: Ioannina, Greece, 2020; pp. 356–362. [Google Scholar] [CrossRef]
- Tripathy, R.; Acharya, U.R. Use of features from RR-time series and EEG signals for automated classification of sleep stages in deep neural network framework. Biocybern. Biomed. Eng. 2018, 38, 890–902. [Google Scholar] [CrossRef]
- Shahbakhti, M.; Beiramvand, M.; Eigirdas, T.; Solé-Casals, J.; Wierzchon, M.; Broniec-Wojcik, A.; Augustyniak, P.; Marozas, V. Discrimination of wakefulness from sleep stage I using nonlinear features of a single frontal EEG channel. IEEE Sens. J. 2022, 22, 6975–6984. [Google Scholar] [CrossRef]
- Hadra, M.; Omidvarnia, A.; Mesbah, M. Temporal complexity of EEG encodes human alertness. Physiol. Meas. 2022, 43, 095002. [Google Scholar] [CrossRef]
- Jain, R.; Ganesan, R.A. Effective diagnosis of sleep disorders using EEG and EOG signals. In Proceedings of the 2024 46th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 15–19 July 2024; IEEE: Zanzibar, Tanzania, 2024; pp. 1–4. [Google Scholar] [CrossRef]
- Tripathy, R.K.; Ghosh, S.K.; Gajbhiye, P.; Acharya, U.R. Development of automated sleep stage classification system using multivariate projection-based fixed boundary empirical wavelet transform and entropy features extracted from multichannel EEG signals. Entropy 2020, 22, 1141. [Google Scholar] [CrossRef] [PubMed]
- García-Martínez, B.; Fernández-Caballero, A.; Alcaraz, R.; Martínez-Rodrigo, A. Application of dispersion entropy for the detection of emotions with Electroencephalographic signals. IEEE Trans. Cogn. Dev. Syst. 2021, 14, 1179–1187. [Google Scholar] [CrossRef]
- Ding, X.W.; Liu, Z.T.; Li, D.Y.; He, Y.; Wu, M. Electroencephalogram emotion recognition based on dispersion entropy feature extraction using random oversampling imbalanced data processing. IEEE Trans. Cogn. Dev. Syst. 2021, 14, 882–891. [Google Scholar] [CrossRef]
- Hu, S.J.; Liu, Z.T.; Ding, X.W. Electroencephalogram emotion recognition using variational modal decomposition based dispersion entropy feature extraction. In Proceedings of the 2021 40th Chinese Control Conference (CCC), Shanghai, China, 26–28 July 2021; IEEE: Wuhan, China, 2021; pp. 3323–3326. [Google Scholar] [CrossRef]
- Kumar, H.; Ganapathy, N.; Puthankattil, S.D.; Swaminathan, R. EEG based emotion recognition using entropy features and Bayesian optimized random forest. Curr. Dir. Biomed. Eng. 2021, 7, 767–770. [Google Scholar] [CrossRef]
- Cai, H.; Liu, X.; Ni, R.; Song, S.; Cangelosi, A. Emotion recognition through combining EEG and EOG over relevant channels with optimal windowing. IEEE Trans. Hum.-Mach. Syst. 2023, 53, 697–706. [Google Scholar] [CrossRef]
- Pusarla, N.; Singh, A.; Tripathi, S.; Vujji, A.; Pachori, R.B. Exploring CEEMDAN and LMD domains entropy features for decoding EEG-based emotion patterns. IEEE Access 2024, 12, 103606–103625. [Google Scholar] [CrossRef]
- Gargano, A.; Scilingo, E.P.; Nardelli, M. The dynamics of emotions: A preliminary study on continuously annotated arousal signals. In Proceedings of the 2022 IEEE International Symposium on Medical Measurements and Applications (MeMeA), Giardini, Naxos, 22–24 June 2022; IEEE: Pisa, Italy, 2022; pp. 1–6. [Google Scholar] [CrossRef]
- García-Martínez, B.; Fernández-Caballero, A.; Alcaraz, R.; Martínez-Rodrigo, A. Assessment of dispersion patterns for negative stress detection from Electroencephalographic signals. Pattern Recognit. 2021, 119, 108094. [Google Scholar] [CrossRef]
- Liu, Z.T.; Xu, X.; She, J.; Yang, Z.; Chen, D. Electroencephalography emotion recognition based on rhythm information entropy extraction. J. Adv. Comput. Intell. Intell. Inform. 2024, 28, 1095–1106. [Google Scholar] [CrossRef]
- García-Martínez, B.; Martínez-Rodrigo, A.; Zangroniz Cantabrana, R.; Pastor Garcia, J.M.; Alcaraz, R. Application of entropy-based metrics to identify emotional distress from Electroencephalographic recordings. Entropy 2016, 18, 221. [Google Scholar] [CrossRef]
- Hadra, M.G.; Maaly, I.A.; Dweib, I. Range entropy as a discriminant feature for EEG-based alertness states identification. In Proceedings of the 2020 IEEE-EMBS Conference on Biomedical Engineering and Sciences (IECBES), Langkawi, Malaysia, 1–3 March 2021; IEEE: Khartoum, Sudan, 2021; pp. 395–400. [Google Scholar] [CrossRef]
- Yadavalli, M.K.; Pamula, V.K. An efficient framework to automatic extract EOG artifacts from single channel EEG recordings. In Proceedings of the 2022 IEEE International Conference on Signal Processing and Communications (SPCOM), Bangalore, India, 11 July 2022; IEEE: Kakinada, India, 2022; pp. 1–5. [Google Scholar] [CrossRef]
- Al-Qazzaz, N.K.; Aldoori, A.A.; Ali, S.H.B.M.; Ahmad, S.A.; Mohammed, A.K.; Mohyee, M.I. EEG signal complexity measurements to enhance BCI-based stroke patients’ rehabilitation. Sensors 2023, 23, 3889. [Google Scholar] [CrossRef]
- Al-Qazzaz, N.K.; Ali, S.H.B.M.; Ahmad, S.A. Recognition enhancement of dementia patients’ working memory using entropy-based features and local tangent space alignment algorithm. In Advances in Non-Invasive Biomedical Signal Sensing and Processing with Machine Learning; Springer: Berlin/Heidelberg, Germany, 2023; pp. 345–373. [Google Scholar] [CrossRef]
- Al-Qazzaz, N.K.; Ali, S.H.B.M.; Ahmad, S.A. Deep learning model for prediction of dementia severity based on EEG signals. Al-Khwarizmi Eng. J. 2024, 20, 1–12. [Google Scholar] [CrossRef]
- Rezaeezadeh, M.; Shamekhi, S.; Shamsi, M. Attention deficit hyperactivity disorder diagnosis using non-linear univariate and multivariate EEG measurements: A preliminary study. Phys. Eng. Sci. Med. 2020, 43, 577–592. [Google Scholar] [CrossRef]
- Lee, J.H.; Choi, Y.S. A data driven Information theoretic feature extraction in EEG-based motor imagery BCI. In Proceedings of the 2019 International Conference on Information and Communication Technology Convergence (ICTC), Jeju Island, Republic of Korea, 16–18 October 2019; IEEE: Seoul, Republic of Korea, 2019; pp. 1373–1376. [Google Scholar] [CrossRef]
- Azami, H.; Rostaghi, M.; Fernández, A.; Escudero, J. Dispersion entropy for the analysis of resting-state MEG regularity in Alzheimer’s disease. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; IEEE: Toronto, ON, Canada, 2016; pp. 6417–6420. [Google Scholar] [CrossRef]
- Varshney, A.; Ghosh, S.K.; Padhy, S.; Tripathy, R.K.; Acharya, U.R. Automated classification of mental arithmetic tasks using recurrent neural network and entropy features obtained from multi-channel EEG signals. Electronics 2021, 10, 1079. [Google Scholar] [CrossRef]
- Dash, S.; Tripathy, R.K.; Dash, D.K.; Panda, G.; Pachori, R.B. Multiscale domain gradient boosting models for the automated recognition of imagined vowels using multichannel EEG signals. IEEE Sens. Lett. 2022, 6, 1–4. [Google Scholar] [CrossRef]
- Xiao, H.; Li, L.; Mandic, D.P. ClassA entropy for the analysis of structural complexity of physiological signals. In Proceedings of the ICASSP 2023–2023 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Rhodes, Greece, 4–10 June 2023; IEEE: London, UK, 2023; pp. 1–5. [Google Scholar] [CrossRef]
- Shahbakhti, M.; Beiramvand, M.; Far, S.M.; Solé-Casals, J.; Lipping, T.; Augustyniak, P. Utilizing slope entropy as an effective index for wearable EEG-based depth of anesthesia monitoring. In Proceedings of the 2024 46th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 15–19 July 2024; IEEE: Kaunas, Lithuania, 2024; pp. 1–4. [Google Scholar] [CrossRef]
- Amalina, I.; Saidatul, A.; Fook, C.; Ibrahim, Z. Preliminary study on EEG based typing biometrics for user authentication using nonlinear features. In Proceedings of the IOP Conference Series: Materials Science and Engineering, Bogor, Indonesia, 15–16 December 2018; IOP Publishing: Bristol, UK, 2019; Volume 557, p. 012035. [Google Scholar] [CrossRef]
- Karmakar, C.; Udhayakumar, R.K.; Palaniswami, M. Distribution entropy (DistEn): A complexity measure to detect arrhythmia from short length RR interval time series. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; IEEE: Melbourne, Australia, 2015; pp. 5207–5210. [Google Scholar] [CrossRef]
- Udhayakumar, R.K.; Karmakar, C.; Li, P.; Palaniswami, M. Effect of embedding dimension on complexity measures in identifying arrhythmia. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; IEEE: Melbourne, Australia, 2016; pp. 6230–6233. [Google Scholar] [CrossRef]
- Udhayakumar, R.K.; Karmakar, C.; Li, P.; Palaniswami, M. Influence of embedding dimension on distribution entropy in analyzing heart rate variability. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; IEEE: Melbourne, Australia, 2016; pp. 6222–6225. [Google Scholar] [CrossRef]
- Li, Y.; Li, P.; Karmakar, C.; Liu, C. Distribution entropy for short-term QT interval variability analysis: A comparison between the heart failure and normal control groups. In Proceedings of the 2015 Computing in Cardiology Conference (CinC), Nice, France, 6–9 September 2015; IEEE: Jinan, China, 2015; pp. 1153–1156. [Google Scholar] [CrossRef]
- Nardelli, M.; Citi, L.; Barbieri, R.; Valenza, G. Characterization of autonomic states by complex sympathetic and parasympathetic dynamics. Physiol. Meas. 2023, 44, 035004. [Google Scholar] [CrossRef]
- Silva, L.E.; Moreira, H.T.; Schmidt, A.; Romano, M.M.; Salgado, H.C.; Fazan, R.; Marin-Neto, J.A. The relationship between nonlinear heart rate variability and echocardiographic indices in Chagas disease. In Proceedings of the 2020 11th Conference of the European Study Group on Cardiovascular Oscillations (ESGCO), Pisa, Italy, 15 July 2020; IEEE: Ribeirão Preto, Brazil, 2020; pp. 1–2. [Google Scholar] [CrossRef]
- Udhayakumar, R.K.; Karmakar, C.; Li, P.; Palaniswami, M. Effect of data length and bin numbers on distribution entropy (DistEn) measurement in analyzing healthy aging. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; IEEE: Melbourne, Australia, 2015; pp. 7877–7880. [Google Scholar] [CrossRef]
- Yan, C.; Liu, C.; Yao, L.; Wang, X.; Wang, J.; Li, P. Short-term effect of percutaneous coronary intervention on heart rate variability in patients with coronary artery disease. Entropy 2021, 23, 540. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Li, P.; Yang, M.; Li, Y.; Li, J.; Zhang, H.; Liu, C. Entropy analysis of heart rate variability in different sleep stages. Entropy 2022, 24, 379. [Google Scholar] [CrossRef]
- Shi, B.; Zhang, Y.; Yuan, C.; Wang, S.; Li, P. Entropy analysis of short-term heartbeat interval time series during regular walking. Entropy 2017, 19, 568. [Google Scholar] [CrossRef]
- Nardelli, M.; Citi, L.; Barbieri, R.; Valenza, G. Intrinsic complexity of sympathetic and parasympathetic dynamics from HRV series: A preliminary study on postural changes. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; IEEE: Pisa, Italy, 2020; pp. 2577–2580. [Google Scholar] [CrossRef]
- Arsac, L.M. Entropy-based multifractal testing of heart rate variability during cognitive-autonomic interplay. Entropy 2023, 25, 1364. [Google Scholar] [CrossRef] [PubMed]
- Aulia, S.; Hadiyoso, S.; Wijayanto, I.; Irawati, I.D. Biometric simulation based on single lead Electrocardiogram signal using dispersion entropy and linear discriminant analysis. Pattern Recognit. 2022, 16, 1359. [Google Scholar] [CrossRef]
- Nicolet, J.J.; Restrepo, J.F.; Schlotthauer, G. Classification of intracavitary electrograms in atrial fibrillation using information and complexity measures. Biomed. Signal Process. Control 2020, 57, 101753. [Google Scholar] [CrossRef]
- Kafantaris, E.; Piper, I.; Lo, T.Y.M.; Escudero, J. Application of dispersion entropy to healthy and pathological heartbeat ECG segments. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; IEEE: Edinburgh, UK, 2019; pp. 2269–2272. [Google Scholar] [CrossRef]
- Singh, R.S.; Gelmecha, D.J.; Sinha, D.K. Expert system based detection and classification of coronary artery disease using ranking methods and nonlinear attributes. Multimed. Tools Appl. 2022, 81, 19723–19750. [Google Scholar] [CrossRef]
- Tripathy, R.K.; Dash, D.K.; Ghosh, S.K.; Pachori, R.B. Detection of different stages of anxiety from single-channel wearable ECG sensor signal using Fourier–Bessel domain adaptive wavelet transform. IEEE Sens. Lett. 2023, 7, 1–4. [Google Scholar] [CrossRef]
- Deka, D.; Deka, B. Investigation on HRV signal dynamics for meditative intervention. In Soft Computing: Theories and Applications; Springer: Berlin/Heidelberg, Germany, 2020; pp. 993–1005. [Google Scholar] [CrossRef]
- Khazaei, M.; Raeisi, K.; Goshvarpour, A.; Ahmadzadeh, M. Early detection of sudden cardiac death using nonlinear analysis of heart rate variability. Biocybern. Biomed. Eng. 2018, 38, 931–940. [Google Scholar] [CrossRef]
- Manis, G.; Bodini, M.; Rivolta, M.W.; Sassi, R. Bubble entropy of fractional Gaussian noise and fractional Brownian motion. In Proceedings of the 2021 Computing in Cardiology (CinC), Brno, Czech Republic, 13–15 September 2021; IEEE: Ioannina, Greece, 2021; Volume 48, pp. 1–4. [Google Scholar] [CrossRef]
- Manis, G.; Sassi, R. Tolerance to spikes: A comparison of Sample and Bubble entropy. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017; IEEE: Ioannina, Greece, 2017; pp. 1–4. [Google Scholar] [CrossRef]
- Rohila, A.; Sharma, A. Detection of sudden cardiac death by a comparative study of heart rate variability in normal and abnormal heart conditions. Biocybern. Biomed. Eng. 2020, 40, 1140–1154. [Google Scholar] [CrossRef]
- Lin, P.L.; Lin, P.Y.; Huang, H.P.; Vaezi, H.; Liu, L.Y.M.; Lee, Y.H.; Huang, C.C.; Yang, T.F.; Hsu, L.; Hsu, C.F. The autonomic balance of heart rhythm complexity after renal artery denervation: Insight from entropy of entropy and average entropy analysis. BioMed. Eng. Online 2022, 21, 32. [Google Scholar] [CrossRef]
- Silva, L.E.V.; Moreira, H.T.; de Oliveira, M.M.; Cintra, L.S.S.; Salgado, H.C.; Fazan, R., Jr.; Tinós, R.; Rassi, A., Jr.; Schmidt, A.; Marin-Neto, J.A. Heart rate variability as a biomarker in patients with chronic Chagas cardiomyopathy with or without concomitant digestive involvement and its relationship with the Rassi score. BioMed. Eng. Online 2022, 21, 44. [Google Scholar] [CrossRef] [PubMed]
- Rohila, A.; Sharma, A. Correlation between heart rate variability features. In Proceedings of the 2020 7th International Conference on Signal Processing and Integrated Networks (SPIN), Noida, India, 27–28 February 2020; IEEE: Roorkee, India, 2020; pp. 669–674. [Google Scholar] [CrossRef]
- Saxena, S.; Gupta, V.K.; Hrisheekesha, P. Coronary heart disease detection using nonlinear features and online sequential extreme learning machine. Biomed. Eng. Appl. Basis Commun. 2019, 31, 1950046. [Google Scholar] [CrossRef]
- Manis, G.; Sassi, R. Relation between fetal HRV and value of umbilical cord artery pH in labor, a study with entropy measures. In Proceedings of the 2017 IEEE 30th International Symposium on Computer-Based Medical Systems (CBMS), Thessaloniki, Greece, 22–24 June 2017; IEEE: Ioannina, Greece, 2017; pp. 272–277. [Google Scholar] [CrossRef]
- Rivolta, M.W. Information theory and fetal heart rate variability analysis. In Innovative Technologies and Signal Processing in Perinatal Medicine: Volume 2; Springer: Berlin/Heidelberg, Germany, 2023; pp. 171–188. [Google Scholar] [CrossRef]
- Gupta, S.; Singh, A.; Sharma, A. CIsense: An automated framework for early screening of cerebral infarction using PPG sensor data. Biomed. Eng. Lett. 2023, 14, 199–207. [Google Scholar] [CrossRef]
- Gupta, S.; Singh, A.; Sharma, A.; Tripathy, R.K. Higher order derivative-based integrated model for cuff-less blood pressure estimation and stratification using PPG signals. IEEE Sens. J. 2022, 22, 22030–22039. [Google Scholar] [CrossRef]
- Zamudio-De Hoyos, J.R.; Vázquez-Flores, D.; Pliego-Carrillo, A.C.; Ledesma-Ramírez, C.I.; Mendieta-Zerón, H.; Reyes-Lagos, J.J. Nonlinearity of Electrohysterographic signals is diminished in active preterm labor. In Proceedings of the Congreso Nacional de Ingeniería Biomédica, Puerto Vallarta, México, 6–8 October 2022; Springer: Toluca, Mexico, 2022; pp. 302–307. [Google Scholar] [CrossRef]
- Reyes-Lagos, J.J.; Pliego-Carrillo, A.C.; Ledesma-Ramírez, C.I.; Peña-Castillo, M.Á.; García-González, M.T.; Pacheco-López, G.; Echeverría, J.C. Phase entropy analysis of Electrohysterographic data at the third trimester of human pregnancy and active parturition. Entropy 2020, 22, 798. [Google Scholar] [CrossRef]
- Nieto-del Amor, F.; Beskhani, R.; Ye-Lin, Y.; Garcia-Casado, J.; Diaz-Martinez, A.; Monfort-Ortiz, R.; Diago-Almela, V.J.; Hao, D.; Prats-Boluda, G. Assessment of dispersion and bubble entropy measures for enhancing preterm birth prediction based on Electrohysterographic signals. Sensors 2021, 21, 6071. [Google Scholar] [CrossRef]
- Diaz-Martinez, A.; Prats-Boluda, G.; Monfort-Ortiz, R.; Garcia-Casado, J.; Roca-Prats, A.; Tormo-Crespo, E.; Nieto-del Amor, F.; Diago-Almela, V.J.; Ye-Lin, Y. Overdistention accelerates electrophysiological changes in uterine muscle towards labour in multiple gestations. IRBM 2024, 45, 100837. [Google Scholar] [CrossRef]
- Nieto-del Amor, F.; Ye Lin, Y.; Garcia-Casado, J.; Díaz-Martínez, M.d.A.; González Martínez, M.; Monfort-Ortiz, R.; Prats-Boluda, G. Dispersion entropy: A measure of Electrohysterographic complexity for preterm labor discrimination. In Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2021), Vienna, Austria, 11–13 February 2021; Curran Associates, Inc.: Red Hook, NY, USA, 2021; pp. 260–267. [Google Scholar] [CrossRef]
- Sasidharan, D.; Venugopal, G.; Swaminathan, R. Complexity analysis of surface Electromyography signals under fatigue using Hjorth parameters and bubble entropy. J. Mech. Med. Biol. 2023, 23, 2340051. [Google Scholar] [CrossRef]
- Sasidharan, D.; Kavyamol, K.; Subramanian, S.; Venugopal, G. Chaotic complexity determination of surface EMG signals. In Proceedings of the Indian Conference on Applied Mechanics, Mandi, India, 14–16 December 2022; Springer: Coimbatore, India, 2022; pp. 323–329. [Google Scholar] [CrossRef]
- Sowmya, S.; Banerjee, S.S.; Swaminathan, R. Assessment of muscle fatigue using phase entropy of sEMG signals during dynamic contractions of biceps brachii. In Proceedings of the 2023 9th International Conference on Control, Decision and Information Technologies (CoDIT), Rome, Italy, 3–6 July 2023; IEEE: Tamil Nadu, India, 2023; pp. 2253–2256. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, C.; Gui, P.; Wang, M.; Zou, T. State Assessment of Rolling Bearings Based on the Multiscale Bubble Entropy. In Proceedings of the 2021 International Conference on Electronics, Circuits and Information Engineering (ECIE), Zhengzhou, China, 22–24 January 2021; IEEE: Beijing, China, 2021; pp. 179–182. [Google Scholar] [CrossRef]
| Symbol | Description |
|---|---|
| Sample from the time series | |
| N | Length of the time series |
| Original time series | |
| m | Embedding dimension |
| d | Time delay |
| Embedded vector | |
| Embedded series from X | |
| Series of symbols | |
| Embedded series of symbols |
| Definition Family | Definition |
|---|---|
| Embedding and Distance-Based | Range Entropy |
| Cosine Similarity Entropy | |
| Diversity Entropy | |
| Distribution Entropy | |
| Symbolic and Ordinal Pattern-Based | Increment Entropy |
| Dispersion Entropy | |
| F-B Dispersion Entropy | |
| Slope Entropy | |
| Symbolic Dynamic Entropy | |
| Complexity Estimation Based on Sorting Effort | Bubble Entropy |
| Multiscale and Hierarchical Definitions | Entropy of Entropy |
| Geometric or Phase-Space Definitions | Phase Entropy |
| Gridded Distribution Entropy | |
| Pattern-Detection Definitions | Attention Entropy |
| As a Citation | In Title | In Keywords | In Abstract | |
|---|---|---|---|---|
| Dispersion Entropy: | 31 | 12 | 16 | 31 |
| Bubble Entropy: | 25 | 6 | 9 | 19 |
| Distribution Entropy: | 22 | 6 | 5 | 20 |
| Increment Entropy: | 8 | 2 | 5 | 8 |
| Phase Entropy: | 9 | 3 | 3 | 9 |
| Slope Entropy: | 6 | 2 | 1 | 4 |
| F-B Dispersion Entropy: | 4 | 1 | 1 | 3 |
| Attention Entropy: | 5 | 1 | 1 | 3 |
| Gridded Distribution Entropy: | 5 | 1 | 1 | 4 |
| Range Entropy: | 2 | 1 | 2 | 2 |
| Entropy of Entropy: | 3 | 1 | - | 3 |
| Symbolic Dynamic Entropy: | 2 | - | - | 1 |
| Cosine Entropy: | 2 | - | - | 1 |
| Diversity Entropy: 1 | - | - | - | - |
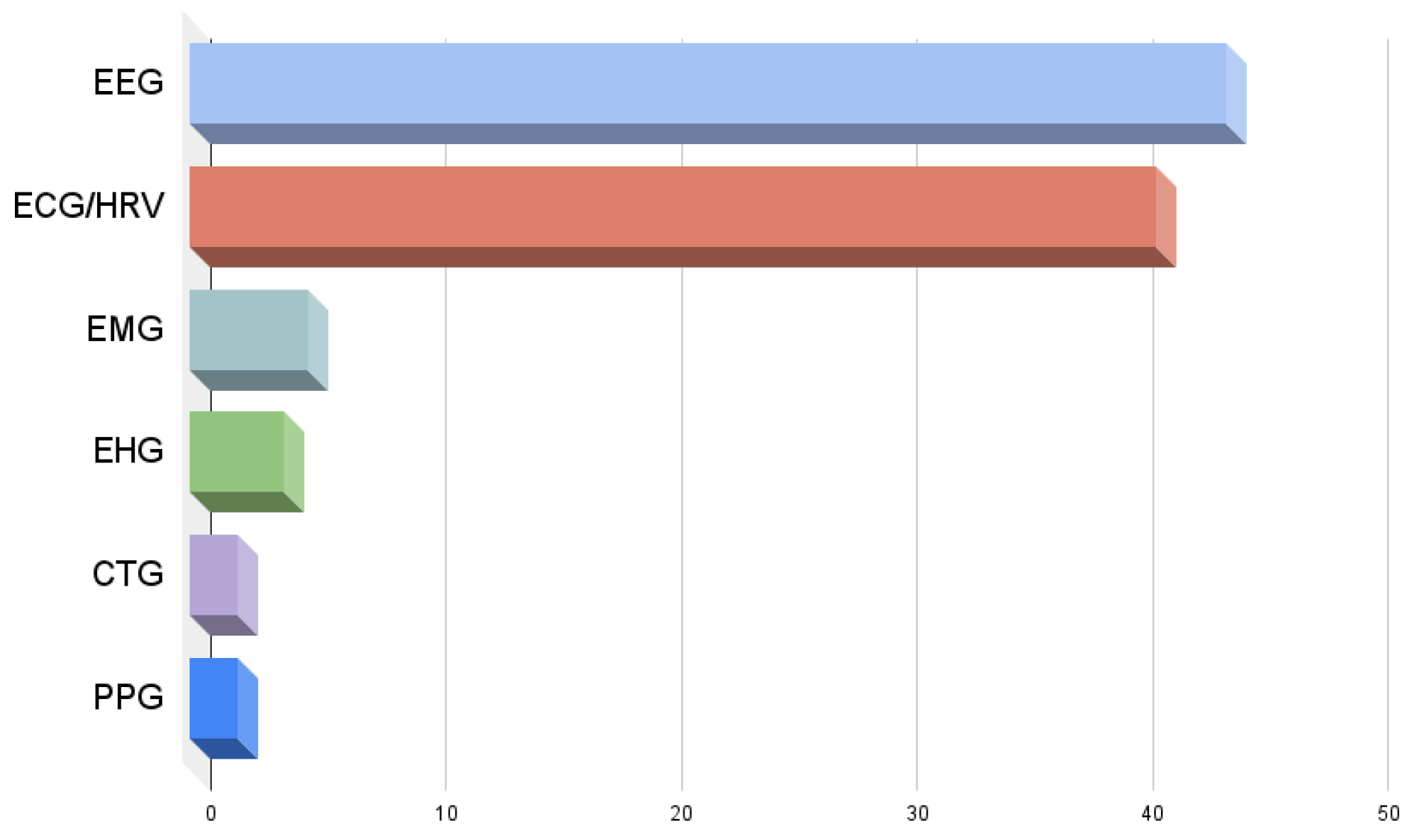
| EEG | ECG/HRV | PPG | EHG | EMG | CTG | |
|---|---|---|---|---|---|---|
| Dispersion Entropy: | 20 | 10 | 1 | 2 | - | - |
| Bubble Entropy: | 8 | 10 | 1 | 2 | 2 | 2 |
| Distribution Entropy: | 5 | 15 | - | - | 1 | - |
| Increment Entropy: | 5 | 5 | - | - | - | - |
| Phase Entropy: | 2 | 4 | 2 | - | 1 | - |
| Slope Entropy: | 4 | 2 | 1 | - | 1 | - |
| F-B Dispersion Entropy: | 3 | - | - | - | - | - |
| Attention Entropy: | 1 | 2 | - | - | - | - |
| Range Entropy: | 2 | - | - | - | - | - |
| Gridded Distribution Entropy: | 2 | 2 | - | - | - | - |
| Entropy of Entropy: | - | 2 | - | - | - | - |
| Symbolic Dynamic Entropy: | 1 | - | - | - | - | - |
| Cosine Entropy: | 1 | - | - | - | - | - |
| Diversity Entropy: | - | - | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Platakis, D.; Manis, G. Review of Recent (2015–2024) Popular Entropy Definitions Applied to Physiological Signals. Entropy 2025, 27, 983. https://doi.org/10.3390/e27090983
Platakis D, Manis G. Review of Recent (2015–2024) Popular Entropy Definitions Applied to Physiological Signals. Entropy. 2025; 27(9):983. https://doi.org/10.3390/e27090983
Chicago/Turabian StylePlatakis, Dimitrios, and George Manis. 2025. "Review of Recent (2015–2024) Popular Entropy Definitions Applied to Physiological Signals" Entropy 27, no. 9: 983. https://doi.org/10.3390/e27090983
APA StylePlatakis, D., & Manis, G. (2025). Review of Recent (2015–2024) Popular Entropy Definitions Applied to Physiological Signals. Entropy, 27(9), 983. https://doi.org/10.3390/e27090983






