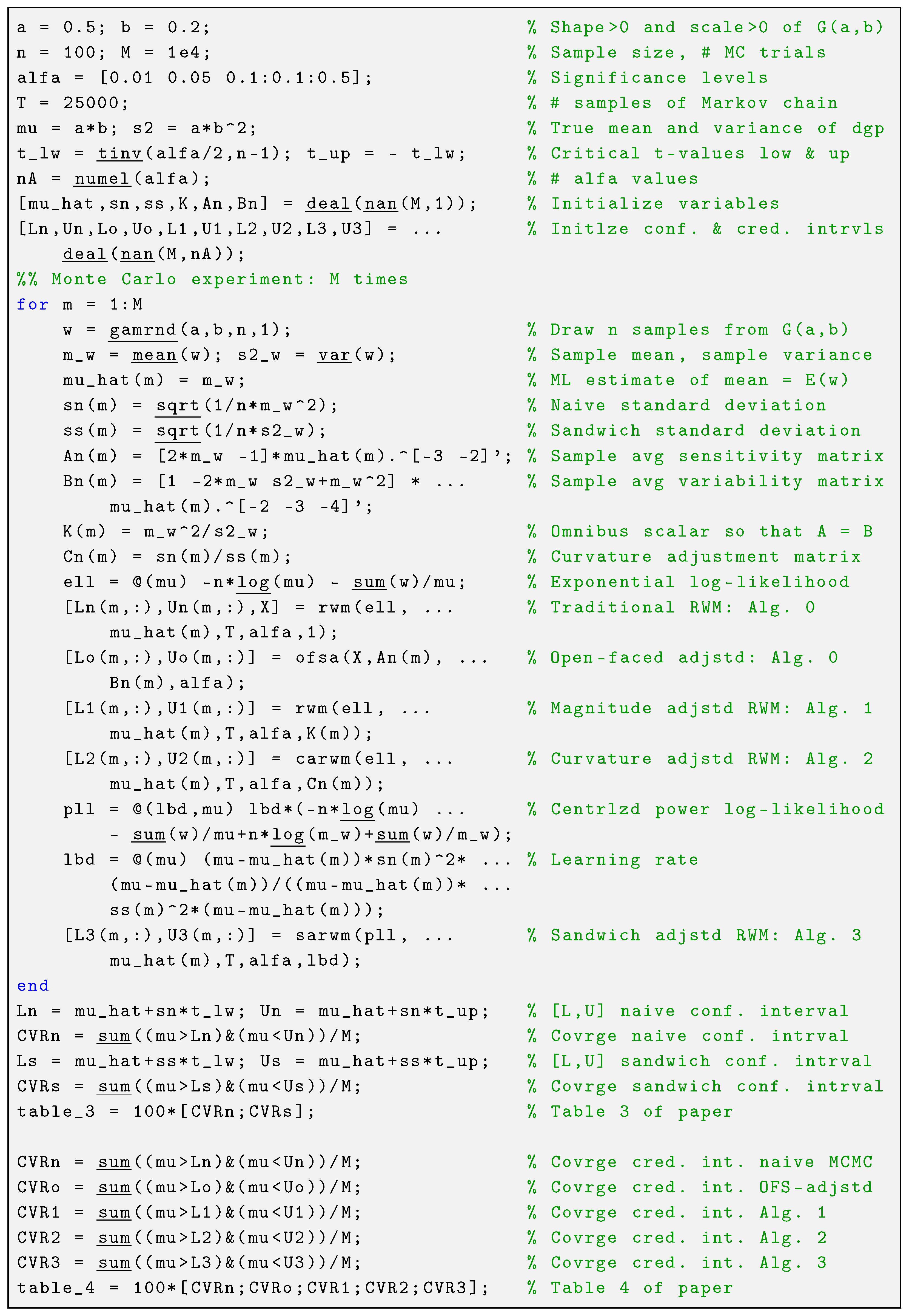
6.1. Case Study 1
We revisit example 1 in
Section 3 and compute the ML estimate
of the mean of the normal distribution model
, and the corresponding values of
,
,
omnibus scalar
, naive
, and sandwich
variances using
,
and
. The ML solution
is simply equal to the sample mean of the data points
drawn from
with
and
and
and
are derived by numerical means using the DERIVESTsuite toolbox of D’Errico [
58]. We repeat this computation for
different realizations of the
n data points.
Table 1 presents the result of this Monte Carlo experiment and lists mean values of
,
,
,
,
and
and their respective standard deviations (in parentheses). The Matlab code is given in
Appendix C.
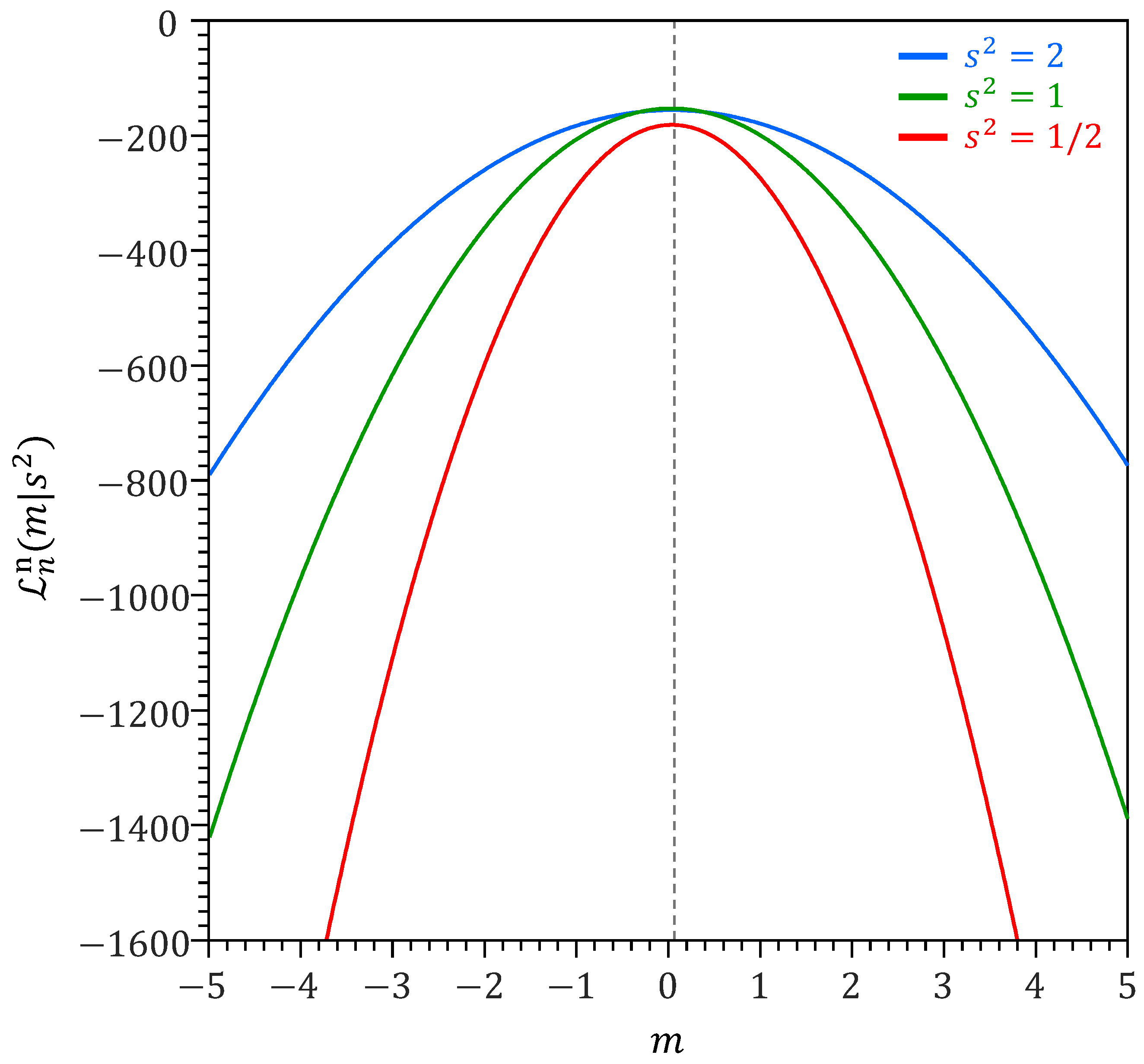
The tabulated results confirm the theory. The ML estimate of the mean is centered around zero and has a standard deviation that approaches the theoretic standard deviation . The ML sensitivity matrix derived from numerical differentiation equals its theoretic value and does not differ between the trials. The variability matrix approaches its theoretic value and has a nonzero standard deviation as is replaced by the sample variance of the ’s. The mean of the omnibus scalar approaches its theoretic value . Thus, we find that and the standard deviation between parenthesis is a result that of the data-generating process is replaced by the sample variance of the ’s. The naive variance estimator is equal to its theoretic value and does not differ between the trials, as n and are fixed. The sandwich variance estimator does not depend on the value of and asymptotically converges to the true variance of the mean of the data-generating process. The standard deviation is the result of the variation in in the Monte Carlo trials.
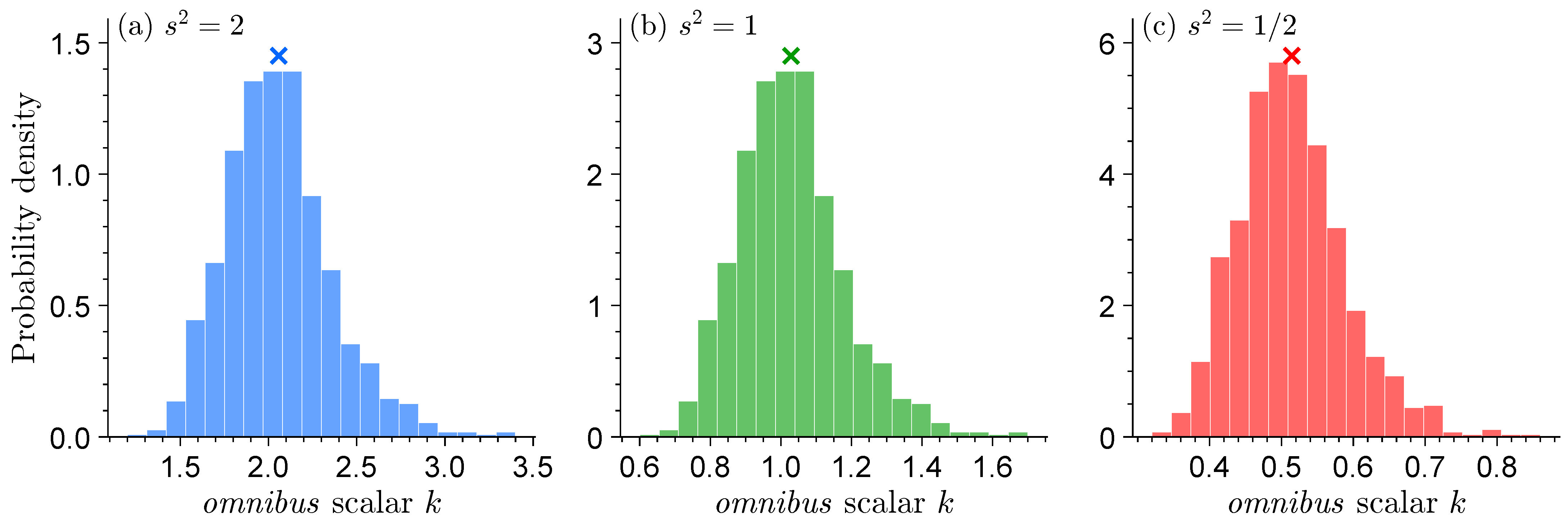
Figure 3 displays the histograms of the
omnibus scalar
k for each of the three normal distribution models. The use of
will retrieve the sandwich variance
.
When , the model underestimates the information contained in the data and the omnibus scalar is greater than one. Vice versa, for we systematically overestimate the informativeness of the data and, as a result, , to slow down learning and produce robust confidence intervals for , the mean of the data-generating process. For , the normal distribution model is equal to the standard normal distribution of the data generating process and .
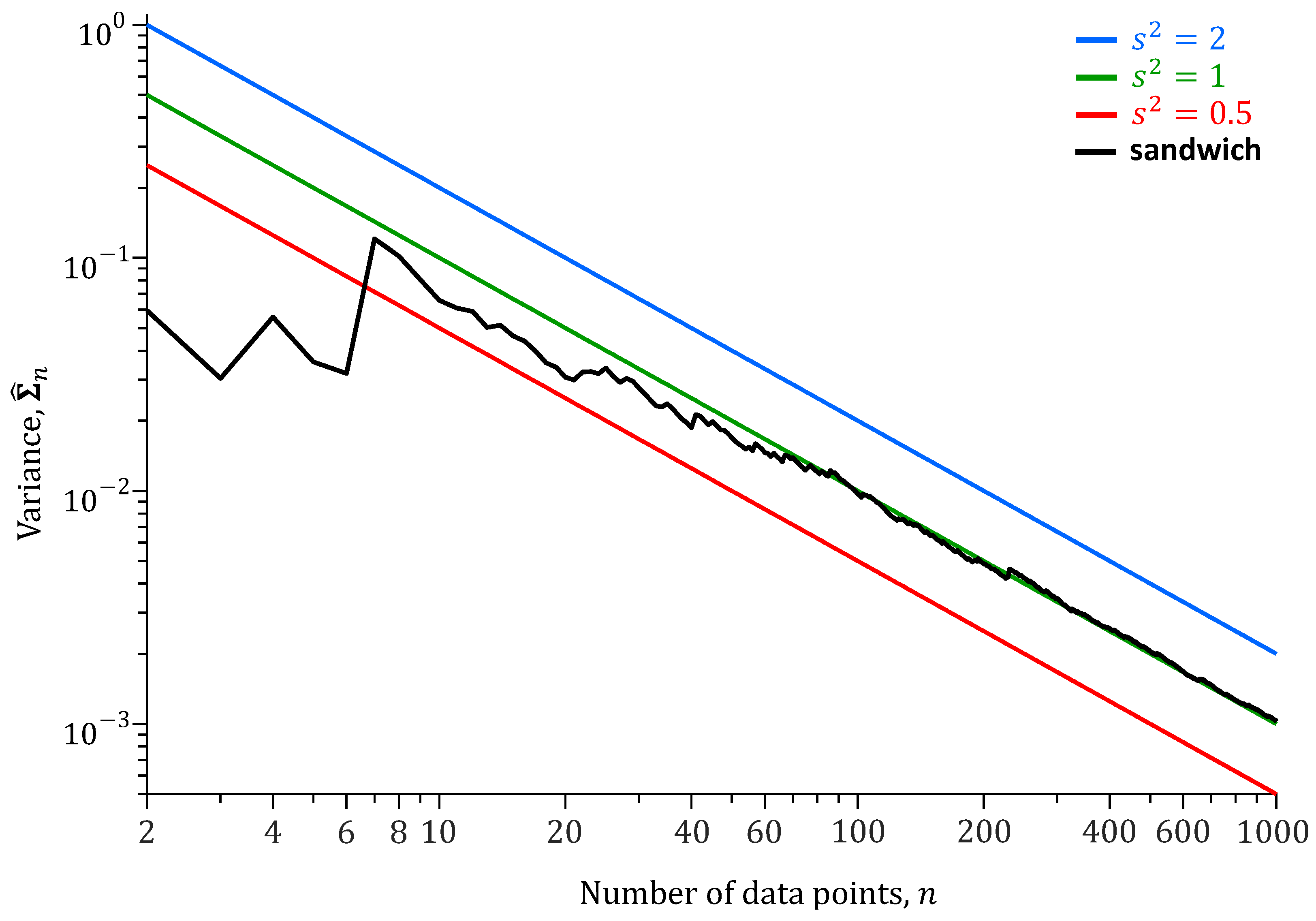
To better understand the relationship between the number
n of data points
and the naive and sandwich variances, we repeat the analysis of
Table 1 for different values of
n.
Figure 4 presents the results of this analysis.
The naive variance decreases linearly on a logarithmic scale with the length n of the training record. The slope of this line is proportional to on a linear scale. The naive variance of m depends on the choice of . The sandwich variance does not depend on and settles on the true variance of m with increasing number of data points .
Table 2 examines the coverage probabilities of the true mean
of the data generating process according to the
% confidence intervals of
derived from the naive and sandwich variance estimators.
The results of
Table 2 demonstrate that the sandwich variance estimator provides adequate confidence intervals of the mean
of the data-generating process, even if the underlying model is misspecified. The sandwich estimator
consistently achieves the correct frequentist coverage probabilities, whereas the naive variance estimator
either over- (
) or under- (
) estimates the coverage probabilities. The confidence intervals are either too dispersed or too sharp.
This concludes our first case study. This study was rather unrealistic in that misspecification was introduced by affixing one of the model parameters to a wrong value. The correct distribution was used, but with a wrong value for one of its parameters, namely the variance . In the next study, we are going to take misspecification one step further and use a different model for inference than was used to generate the data.
6.2. Case Study 2
Our second case study is another analytic exercise, but one that better reflects practice as the parametric form of our model differs from that of the data-generating process. We draw
n measurements
from a gamma distribution
with pdf
where
and
are shape and scale parameters, respectively, and
is the Gamma function. Now, suppose our model for
is not a gamma but an exponential distribution
with location parameter
and pdf
Note that it is not uncommon to parameterize
with a rate parameter
instead. The likelihood function for a single observation
is now equal to
and the log-likelihood
becomes
Appendix D derives analytic expressions for the naive and sandwich variance estimators of the mean of the exponential distribution. We yield
where
and
are the sample mean and sample variance of the data points,
. According to Equation (
A12), the
omnibus scalar
k is now equal to
, whereas its theoretical value, derived in Equation (A13), corresponds to the shape parameter
a of
.
Table 3 confirms again the erroneous description of the confidence intervals by the naive variance estimator. The
% confidence intervals are too sharp and underestimate the theoretic coverage probabilities of the mean
of the data generating process. This overconditioning is a result of misspecification and, thus, due to a misalignment of the sensitivity and variability matrices. In contrast to other methods, the coverage probabilities of the sandwich estimator align much more closely with theoretical expectations. The estimates are not perfect, as a result of the symmetry assumption used in constructing confidence intervals for
. This assumption is not valid for the exponential distribution and is further exacerbated when the sample size
n is small. To mitigate this latter effect, we chose
in our Monte Carlo experiments. To address the asymmetry, one could construct non-symmetric confidence intervals by identifying the shortest interval for
that contains the true mean with probability
. However, doing so would require knowledge of the posterior distribution of
, which is generally not available in frequentist settings. Importantly, the variance of the ML estimates
across the
M Monte Carlo trials matches the theoretical sandwich variance,
. This confirms that the only correct confidence intervals of
are those derived from the sandwich estimator.
Table 4 documents the nominal coverage probabilities of the credible regions obtained from MCMC simulation with the DREAM
(ZS) algorithm using the log-likelihood
of Equation (
A10) (=naive estimator), OFS-adjusted naive posterior samples of Equation (
11), magnitude-adjusted log-likelihood
with
omnibus scalar
k of Equation (
10) (=Algorithm 1), curvature-adjusted log-likelihood
of Equation (
15) (=Algorithm 2), and centralized power log-likelihood
of Equation (
21) (=Algorithm 3).
The curvature-adjustment matrix
is a scalar in this case, and according to Equations (
18) and (
29), we yield
, where
is the reciprocal of the sample standard deviation of the
’s. For OFS-adjustment, we substitute the expressions for
and
of Equation (
A7) into Equation (
12) and yield
. The tabulated values confirm that
The asymptotic covariance matrix of the Metropolis algorithm is a single slice of bread. The 100
% credible intervals are in agreement with the frequentist confidence intervals of the naive variance estimator,
in
Table 3 and underestimate the theoretic coverage probabilities.
The OFS adjustment of Equation (
11) enlarges the spread of the naive posterior samples but the coverage probabilities of the so-obtained sandwich credible regions underestimate their counterparts of the sandwich estimator in
Table 3.
The three MCMC recipes discussed in this paper successfully join a single slice of bread
to the open-faced sandwich
to produce the sandwich variance
. The coverage probabilities of the 100
% credible regions of Algorithms 1–3 match those of the sandwich estimator in
Table 3.
The tabulated values for Algorithm 3 are the first proof that the centralized power log-likelihood function
of Equation (
21) works in practice. This inspires confidence that we can sample the sandwich distribution without using matrix square roots.
The OFS adjustment is computationally appealing and enlarges the spread of the naive posterior samples, yet the so-obtained credible regions underestimate the theoretical coverage probabilities. Magnitude, curvature, and kernel adjustment of the log-likelihood function all appear viable methods for sandwich-adjusted MCMC simulation. There are important differences between these three sampling methods and the practical consequences of this are better illustrated with a multivariate target distribution.
Having completed the above exercise, we now replace
with alternative distributions for the data-generating process.
Figure 5 shows histograms of the
omnibus scalar
k when the data-generating process is (a)
, (b)
, (c)
, (d)
, and (e)
. For comparability, the scale, shape, and/or location parameters of each distribution are chosen such that
and
. The theoretical value of the omnibus scalar for each distribution is
.
The histograms of appear remarkably similar across the different distributions. This confirms that our inferences for are robust and do not depend on the distribution of the data-generating process. The marginal distributions of the omnibus scalar center on the theoretic value of and display a small right tail. The dispersion of is a result of sample size and will disappear if we set n much larger in the Monte Carlo trials.
We now move on to our third and last case study. This will involve the use of real-world data and a multivariate posterior distribution.
6.3. Case Study 3
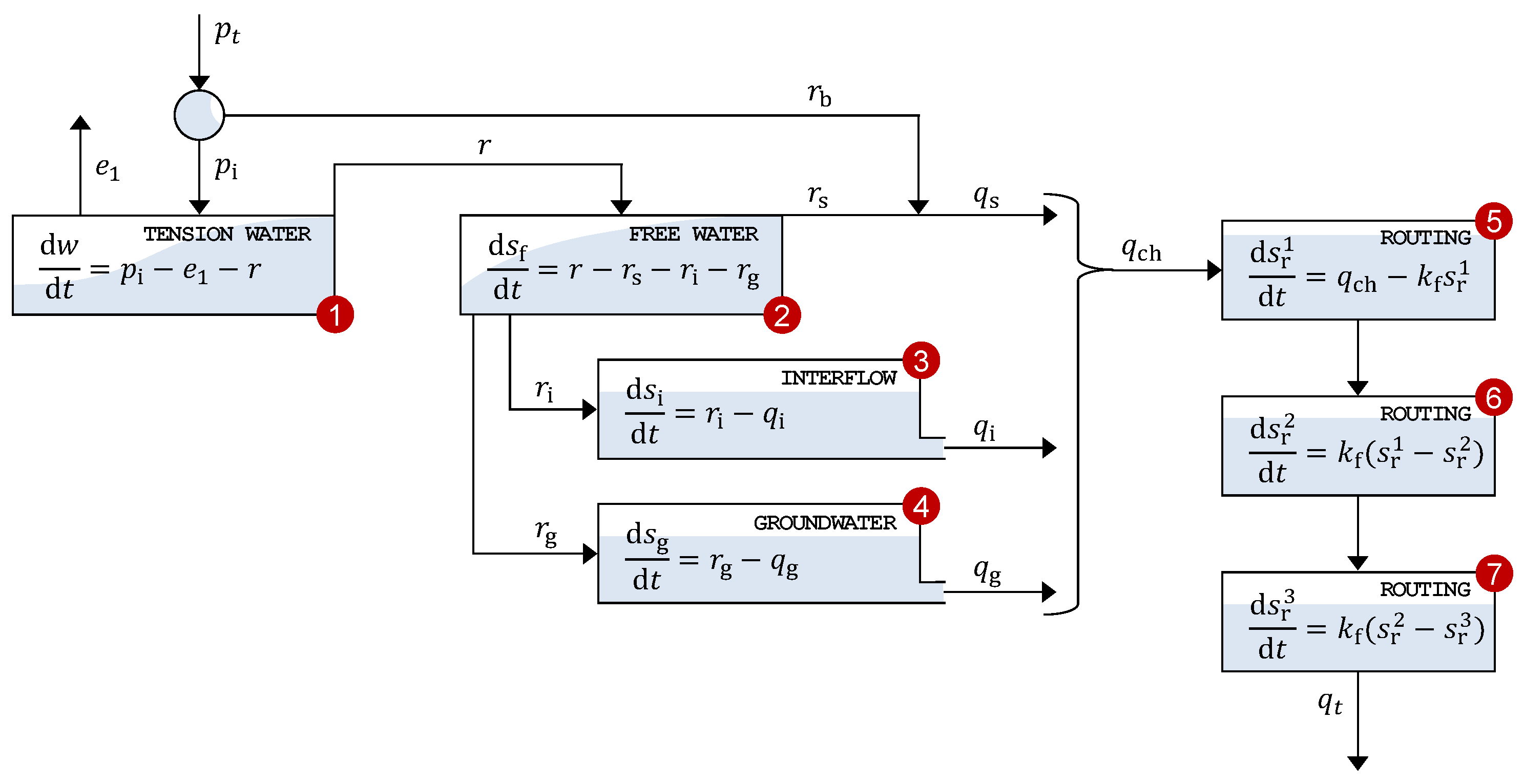
Our third and final case study examines the streamflow response of the Leaf River near Collins, MS, USA. The precipitation–discharge transformation is simulated using the Xinanjiang conceptual watershed model originally developed by Zhao and Zhuang [
65]. We adopt the implementation of Jayawardena and Zhou [
75] and Knoben et al. [
76], augmented with a pan evaporation parameter and three linear routing reservoirs. This configuration comprises seven control volumes that conceptually represent water storage and routing.
Appendix E provides a detailed description of the Xinanjiang model structure, including the control volumes, state variables, flux relationships, and routing scheme used to convert areal average precipitation into total channel inflow and river discharge. The model equations are solved using a mass-conservative, second-order integration method with adaptive time stepping, ensuring both numerical stability and accuracy. A one-year spin-up period removes the influence of state variable initialization.
Table A2 lists the 14 parameters of the Xinanjiang model to be estimated from streamflow measurements. For inference, we express the Xinanjiang model as the vector-valued regression
where
is the
vector of discharge observations,
signifies the parameter vector,
is the
matrix of exogenous variables containing daily areal-average rainfall and potential evapotranspiration, and
is the
vector of discharge measurement errors. We assign a uniform prior
over the bounds given in
Table A2 and use the standardized skewed-
t (SST) density of Scharnagl et al. [
77] to evaluate agreement between observed and simulated streamflows
where
is the
tth studentized streamflow residual,
, denotes the signum function, and scalars
and
are shift and scale constants, respectively, which depend on the degrees of freedom
, the skewness parameter
, and the first and second absolute moments
and
of the SST density [
57,
77]. The total likelihood
for a
n-record of studentized residuals
is now equal to
where the prefactor
is
The measurement error standard deviation
of the
tth streamflow observation
is modeled as a linear function of the simulated discharge
under model parameters
where the intercept
(mm/d) is fixed at a small positive value, and the slope
is determined offline so as to enforce unit variance of the studentized raw residuals
. The slope is obtained via an iterative root-finding procedure described in detail by Vrugt et al. [
57]. With this variance model, the Student–
t log-likelihood becomes
To facilitate both pairwise and parameter-wise comparisons of the
sensitivity
and variability
matrices, we apply the affine rescaling
which maps the Xinanjiang parameters
and nuisance variables
onto the unit hypercube. Inference is then conducted on the normalized parameters
and normalized nuisance variables
. Prior to Xinanjiang model execution,
is transformed back to the original parameter scales using the lower and upper bounds in
Table A2. The prior distributions for the degrees of freedom and skewness parameters are uniform with support
and
, respectively.
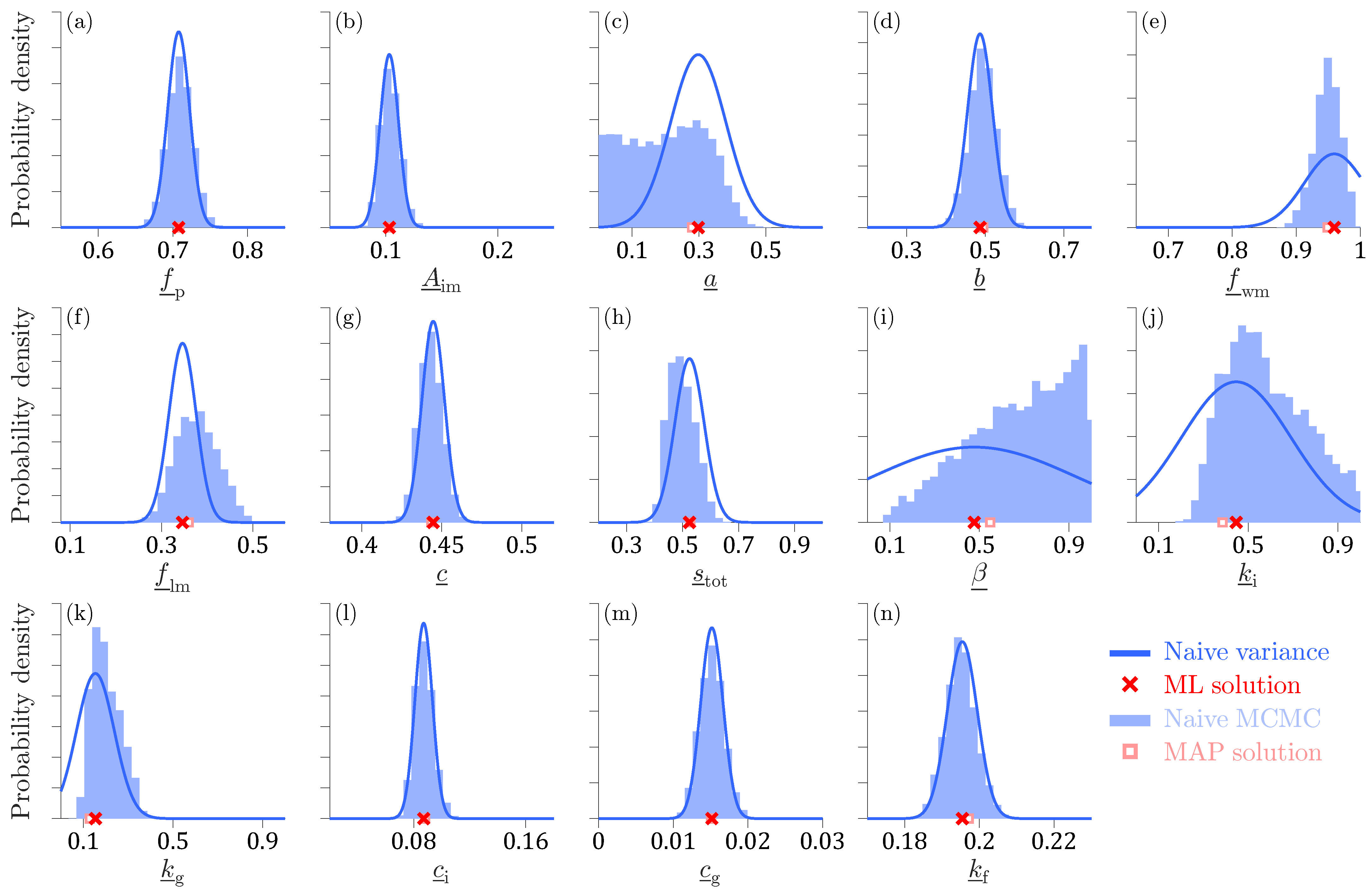
Figure 6 shows histograms of the marginal posterior distributions of the normalized Xinanjiang model parameters obtained using the DREAM
(ZS) algorithm.
The Markov chain sample with highest value of (red square) coincides almost perfectly with the ML solution (red cross) of the frequentist estimator separately obtained by maximizing the Student t likelihood using a gradient-based optimization method. For all Xinanjiang model parameters except the tension water inflection parameter a and the free water shape parameter , the MCMC-sampled marginal posterior distributions are unimodal, bell-shaped and centered around the ML solution. In contrast, the marginal posterior distribution of a is approximately uniform on the interval between 0–, whereas the density function of has a trapezoidal shape. The MAP values of these two parameters do not coincide with distinct posterior peaks, yet are in close vicinity of their ML estimates.
Most of the MCMC-sampled posterior histograms are in close agreement with the normal marginal distributions (blue lines) derived from the naive variance of the frequentist estimator where the sensitivity matrix is computed from the second-order partial derivatives of the Student t likelihood function. This frequentist estimator assumes model linearity and a symmetric Gaussian distribution around the ML estimate. In contrast, the MCMC method approximates the marginal distributions of the parameters from a large sample of posterior realizations. This enables MCMC to account for nonlinear model relationships and represent arbitrary posterior shapes, including skewed and heavy-tailed distributions. As a result, frequentist and Bayesian estimates of parameter uncertainty may differ. In the literature, this distinction is often framed in terms of linear versus nonlinear confidence intervals. However, in the Bayesian context, the more appropriate term is credible intervals, which reflect the probabilistic interpretation of uncertainty inherent to Bayesian inference.
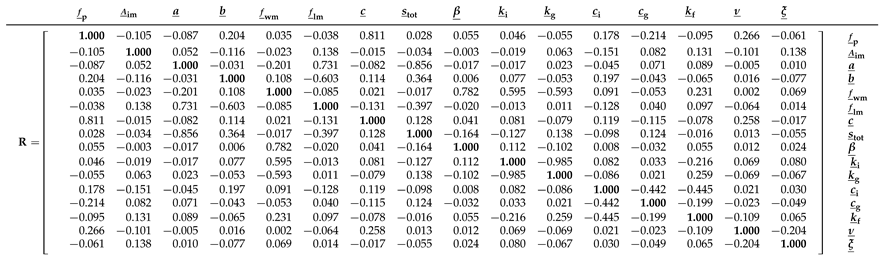
Table A3 in
Appendix F shows that most Xinanjiang parameters exhibit only weak correlations. Notable exceptions are the recession parameters
and
of the interflow and groundwater reservoirs, respectively, which display a very strong correlation (
), followed by a correlation of
between the tension water inflection parameter
a and the total soil moisture storage
, and
between the fraction of tension water storage
and the free water distribution shape parameter
. The generally low posterior correlation coefficients account in part for the close agreement between the naive posterior histograms of the Xinanjiang parameters and the normal marginal distributions derived from the frequentist estimator.
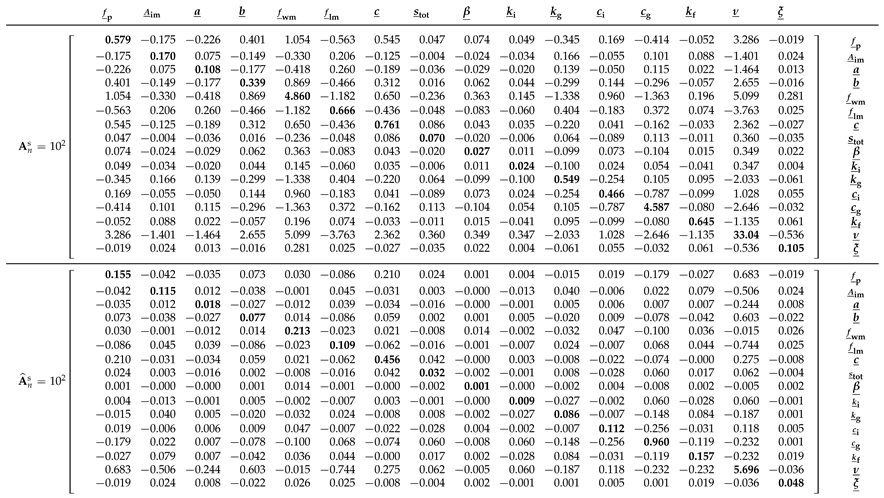
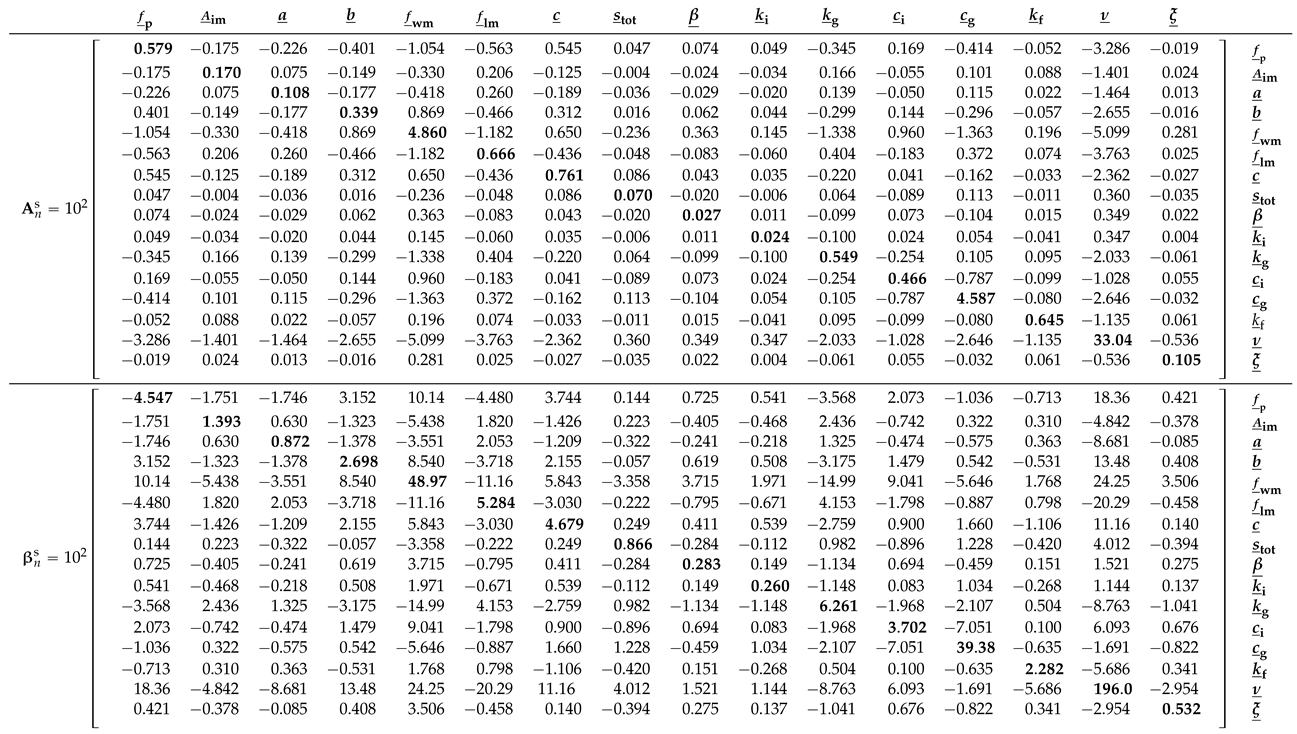
Before examining the posterior Xinanjiang parameter distributions obtained from Algorithms 1–3, we first take a closer look in
Table 5 at the bread and meat matrices of the Student
t likelihood
. Comparing these two matrices offers insight into the magnitude of the sandwich correction.
The main diagonal entries of
and
are in relatively poor agreement. The bolded entries of
are nearly an order of magnitude larger than their counterparts of
. This gives rise to a value of
for the
omnibus scalar of Pauli et al. [
31] in Equation (
10). This value is far removed from the desired value of
under correct specification. In
Section 9, we formulate several other quantitative measures of (dis)similarity between the bread and meat matrices. This includes the Frobenius norm of the naive and sandwich variance matrices in Equation (
35). The norm exceeds
, indicating substantial misspecification and underscoring the need for the sandwich estimator to robustly quantify Xinanjiang parameter uncertainty.
In
Table A4 of
Appendix G we compare the frequentist bread matrix
with the inverse of the covariance matrix of the DREAM
(ZS)-sampled naive posterior realizations. The MCMC-derived bread matrix is in reasonable agreement with
, consistent with the close correspondence observed in
Figure 6 between the frequentist characterization of naive parameter uncertainty and the normal posterior histograms sampled by the DREAM
(ZS) algorithm for most parameters. The marginal posterior distributions of the tension water inflection parameter
a and the free water shape parameter
deviate noticeably from normality, which explains in part the relatively large differences in their diagonal elements of the bread matrices of the frequentist and MCMC methods. The largest discrepancy is observed for the parameter
, whose MCMC-derived bread matrix value on the main diagonal is
, approximately 40 times smaller than the corresponding value of
from the frequentist estimator. The culprit may be the prior distribution, which truncates the posterior distribution of
at unity but does not affect the normal approximation underlying the frequentist characterization of naive parameter uncertainty. Thus, in summary, good agreement between the linear (frequentist-based) and nonlinear (Bayesian sample-based) estimates of the sensitivity (bread) matrix suggests that the posterior distribution is approximately Gaussian, and that the multinormal frequentist description of the ML uncertainty is consistent with the fully Bayesian approach.
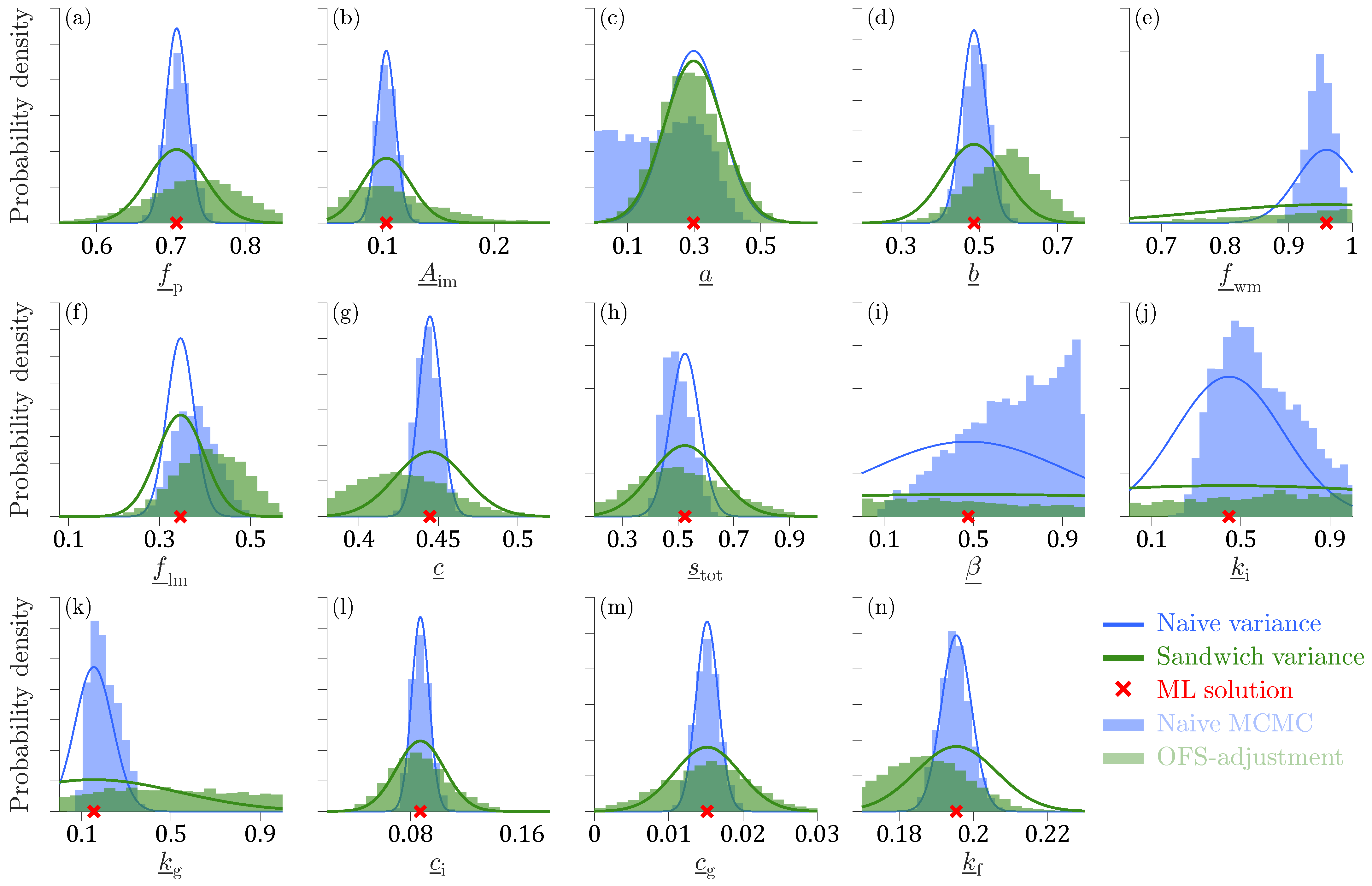
Figure 7 presents histograms of the OFS-adjusted posterior samples of the Xinanjiang parameters and degrees of freedom
of the Student
t likelihood function
. The OFS-adjusted posterior samples are derived from Equation (
11) using
where the matrix square roots,
and
, are computed according to Equation (
13) using singular value decomposition.
The OFS-adjustment enhances substantially the dispersion of the posterior samples for all Xinanjiang parameters but the tension water inflection parameter
a. The histograms of the OFS-adjusted posterior samples (green bars) stretch far beyond the normal marginal distributions (blue lines) derived from the sensitivity matrix
of second-order partial derivatives of the Student
t log-likelihood function w.r.t. the ML solution
and the naive (blue) histograms of the Xinanjiang parameters. The culprit is model misspecification and, consequently, a poor alignment of the sensitivity
and variability
matrices. The OFS-derived sandwich histograms of the Xinanjiang parameters are in reasonable agreement with the normal marginal distributions (green lines) of the frequentist estimator of
. Note that the OFS-adjusted sandwich density functions for
,
, and
are visibly lower than their corresponding frequentist densities. This discrepancy arises because the OFS transformation in Equation (
11) does not honor the unit interval of the normalized Xinanjiang parameters. Infeasible parameter values lower the probability density of the adjusted posterior samples within the admissible range. Last but not least, for several parameters, the OFS adjustment of Equation (
11) altered the location of the mode (peak) of the sandwich distribution. The most notable shifts occurred for
,
b,
,
c,
, and
. Such changes are somewhat counterintuitive and arise in part from the non-uniqueness of the matrix square roots
and
used in the adjustment.
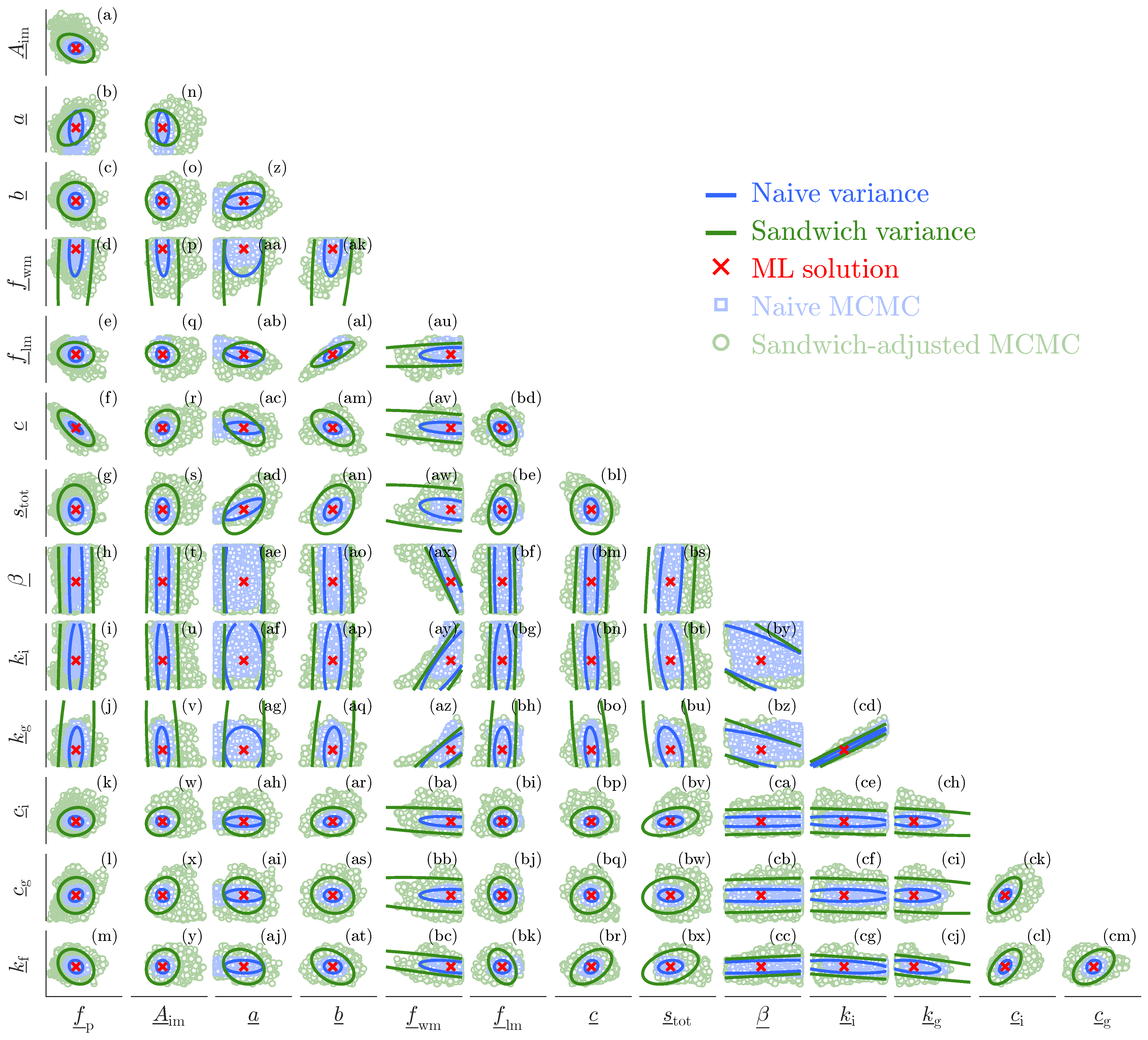
Figure 8 presents a matrix plot of the bivariate 95% confidence (lines) and credible (dots) regions of all pairs of Xinanjiang parameters. The blue area corresponds to the naive variance whereas the green area is associated with the sandwich-adjusted posterior samples of Algorithm 3.
The bivariate scatter plots offer a clearer depiction of the naive and sandwich uncertainty estimates for the Xinanjiang parameters. The following conclusions can be drawn.
The naive Bayesian 95% credible regions (blue squares), as sampled by the DREAM
(ZS) algorithm are in strong agreement with the frequentist 95% confidence ellipsoids derived from the naive variance estimator. There are some notable exceptions, particularly in the bivariate scatter plots involving parameter
a, where the MCMC-sampled naive confidence regions exceed the frequentist ellipsoids. This is a well-known phenomenon that highlights the distinction between linear and nonlinear confidence (or credible) regions [
49,
78,
79,
80].
The 95% credible regions of the sandwich-adjusted posterior samples (green dots) extend well beyond the sandwich ellipsoids (green lines) of the frequentist estimator. These linear sandwich confidence regions substantially underestimate the true parameter uncertainty, and appear woefully inadequate for accurately characterizing Xinanjiang discharge uncertainty.
For most parameter pairs, the MCMC-derived sandwich credible regions are unimodal and well described by a bivariate normal distribution.
The sandwich credible regions of the Xinanjiang parameters are much larger than their naive counterparts. This is a result of misspecification and confirms that the sensitivity (bread) matrix of the Student t likelihood function substantially overestimates the information content of the discharge observations. The only valid currency of discharge data informativeness under model misspecification is the Godambe information, as expressed by the sandwich credible regions. The enlarged parameter uncertainty should yield the appropriate parameter coverage probabilities.
Substantial differences between linear and nonlinear confidence regions, such as those observed for the tension water inflection parameter a, often signal problems in model formulation. Other indicators of model misspecification include parameters whose MAP estimates occur at or near the bounds of their prior ranges. Although the Xinanjiang model does not exhibit this behavior for the Leaf River dataset, practical experience with other conceptual hydrologic models suggests that such issues are more common than rare. When a MAP estimate lies close to a parameter bound, the local curvature of the log-likelihood becomes poorly defined, making it difficult or impossible to compute a stable Hessian (bread) matrix. This, in turn, undermines the validity of asymptotic approximations in frequentist inference, such as the ML sandwich estimator used herein.
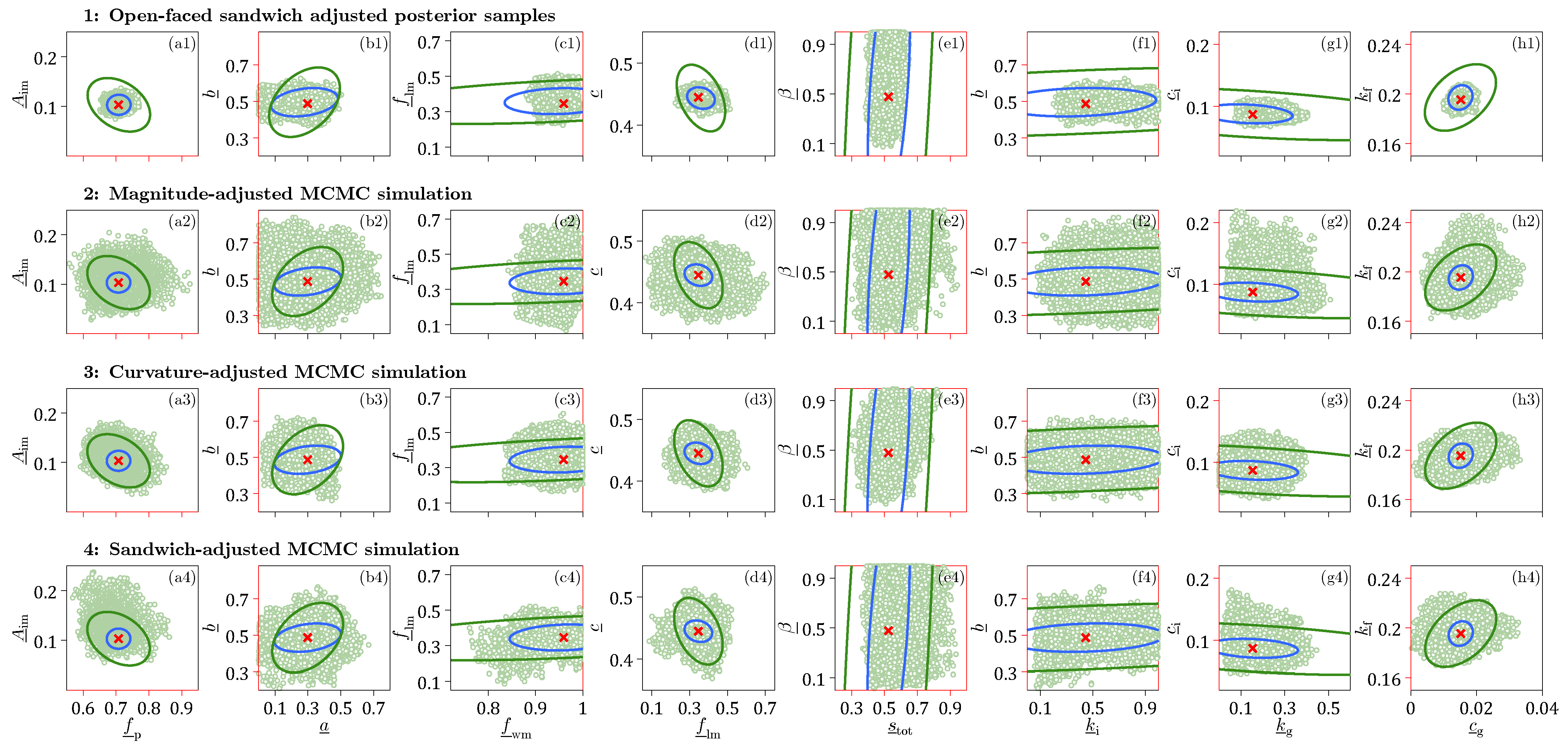
Before turning our attention to Xinanjiang discharge uncertainty, we first examine in
Figure 9 bivariate scatter plots of the OFS-adjusted posterior samples and their counterparts obtained from magnitude-, curvature-, and sandwich-adjusted MCMC simulation. For a direct comparison of the different methods, the same
x- and
y-axis limits are used for all four graphs in each column. We focus our attention on only a subset of the Xinanjiang parameter pairs.
The results in
Figure 9 highlight several interesting observations.
We cannot prove that the sandwich credible regions are more accurate, as the pseudo-true values of the Xinanjiang model parameters that generated the observed discharge record are unknown. Instead, we rely on statistical theory, which establishes the sandwich estimator as the only valid asymptotic descriptor of data informativeness, and, consequently, parameter uncertainty under model misspecification.
Figure 10 shows posterior predictive bands for simulated streamflow from Xinanjiang over a representative segment of the six-year training period, obtained by propagating posterior draws of
through the model.
The sandwich variance estimator substantially widens the parameter uncertainty induced intervals for simulated streamflow from the Xinanjiang model, as evident in the right-hand panels. The 99% intervals expand markedly, especially near peak flows. Quantitatively, the 99%, 95%, 90% and 68% streamflow intervals based on sandwich parameter uncertainty contain
%,
%,
% and
% of the discharge observations, respectively, compared with
%,
%,
% and
% under the naive variance estimator. Thus, the naive 99% intervals achieve roughly the same coverage (≈13%) as the sandwich 68% intervals. In terms of width, the sandwich intervals are about twice as wide at low flows and roughly three times as wide at the hydrograph peaks (see
Figure A2 in
Appendix H).
Finally, we examine the discharge residuals obtained from the ML parameter values of the Xinanjiang model.
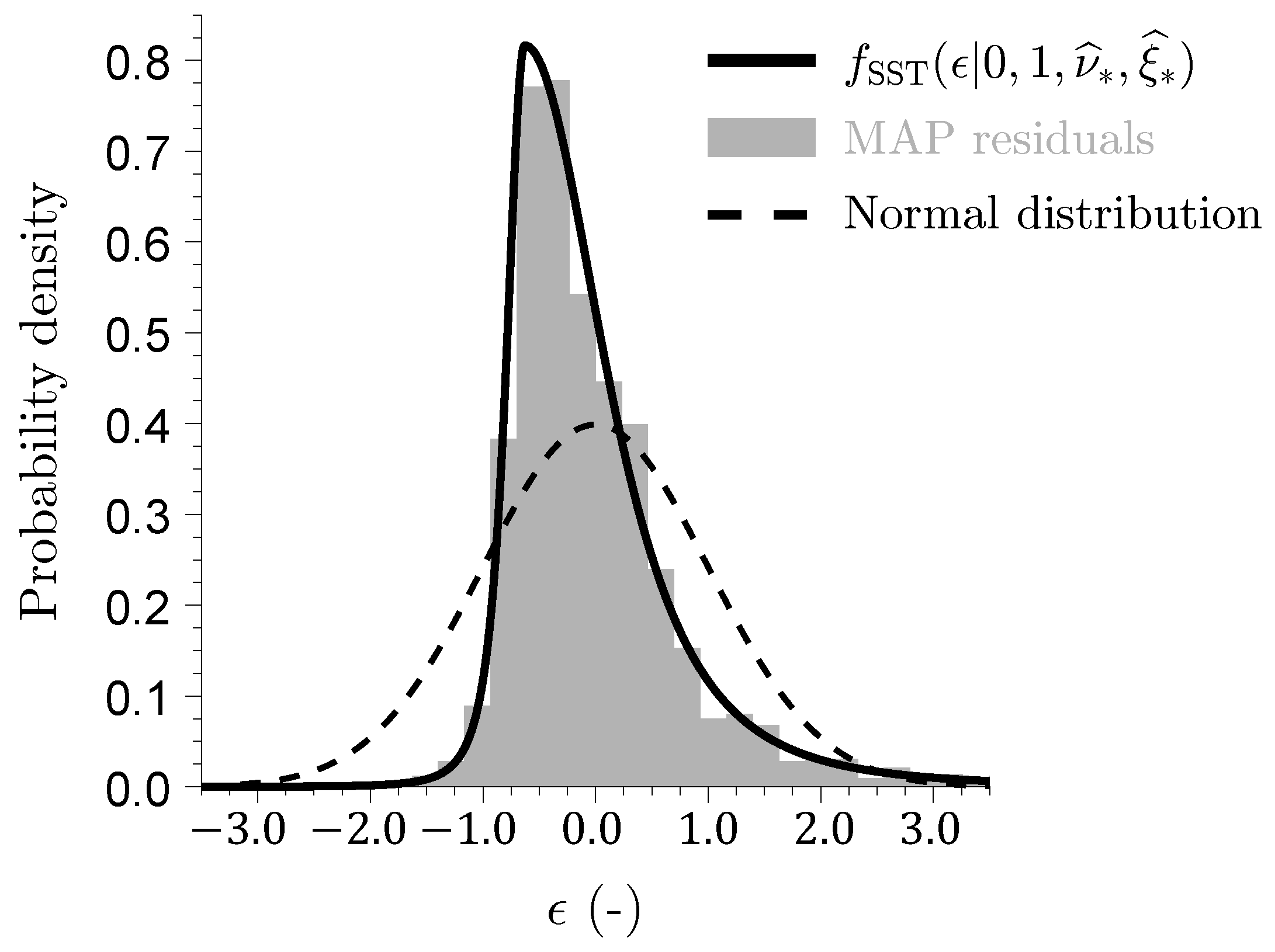
Figure 11 shows a histogram of the studentized discharge residuals
, with gray bars normalized to represent a probability density estimate such that the total area under the bars amounts to one. We also plot the SST density
of Equation (
30) using the ML values of
and
.
The histogram of the discharge residuals is in excellent agreement with the SST density. The studentized residuals
follow a Student
t distribution with
degrees of freedom and skewness
. This degrees of freedom is much smaller than one would expect from the sample size
and the number of parameters
alone. This result once again confirms that the discharge residuals follow a Laplacian or double exponential distribution [
11,
57]. A skewness of
indicates that the distribution of MAP discharge residuals is right-skewed. Consequently, the mode (peak) of the distribution of the studentized streamflow residuals is located at
, to the left of the median value of
, which itself is smaller than the mean studentized residual of approximately
. This mean value points to a negative bias in the Xinanjiang model, indicating a tendency, on average, to underestimate measured streamflows. The magnitude of this bias is around
mm/d or
% of the mean measured discharge of
mm/d.
The SST density with low degrees of freedom exhibits both a sharper peak near its mean and heavier tails compared to the normal distribution (dotted line). This makes the Student t likelihood more robust to outliers and is well suited for inverse modeling of discharge data with the occasional large streamflow residuals. The largest residuals are typically attributable to precipitation measurement errors and are less governed by structural limitations and/or deficiencies of the hydrologic model. However, the sandwich estimator cannot distinguish between these two error sources. Both count as misspecification.