3. Simulation Study
The simulation setup is designed to emulate high-dimensional regression problems with strong collinearity and different error distributions. Such settings are commonly found in econometric modeling, genomics, environmental studies, and machine learning applications. The synthetic datasets reflect these structures in a controlled way, allowing for a focused evaluation of different methods (estimation and aggregation) under ill-conditioning and data heterogeneity scenarios. All the code is developed in MATLAB (R2019b version) [
17] by the authors.
The general context of the simulation is as follows: the two collinearity scenarios are intended to illustrate models with low and high collinearity found in practice; the vector of parameters illustrates scenarios “close” to zero, positive and negative, very common in practice; the different error distributions reflect variations in noise; the size of the models is still computationally feasible for the design of the simulations and allows for the possibility of sampling; the two parameter supports for GME and W-GME reflect reduced and high information by the user regarding the model parameters; the supports for the errors, as well as the number of points in the supports, are the usual ones in the literature. Finally, the number of groups and the number of observations per group try to reflect the purpose of aggregation (obtaining accurate information with minimum computational effort and observing a small part of the population under study).
3.1. Simulation Settings
A linear regression model is considered, with a number of observations of 30,000 and a number of explanatory variables of 10 (N = 30,000 and ). Two (30,000 × 10) matrices of explanatory variables are simulated, corresponding to matrices with two different condition numbers, representing two distinct collinearity scenarios: cond() = 10, for cases of near absence of collinearity; and cond() = 20,000, for cases where high collinearity is present in the data. The matrices were initially constructed using standard normal distributions, and then the singular value decomposition was used to algebraically adjust the matrices to the desired condition numbers. The MATLAB functions available imposed a restriction on the size of the matrices that could be used, and the values = 30,000 × 10 represented the maximum feasible under these conditions. Anyway, the size of the original dataset in the simulation study (whether thousands or millions) is not relevant, as we will always work on estimating parameters in models with small datasets extracted by sampling. The simulation framework assumes a linear relationship between explanatory variables and the dependent variable, with errors following known distributions. These assumptions were chosen deliberately in order to provide a controlled environment where the effects of ill-conditioning, choice of the estimators, and aggregation methods could be systematically isolated and evaluated. However, it is also important to note that data scientists dealing with huge amounts of data frequently use linear models as a simplified view of the reality and these are actually very satisfactory models quite often.
The number of parameters in the regression model will correspond to the number of explanatory variables and, consequently, the parameter vector will be a vector, defined as , not containing the constant term. Furthermore, three distinct types of error are considered for the given datasets: errors modeled by a normal distribution with zero mean and unit standard deviation, , to represent scenarios with relatively low noise; errors based on a t-Student distribution with three degrees of freedom, , to represent scenarios with significant but moderate noise; and errors following a Cauchy distribution with location parameter zero and scale parameter two, , to represent scenarios with high noise. In this way, three distinct vectors (30,000 of random perturbations are obtained, resulting in six vectors (30,000 of noisy observations, combining the two different condition numbers with the three vectors of random perturbations, to obtain a wide variety of noisy observation vectors with different characteristics.
The reparameterizations imposed by the GME and W-GME estimators are carried out by defining the matrix , which contains the supports of the errors, with symmetric supports centered at zero, using the 3-sigma rule, where sigma is approximated by the empirical standard deviation of the noisy observations, and the support is always the same for each unknown perturbation. Consequently, the support vectors of the errors will take the form for all 30,000, being equally spaced, with points used to form these supports. Additionally, two distinct supports for the parameters are considered, both symmetric and centered at zero, so these vectors will also be equally spaced, considering points to form these supports (the same for each of the unknown components of the parameter vector of the model). Thus, two distinct matrices are defined, containing the supports for the parameters: one with for all , to reflect scenarios where there is some prior knowledge about the range in which the parameters may be found, allowing for a reduction in the amplitude of the parameter supports; and another with for all , to reflect scenarios where the available prior knowledge is insufficient. As a clarification remark, the support vectors are actually given by and , but, to simplify notation, they will be mentioned henceforth as and . The objective is to understand (albeit preliminarily, as this is not the main aim of the work) how the amplitude of the parameter supports affects the results of the GME and W-GME estimators.
Completing with the necessary specifications regarding the aggregation methods, random sampling with replacement was performed considering two values for the number of groups, namely, and , with the aim of analyzing whether increasing the number of groups in each aggregation method causes significant differences in the results obtained, and two values for the number of observations per group, Obs and Obs , to assess whether the increase in observations in each group, that is, the increase in information available in each group, produces relevant changes in the performance of each of the aggregation methods.
In summary, a total of 48 scenarios will be analyzed, referring to the combination of the two matrices of explanatory variables (two condition numbers) and the three error vectors (three error distributions), generating a total of six different datasets, associated with the two supports for the parameters, the two values for the number of groups in the aggregation methods, and the two values for the number of observations per group, forming a total of eight variants of structures. Additionally, a key point of this simulation study will also be to study the variance of the results, as there is a random sampling process involved, to understand whether the results obtained are directly associated with the aggregation method used or if it is a consequence of a less informative sampling process. For this purpose, Monte Carlo experiments with 10 replicas for each scenario are conducted, with the objective of exploring the sampling behavior of the situations under study.
Once the datasets are obtained, the model coefficients are estimated using the previously mentioned aggregation techniques, bagging, magging, and neagging, varying the estimators used in each aggregation procedure, namely, the OLS, GME, and W-GME estimators. For each of the 48 Monte Carlo experiments related to the scenarios under analysis, the regression coefficient estimates of each considered aggregation method are calculated in each of the 10 replicas, using the different estimators discussed. These estimates will be defined by , expressing the regression coefficients estimated after aggregation. The final regression coefficient estimates, that is, those obtained after conducting the Monte Carlo experiment, relative to each of the aggregation procedures, using the different estimators considered in this study, are calculated by averaging the 10 values obtained from each replica, being referred to as , expressing the final estimated regression coefficients after aggregation (mean of the replicas). Henceforth, when presenting the results of this study, the general abbreviation ‘aggr’ will be replaced by the following abbreviations: bOLS to represent the bagging aggregation procedure with OLS coefficient estimates; bGME to represent the bagging procedure with GME estimates; bW-GME to represent the bagging procedure with W-GME estimates; mOLS to represent the magging procedure with OLS estimates; mGME to represent the magging procedure with GME estimates; mW-GME to represent the magging procedure with W-GME estimates; nGME to represent the neagging procedure with GME estimates; and nW-GME to represent the neagging procedure with W-GME estimates.
3.2. Evaluation Metrics Based on Prediction and Precision Errors
For each of the Monte Carlo experiments related to the scenarios under analysis, the prediction and precision errors associated with each of the obtained are calculated for all 10 replicas. The prediction error is derived from the Euclidean norm of the difference between the vector of predicted observations using the model (multiplication of the matrix of explanatory variables by the estimated regression coefficients after aggregation) and the simulated vector of noisy observations, that is, . The precision error is calculated through the Euclidean norm of the difference between the vector of estimated regression coefficients after aggregation and the original parameter vector considered in the simulation, that is, . These two complementary metrics are used to assess the effectiveness of each aggregation strategy: precision error (a standard metric in regression analysis under simulation; reflects the overall stability of the parameter estimates across replications) and prediction error (a standard metric in regression analysis; reflects the practical predictive accuracy of the estimated model).
The prediction error 1 and precision error 1 are obtained by averaging the 10 prediction and precision errors obtained in each replica, thus obtaining and , respectively, and the prediction error 2 and precision error 2 are determined by implementing the formulas for prediction and precision errors, but using the final estimated regression coefficients after aggregation (mean of the replicas), , obtaining the values and , respectively.
The variances of the results are calculated using the usual variance formula, that is, the mean of the sum of the squares of the differences between each element and its mean. Therefore, the variance of
, the variance of the prediction error, and the variance of the precision error are
respectively, with
R corresponding to the number of replicas, 10. While formal hypothesis testing is not applied in this study, the use of replicas per scenario, variance estimates, and boxplot visualizations offer a preliminary assessment of the stability and dispersion of results across different techniques (estimation and aggregation).
4. Results and Discussion
The results presented in this work aim to highlight the general trends of this simulation study. To better understand and analyze the results obtained, summary tables of precision errors 1 and 2 for all the previously mentioned scenarios are presented in
Table 1,
Table 2 and
Table 3. Data visualization was carried out using MATLAB for the boxplots and R (ggplot2 package) for the heatmaps.
Additionally, comparative graphs of precision errors between the different methods are presented, when the number of groups and the number of observations per group are modified, in order to evaluate the impact of these factors on the studied aggregation methods, with the respective estimators considered. Due to the extent of this simulation study, given the high number of presented scenarios and subsequent vectors of estimates and variances obtained (eight vectors per scenario, relative to each of the aggregation procedures with the different estimators considered), only the results of certain scenarios are highlighted. These scenarios aim to establish a representative set of the total scenarios considered in the study. The remaining are available upon request to the authors.
The values of the prediction errors 1 and 2 resulting from these selected scenarios, and the variances associated with these results, are found in
Table 4,
Table 5 and
Table 6. Note that values presented as 0.00 do not represent a null value, but, rather, a value less than 0.01. Additionally, boxplots of the regression coefficient estimates and precision error for these scenarios are presented for a graphical analysis (among other aspects) of the variances relative to these results.
Analyzing the precision errors obtained in the scenarios
,
Table 1, in situations of low collinearity (cond(
) = 10), it is observed that
is generally lower when one of the estimators based on the ME principle is applied. Although the OLS estimator usually performs well in data with low collinearity, the estimates obtained by procedures using the GME and W-GME estimators stand out compared to the estimates obtained by the bOLS and mOLS procedures. The bGME, bW-GME, mGME, and mW-GME are the methods that distinguish themselves by their good performance. However, the performance of the bagging method with the GME and W-GME estimators tends to worsen with the increase in the width of the coefficient supports, highlighting the better performance of magging with the use of these estimators. Additionally, the performance of mW-GME generally surpasses that of mGME in scenarios where
. The nGME also presents one of the best performances, but only in the situation where the support for the parameters is narrower (
).
Now, analyzing a scenario with high collinearity (cond() = 20,000), a significant increase in is observed in aggregation methods using the OLS estimator, while the methodologies that stood out previously in the scenario with low collinearity continue to behave similarly. This indicates that the presence of collinearity does not seem to affect the results provided by the previously mentioned aggregation methods, namely, those using estimators based on the ME principle. As the performance of the OLS estimator is generally affected by the degree of collinearity in the data, and the GME estimator (and, by extension, the W-GME estimator) is suitable for handling data affected by collinearity, these results are not unexpected. Again, the magging aggregation method with the GME and W-GME estimators stands out for its performance, and the behaviors observed for low collinearity scenarios are also observed in these cases. However, with the increase in G and Obs, the performance of bGME and bW-GME improves once more.
Note that, when evaluating the quality of the obtained estimates (precision error 2), the scenarios with a wider range of parameter supports, a larger number of groups, and a greater number of observations exhibit the most suitable estimates for the regression coefficients of the presented problem, with the use of bGME, bW-GME, and nGME, regardless of the degree of collinearity present (but more visibly with higher collinearity).
When
, the performance of bGME and bW-GME decreases compared to the previous case. From
Table 2, it can be seen that the methods that consistently present the lowest
are mGME and mW-GME. The bagging and neagging aggregation methods generally show a decrease in their performance compared to the previous table, suggesting that these methods are more affected by the presence of larger perturbations in the data. The effects caused by collinearity and the existence of more adverse perturbations are noticeable in the aggregation methods that apply the OLS estimator.
Finally, analyzing the case where
,
Table 3, the conclusions are similar to the case
. The bagging and neagging aggregation methods show poor performance, especially when using the OLS estimator, and the magging aggregation method is recognized for its performance when using the GME and W-GME estimators. However, mW-GME reveals a predominantly dominant behavior in terms of precision error compared to mGME (with similar performance or insignificant differences in cases where it does not stand out), particularly in scenarios with
.
Subsequently, it is necessary to analyze to what extent the increase in the number of groups (G) and observations per group (Obs) can affect the performance of the various aggregation methods using the considered estimators. Comparing the same support, , with cond() = 10 and cond() = 20,000, it is observed (illustrative graphical representions highlighting this observation can be requested from the authors) that bOLS demonstrates worse performance when Obs is smaller, and is lower when this number increases. The increase in G seems to improve the performance of bOLS, except in the case , which shows an atypical effect on the precision error of bOLS, possibly due to the pronounced presence of noise.
These results are expected due to sampling and inferential statistical theory. The mOLS method seems to follow a similar behavior, where both the increase in G and the increase in Obs lead to better performance, even in cases where .
However, the estimators based on the ME principle do not appear to follow this behavior, remaining approximately constant with the increase in G and the addition of more observations per group, being more unequivocal in the bagging and magging aggregation methods. The invariability of values concerning the increase in the number of groups/observations is a significant advantage in using the bagging and magging methods that use the GME and W-GME estimators. As there is no need for a larger number of groups or larger datasets to obtain suitable estimates for the problem in terms of precision error, it is possible to reduce the computational burden of implementing these aggregation procedures.
In summary, as observed in
Table 1,
Table 2 and
Table 3, the best precision error results are obtained through bGME, bW-GME, mGME, and mW-GME in scenarios of errors modeled by a normal distribution, and by mGME and mW-GME in scenarios modeled by a t-Student or Cauchy distribution.
Analyzing the same support , with cond() = 10 and cond() = 20,000, the conclusions for the aggregation methods employing the OLS estimator remain substantially the same as before (illustrative graphical representations highlighting this observation can be requested from the authors) for all considered error distributions. However, with the increase in the parameter supports’ range (and despite scenarios where , in which remains approximately constant for bGME, bW-GME, mGME, and mW-GME), when and , all aggregation procedures with estimators based on the ME principle show a decrease in performance as Obs increases (contrary to the results obtained for the narrower support). Except for the neagging method in the case of , this decrease appears to be more pronounced with the increase in the number of observations and remains unchanged with the increase in the number of groups. In the case of the neagging method, this effect might indicate that aggregating estimates with the knowledge of the proportion of the information content of various groups is more advantageous compared to aggregating with the proportion of the information content of larger groups, as groups with many observations do not necessarily imply a higher state of knowledge or a lower state of uncertainty.
Lastly, considering the comparison of situations with the same condition number, and naturally excluding methods that used the OLS estimator, it can be said that the increase in the parameter supports’ range, from
to
, caused a significant decrease in the performance of all aggregation procedures, except for mGME and mW-GME (excluding cases with lower G and higher Obs). This analysis is in agreement with the discussion of
Table 1,
Table 2 and
Table 3.
One of the most characteristic aspects of this simulation study was the similar performance of prediction errors for all analyzed scenarios, with differences only occurring with changes in the distribution of random disturbances. The most pronounced difference in the values of was associated with bOLS, which exhibits the highest prediction error value and associated variance. However, is similar for all other aggregation methodologies and their estimators used, even when changing the level of collinearity present in the explanatory variables, the supports of the unknown parameters, and the number of groups and observations per group. The only exception is in the neagging aggregation methodologies when the supports’ range is broader, producing higher prediction errors and greater associated variance, except when . However, the magging aggregation method consistently shows low prediction error and associated variance across all analyzed scenarios.
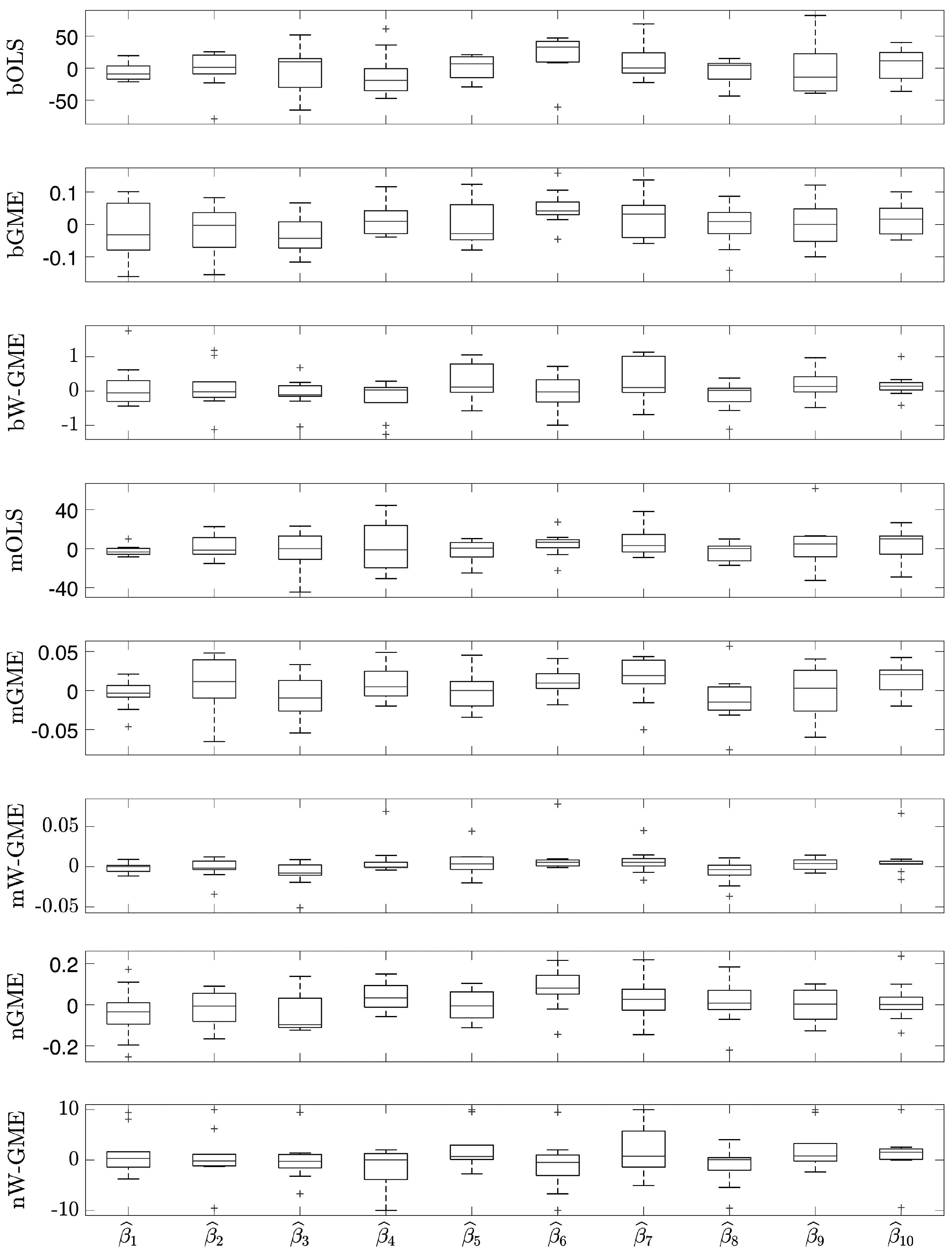
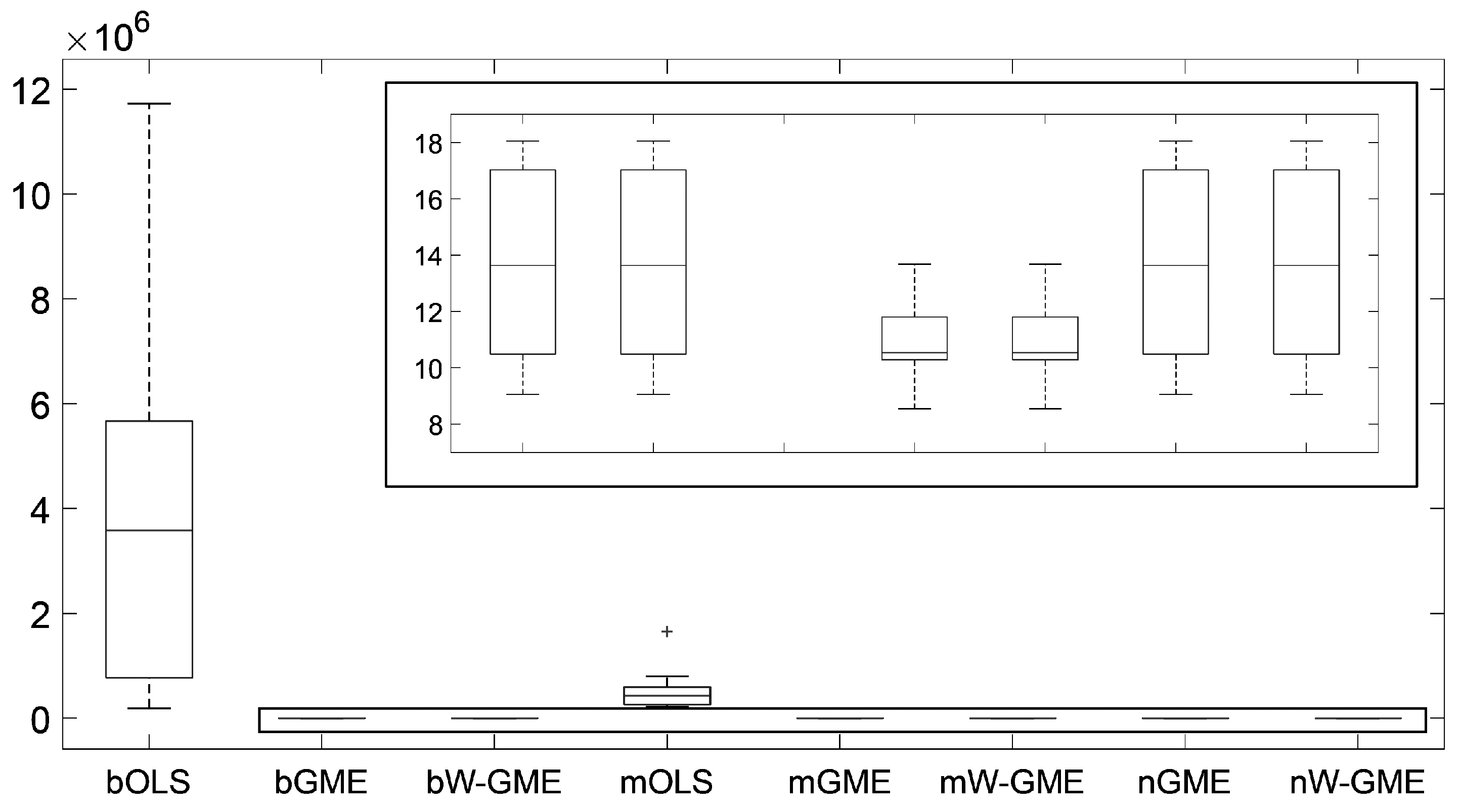
In this initial scenario, the estimates provided by bGME, bW-GME, mGME, mW-GME, and nGME in
Table 4 show a high level of shrinkage towards the center of their supports and reduced variance in the results—the GME estimator can be considered a shrinkage estimator. As the support limits are narrow, they exert significant pressure on the estimates, as decreasing the support limits diminishes the impact of the data and increases the impact of the supports. Despite this, due to the prior information provided to the method, they yield better results than the conventional OLS estimator in the context of aggregation. The analysis of the boxplots associated with these estimates shows very little variation, indicating that these procedures are more stable than the other procedures analyzed, as evidenced in
Figure 1. Additionally, this behavior is identified in
Figure 2, as the widths of the box plot boxes for the precision errors of these procedures are much smaller than those of other methods, indicating reduced variation in the results (please note the different scales inside some figures and between figures throughout this work).
Given this, it can be said that the bagging and magging aggregation methods exhibit more stable estimates for this scenario, but only when applied with the GME and W-GME estimators, as well as the neagging method when applied with the GME.
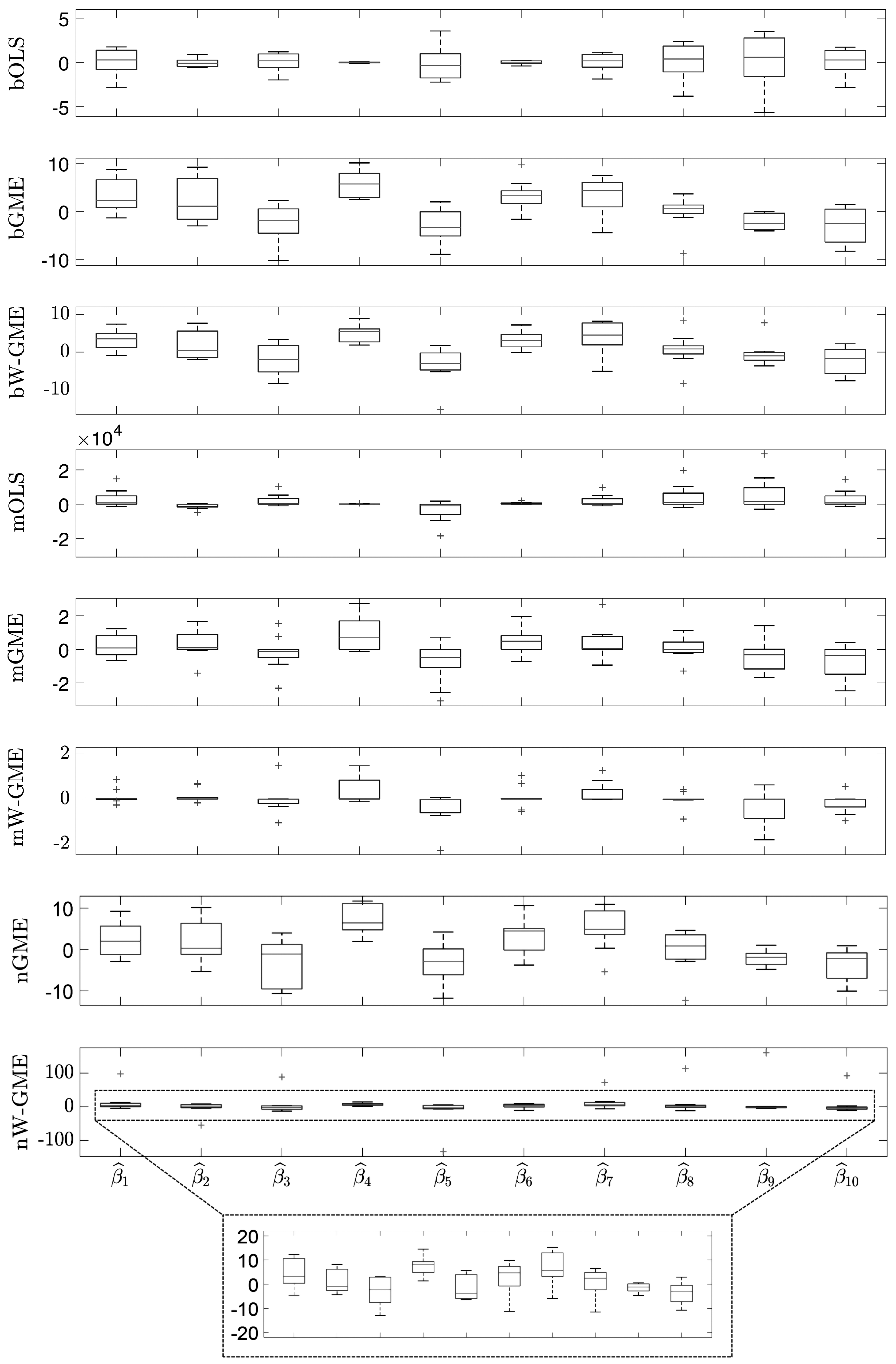
When comparing the same scenario of normal errors, but with a higher condition number and an increase in the number of groups, in the number of observations, and in the amplitude of the support for the errors (cond(
) =20,000,
, G = 20 and Obs = 100), in
Table 4, the estimates of the methods previously discussed (namely, bGME, bW-GME, mGME, mW-GME, and nGME) show a low precision error, but only mGME and mW-GME present estimates with less variation compared to the other procedures, as observed in
Figure 3. Additionally, the same is observed in
Figure 4, where a reduced variance corresponding to these methods in relation to precision error is noted. Thus, the methods highlighted in these scenarios are mGME and mW-GME, with the lowest values of
and associated variances, indicating more stable estimates.
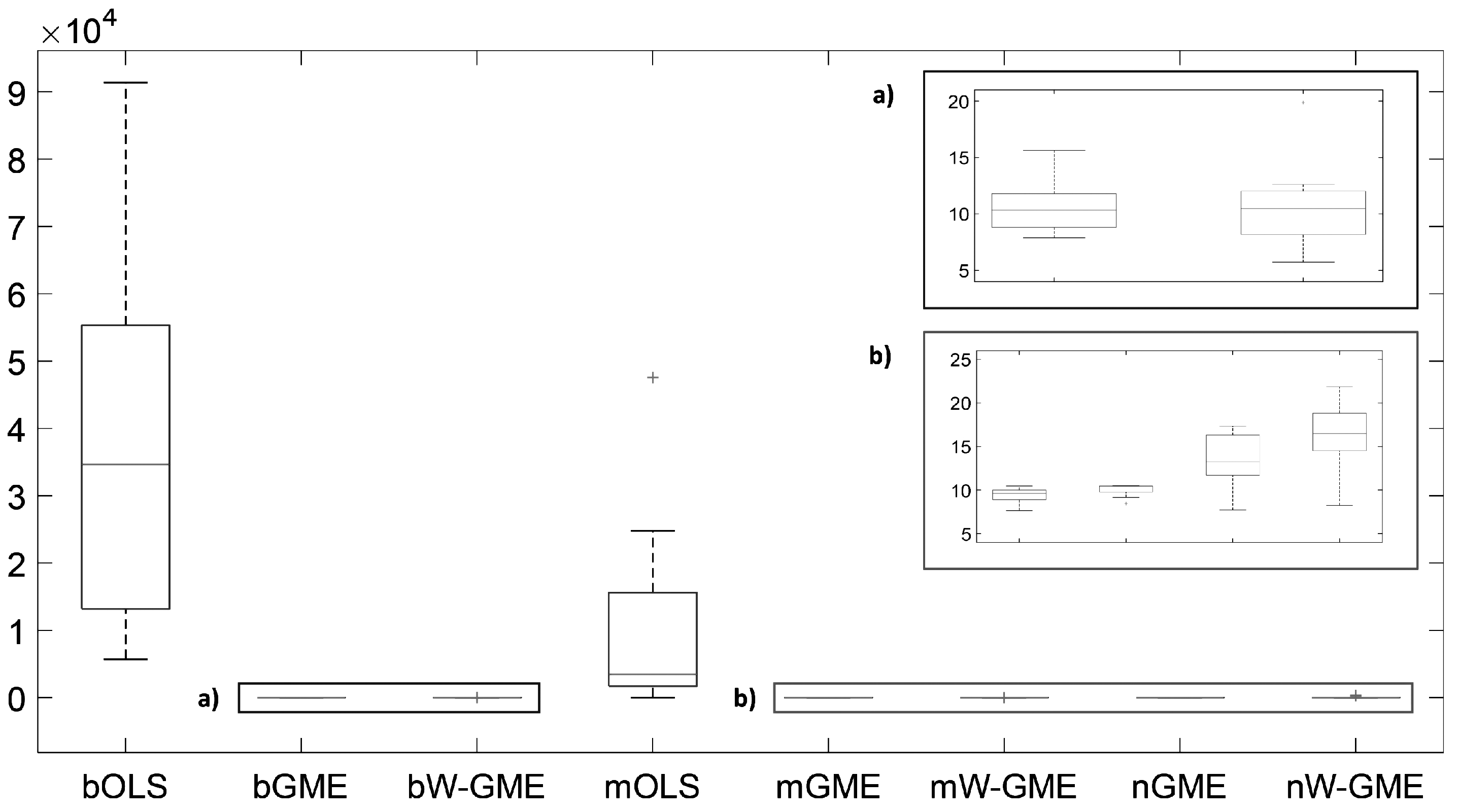
In the scenario
, with a lower condition number, G, and Obs, but
, in
Table 5, the standout procedure is the magging aggregation methodology using the W-GME estimator due to the reduced
and associated variances (see
Figure 5). As previously discussed, in this scenario, a decrease in prediction performance of bOLS, nGME, and nW-GME is already visible, along with a general increase in
in the remaining methods, compared to the scenarios with
.
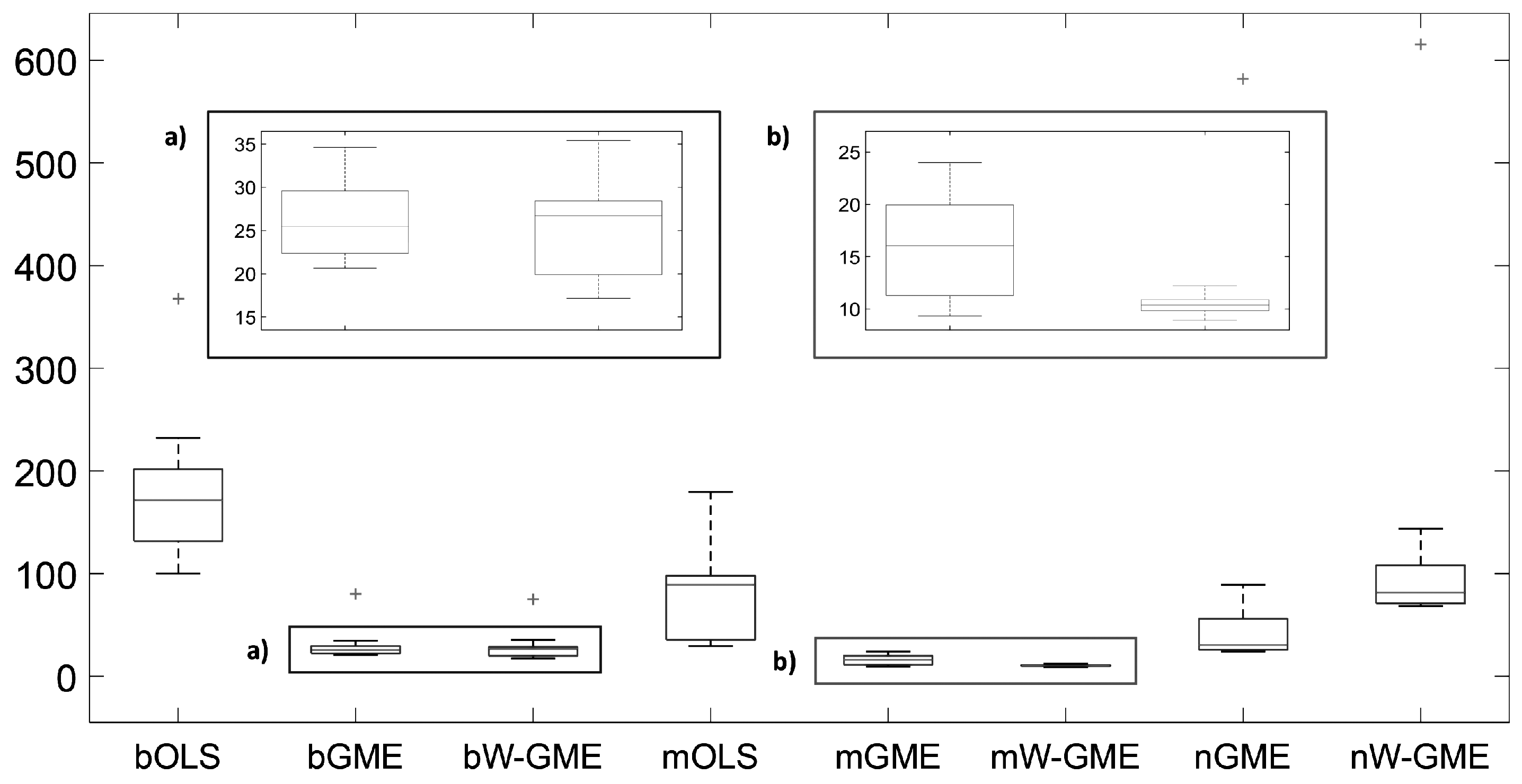
With the reduction in the amplitude of the support to
, and the increase in the condition number, in G, and in Obs, the aggregation methods with estimators based on the ME principle regain prominence, as can be seen from
Table 5, both in terms of precision and reduced variance. In particular, the magging method exhibits the best results in terms of
and reduced variation in the estimates (see
Figure 6). Another evident characteristic is the practically constant prediction error value corresponding to all methods (although higher than in the
scenarios).
Now, analyzing the situation of
, with values of G = 20 and Obs = 50 and
, the estimates with lower precision error and low variance values come from mW-GME, as can be confirmed in
Table 6, further supported by the analysis of
Figure 7. However, despite showing low variance, the estimates are excessively centered around zero. Due to the more pronounced presence of noise in this scenario, the standard deviation of the random disturbances, estimated by the standard deviation of the noisy observations, was eventually very high, which may have resulted in obtaining wider error supports. Additionally, the supports for the parameters also have a large amplitude, increasing the impact of the data.
To present a comparison with the previous scenario, consider now the situation where
, G decreases to 10, and Obs increases to 100, as described in
Table 6. Again, the methodologies mGME and mW-GME show good performance, with reduced associated variances and corresponding precision errors with lower variation values, as confirmed in
Figure 8.
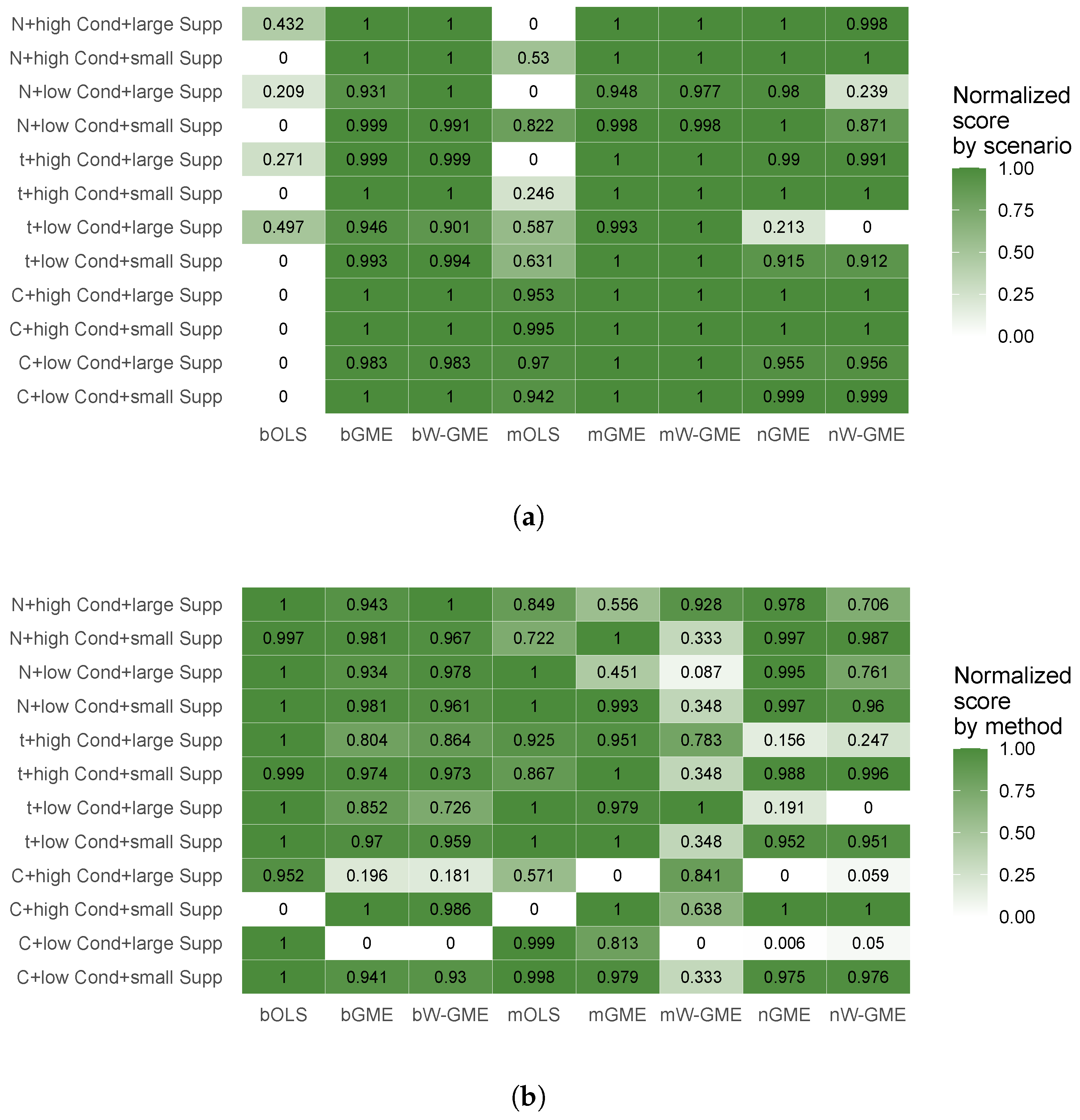
To highlight how different simulated conditions affect performance and to summarize the most relevant relationships, two heatmaps were created. For illustrative purposes, the simulated condition with 10 groups and 50 observations per group (G = 10, Obs = 50) was selected. An inverse min–max transformation was applied to the precision error values (type 2 error was chosen as it represents the precision error of the final aggregated estimate), resulting in a normalized performance score, ranging from 0 (worst) to 1 (best).
Figure 9 presents normalized performance scores, with normalization applied independently in two complementary ways: by scenario and by aggregation/estimation method. Panel (a) allows a comparative assessment of methods within each scenario, but not across scenarios. Conversely, in panel (b), the scores enable comparison of scenarios within each method, but not across methods.
As a final technical remark, the method designed for determining the parameter
is a data-driven method, but different from that suggested in the work of [
14], which was the least squares cross-validation (LSCV) method. Due to computational complexity, LSCV is not suitable for big data problems, as it implies thecalculation of
N estimates of
using the W-GME estimator, for each value of the
parameter to be tested, for each sample of size
N. A new method was used in this work for determining this parameter, based on the Holdout methodology, which firstly involves dividing the entire dataset into two mutually exclusive subsets (the training set,
, and the test set,
). For each value of the
parameter to be tested, the
is then estimated, in the training set, using the W-GME estimator, and the prediction error is calculated using the test data, as follows:
This procedure is executed for any set of possible values of that one wishes to test, and, in the end, the value of that minimizes the prediction error, , is selected. Although this method aims to determine a reasonable estimate for the parameter , and not necessarily the best estimate, it is believed to be sufficient to obtain adequate results, with a much lower computational cost than with LSCV. Using this approach, all values in the range with a spacing of 0.01 were tested for the parameter .
It was possible to observe the absence of any kind of pattern in the analyzed scenarios, indicating that the weighting parameter was being selected based on the data. As in the aggregation context, it is necessary to calculate a parameter for each group, it was possible to observe the value of adjusting to the data subsets according to the implemented method (it is not possible to present a parameter value for each scenario. In the aggregation context, an estimate of the parameter is calculated per group, using the W-GME estimator. Thus, the value of the parameter refers to each set of observations (group). Graphical representations available upon request from the authors). As noted earlier, the combination of the magging aggregation procedure with the estimation through W-GME, with this new method of selecting the parameter , results in the best outcomes in terms of precision error and low associated variances, among all the aggregation procedures considered with the different estimators discussed. From this, it can be concluded that the new method, although simple, works effectively in determining a reasonable value for the parameter , allowing the testing of a variety of values, as many as desired.
5. Conclusions
The effectiveness of aggregation methods in solving big data problems is, indeed, remarkable. The parameters of ill-posed linear regression models (particularly, ill-conditioned) in a big data context can be estimated stably through the info-metrics approach. The principle of maximum entropy developed by [
2,
3] is the theoretical basis for solving ill-posed problems, but the info-metrics approach developed by [
4,
5] allowed its generalization to more complex ill-posed problems, which are very common in various scientific fields.
The objective of this work was to understand which methodologies are most suitable for big data problems in linear regression models affected by collinearity. The first major conclusion was that the performance of aggregation methods critically depends on the estimator used in the groups obtained by sampling to obtain the respective regression coefficient estimates. The most significant differences highlighted by the simulation work were the diversity of estimates obtained by each aggregation procedure (with the respective applied estimators) and their corresponding precision errors. The prediction error shows significant differences only when the distribution associated with the error component is modified. On the other hand, when the supports for the regression coefficients have a smaller range, assuming that the true value of is contained within these supports, the choice of the number of groups and the number of observations per group is indifferent in the use of the bagging and magging aggregation methods, using the GME and W-GME estimation methods. This is because the results are approximately similar, presenting reduced precision errors and associated variances, indicating stable estimates. To reduce computational load, a low number of groups and observations per group is sufficient to obtain consistent estimates in the previously described scenario. This is undoubtedly another important result of this work.
The excellent performance of the magging aggregation method using the W-GME estimator was perhaps the most relevant discovery in this research work. When there is no prior information about the supports of unknown parameters and the error distribution, the magging aggregation method with the W-GME estimator shows superior performance in all studied scenarios compared to other aggregation procedures with various estimators used. In particular, the W-GME estimator generally provides better results in terms of precision error (when combined with the magging aggregation method), especially when the supports are wider. This result is quite promising, as the GME estimator, which usually performs well with data affected by collinearity, behaves worse in this circumstance of wider supports (as expected). If the W-GME estimator, being one of its extensions, can handle this problem, it becomes a very attractive estimation method.
Beyond the promising results in the simulation study, the proposed strategies for estimation and aggregation have strong potential for application in diverse real-world domains. These include econometrics, genomics, environmental sciences, and machine learning, where data are often noisy and ill-conditioned. The good performance of GME and W-GME estimators under collinearity and different distributional contexts for the errors, combined with an appropriate aggregation procedure, make them attractive for inference purposes under big data scenarios.
Given the small number of groups and observations per group needed to obtain good results, the computational cost is not a relevant factor, although it is important to keep in mind that the GME and W-GME estimators are slightly slower (constrained nonlinear optimization). Of course, if necessary, parallel computing can be easily implemented, making processing times even shorter.
This work also alerts us to the risk of using the OLS estimator, knowing that data scientists frequently use linear models as a simplified view of the reality in big data analysis, and the OLS estimator is routinely used in practice. Under low collinearity scenarios (condition number of 10), it was verified that the traditional OLS estimator with bagging performs well; however, this strategy tends to become very unstable under high collinearity scenarios (condition number of 20,000), where magging with W-GME stands out in terms of higher performance.
It is important to note some limitations and new avenues of research uncovered by this work. Further research needs to include a broader set of aggregation procedures and estimation methods, which could enhance the comparative framework and potentially offer a more comprehensive view of the context-specific advantages or limitations of each technique (estimation and aggregation), which may not be yet completely revealed [
18,
19,
20]. While the design adopted in this work enabled a systematic and controlled comparison of different methodologies under ill-posed linear regression models, future work will aim to extend the simulation study to account for more realistic scenarios, including nonlinear relationships, heteroscedasticity, skewed distributions for the explanatory variables, and other forms of model misspecification, as well as to test the methods’ performance on real-world datasets.
In summary, as previously mentioned, the identification of the excellent performance of the magging aggregation method, suitable for non-homogeneous data circumstances, using the W-GME estimator, which uses weights in the objective function of the optimization problem to seek a better balance between precision and prediction, was a relevant discovery that could contribute to the analysis of large volumes of information, which is an urgent need in many fields of human activities. Although the entropy-based framework of neagging provides an interpretable aggregation mechanism, its behavior may vary depending on group structure, data quality, and the stability of individual estimators. A deeper exploration of these factors will be fundamental to broadening its applicability. We hope that this work contributes not only by benchmarking methods under controlled conditions, but also by encouraging further research into stable and information-driven aggregation techniques that can meet the challenges posed by modern data environments.