Characterizing the Impact of Communication on Cellular and Collective Behavior Using a Three-Dimensional Multiscale Cellular Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Mathematical Model
2.1.1. Cells as Boolean Networks
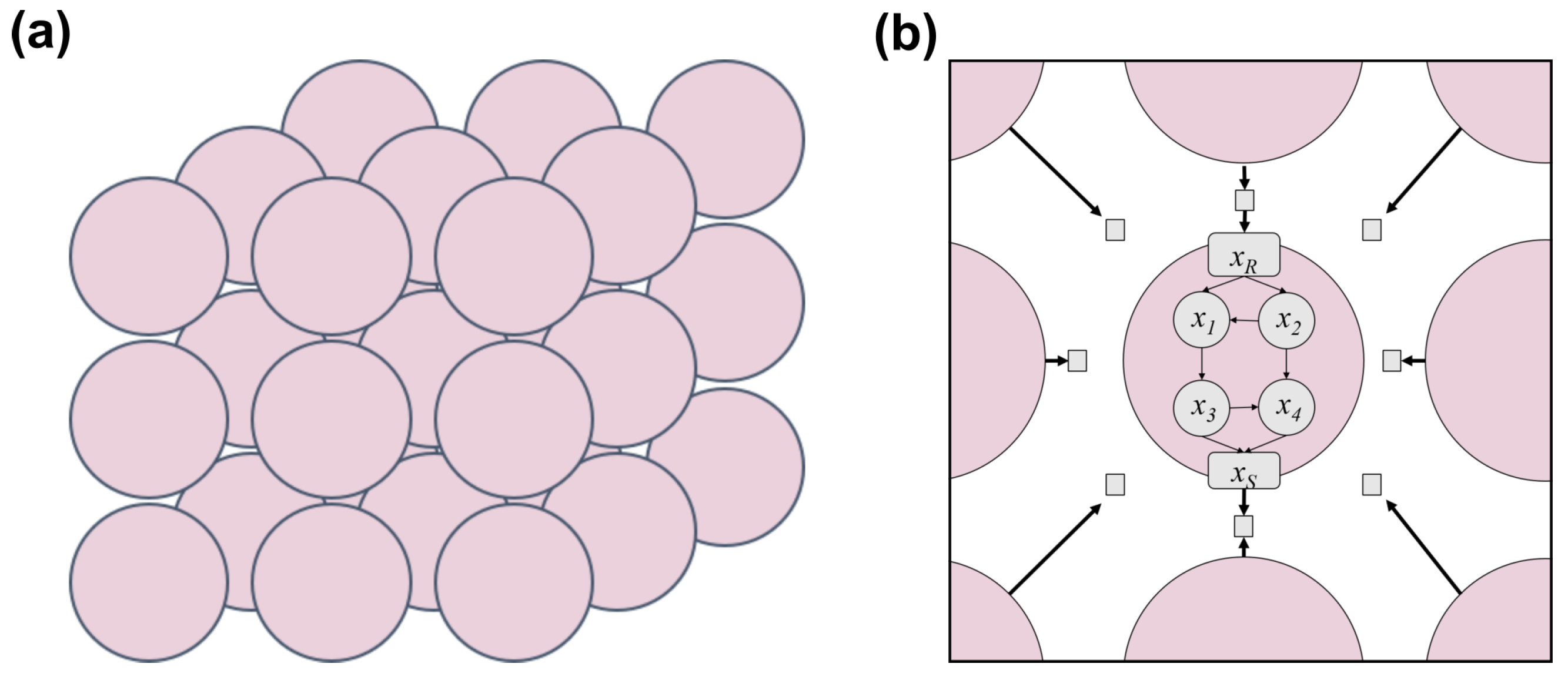
2.1.2. Tissue Architecture
2.1.3. Cell–Cell Communication via Diffusible Molecules
2.1.4. Simulation Framework
3. Experiments and Results
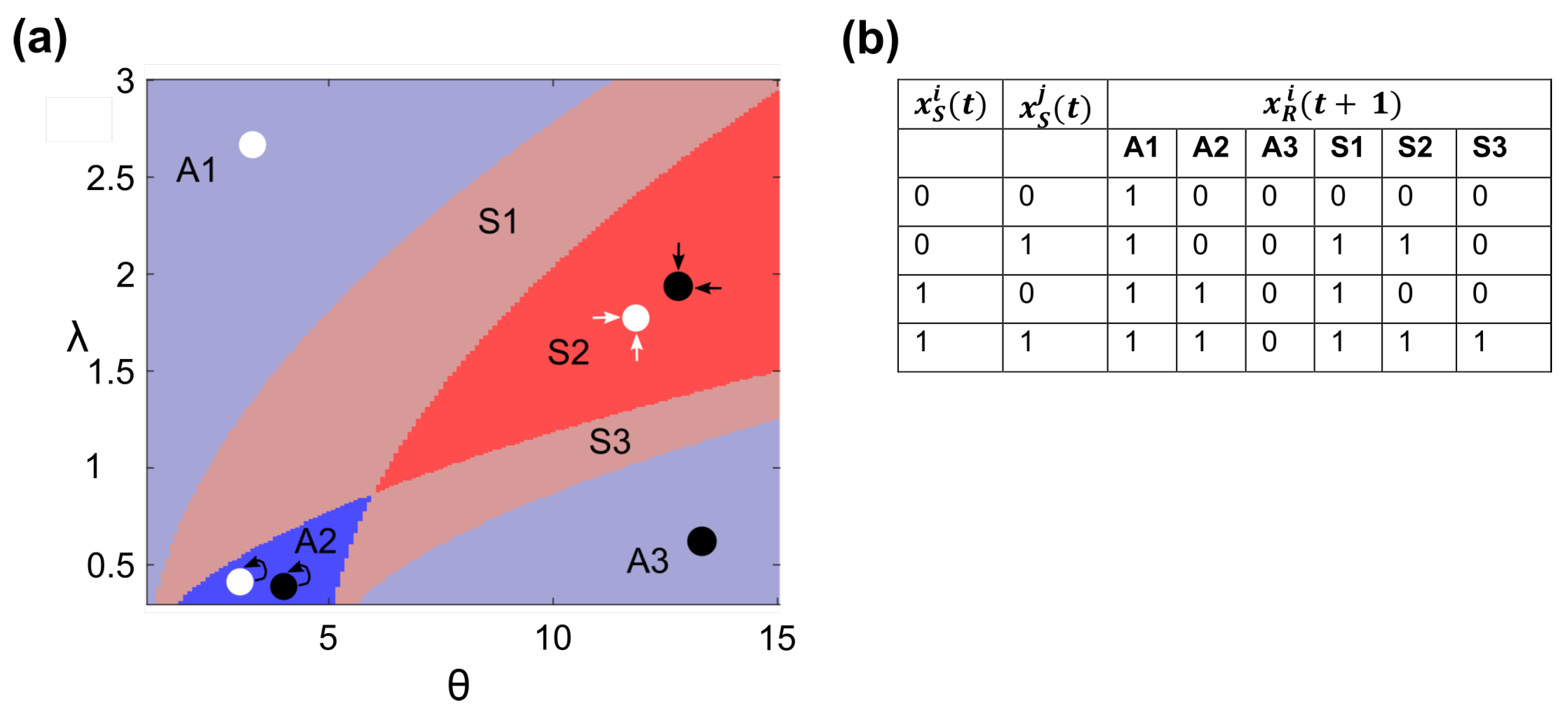
3.1. Changing Interaction Distance and Receptor Activation Threshold Can Change the Mode of Communication
- All cells’ signal nodes are OFF: ;
- Cell i’s signal node is OFF and all other cells’ signal nodes are ON: ;
- Cell i’s signal node is ON and all other cells’ signal nodes are OFF: ;
- All cells’ signal nodes are ON: .
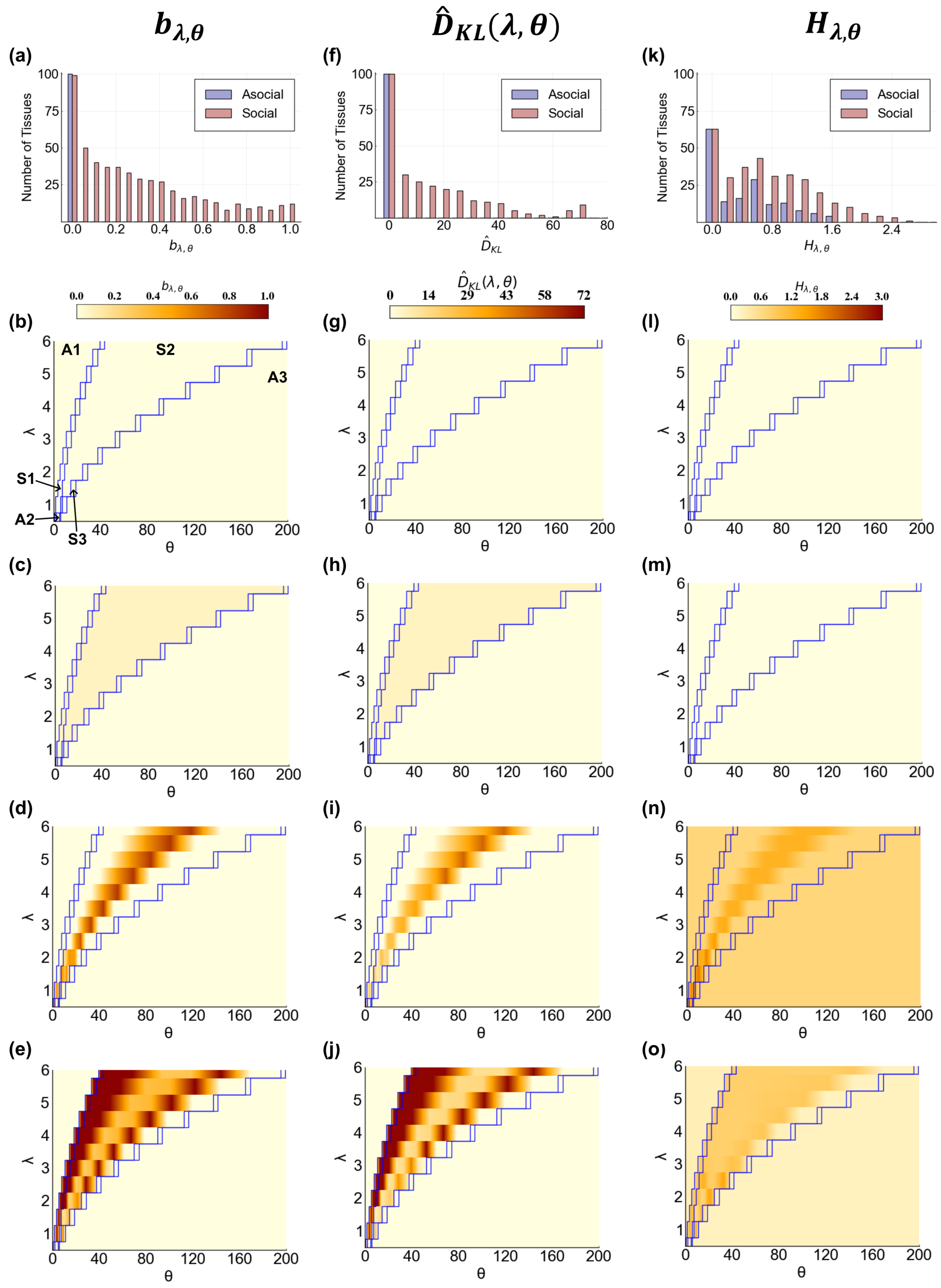
3.2. Cellular and Tissue Behavior Are Distinct between Asocial and Social Behavior
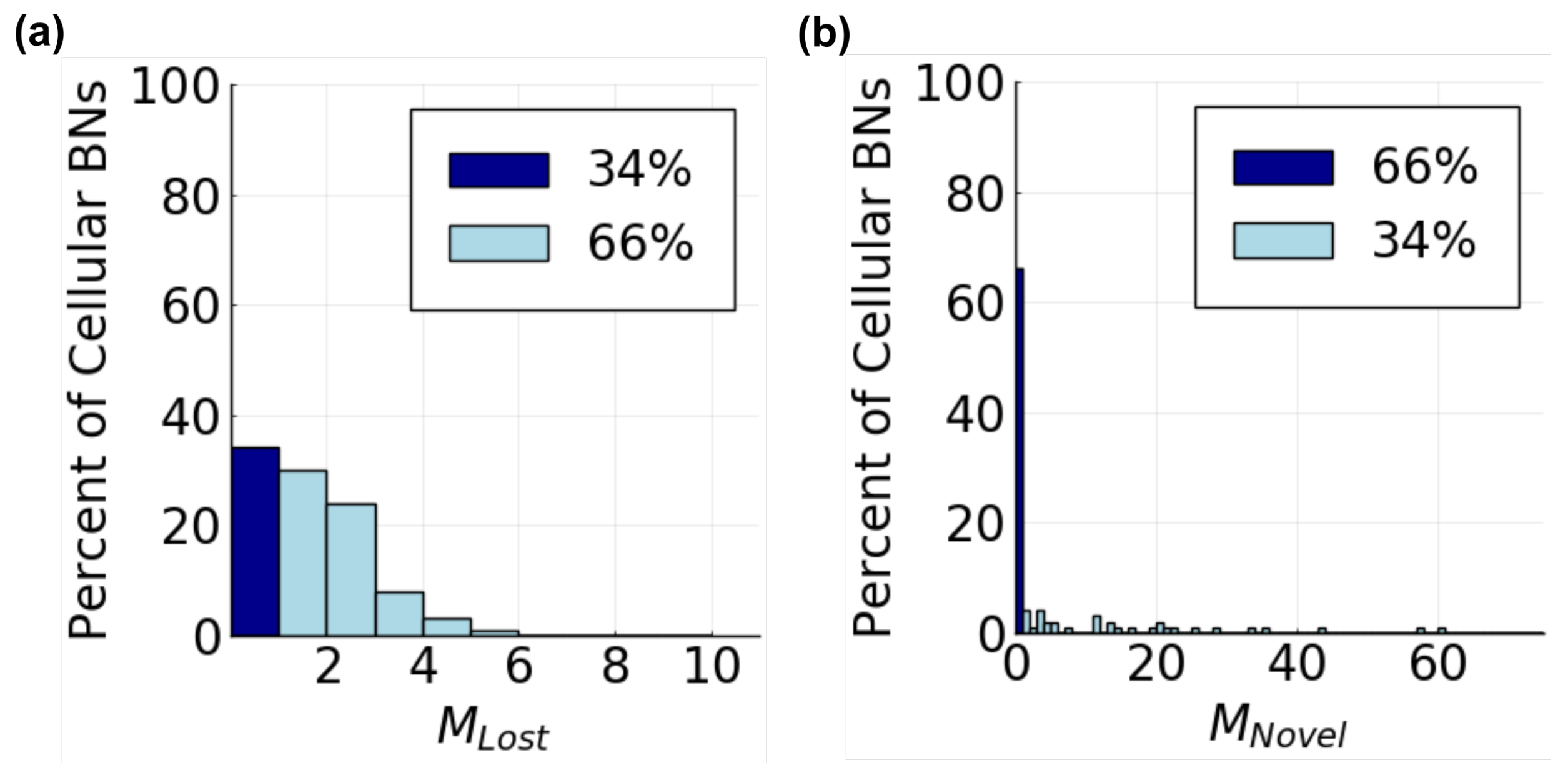
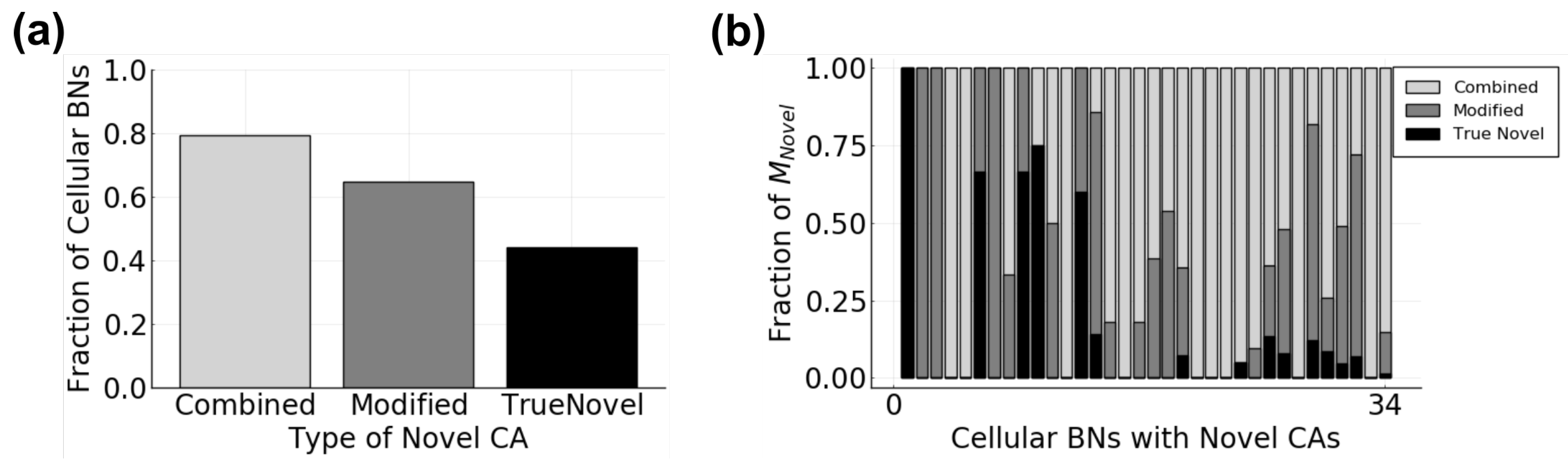
3.2.1. Cellular Behavior
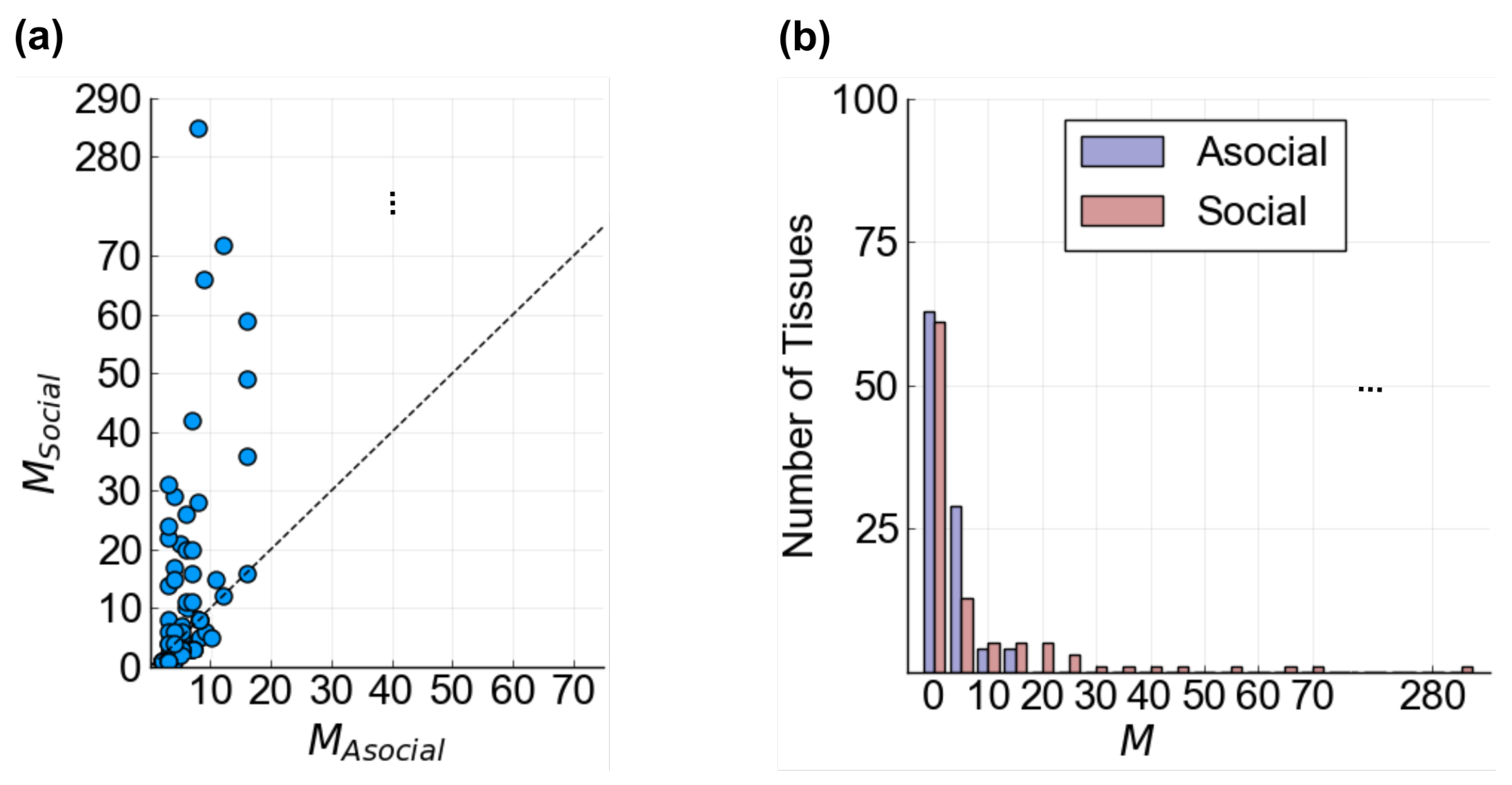
3.2.2. Tissue Behavior
- Does the composition of a tissue change as communication is added?
- Does the diversity of a tissue depend on communication?
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Analysis of Social Boundary Curves
References
- Prindle, A.; Liu, J.; Asally, M.; Ly, S.; Garcia-Ojalvo, J.; Süel, G.M. Ion Channels Enable Electrical Communication in Bacterial Communities. Nature 2015, 527, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Larkin, J.W.; Zhai, X.; Kikuchi, K.; Redford, S.E.; Prindle, A.; liu, J.; Greenfield, S.; Walczak, A.M.; Garcia-Ojalvo, J.; Mugler, A.; et al. Signal Percolation within a Bacterial Community. Cell Syst. 2018, 7, 137–145.e3. [Google Scholar] [CrossRef]
- Rutherford, S.T.; Bassler, B.L. Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef]
- Williams, P.; Camara, M.; Hardman, A.; Swift, S.; Milton, D.; Hope, V.J.; Winzer, K.; Middleton, B.; Pritchard, D.I.; Bycroft, B.W. Quorum Sensing and the Population-dependent Control of Virulence. Philos. Trans. R. Soc. Lond. Ser. Biol. Sci. 2000, 355, 667–680. [Google Scholar] [CrossRef]
- Long, Z.; Quaife, B.; Salman, H.; Oltvai, Z.N. Cell-cell Communication Enhances Bacterial Chemotaxis Toward External Attractants. Sci. Rep. 2017, 7, 12855. [Google Scholar] [CrossRef]
- Bischofs, I.B.; Hug, J.A.; Liu, A.W.; Wolf, D.M.; Arkin, A.P. Complexity in Bacterial Cell–cell Communication: Quorum Signal Integration and Subpopulation Signaling in the Bacillus subtilis Phosphorelay. Proc. Natl. Acad. Sci. USA 2009, 106, 6459–6464. [Google Scholar] [CrossRef] [PubMed]
- López, D.; Vlamakis, H.; Losick, R.; Kolter, R. Paracrine Signaling in a Bacterium. Genes Dev. 2009, 23, 1631–1638. [Google Scholar] [CrossRef]
- van Gestel, J.; Vlamakis, H.; Kolter, R. From Cell Differentiation to Cell Collectives: Bacillus subtilis Uses Division of Labor to Migrate. PLoS Biol. 2015, 13, e1002141. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.S.; Golden, J.W. Heterocyst Pattern Formation Controlled by a Diffusible Peptide. Science 1998, 282, 935–938. [Google Scholar] [CrossRef]
- Callahan, S.M.; Buikema, W.J. The Role of HetN in Maintenance of the Heterocyst Pattern in Anabaena sp. PCC 7120. Mol. Microbiol. 2001, 40, 941–950. [Google Scholar] [CrossRef]
- Vlamakis, H.; Aguilar, C.; Losick, R.; Kolter, R. Control of Cell Fate by the Formation of an Architecturally Complex Bacterial Community. Genes Dev. 2008, 22, 945–953. [Google Scholar] [CrossRef]
- Ben-Jacob, E.; Cohen, I.; Shochet, O.; Tenenbaum, A.; Czirók, A.; Vicsek, T. Cooperative Formation of Chiral Patterns during Growth of Bacterial Colonies. Phys. Rev. Lett. 1995, 75, 2899–2902. [Google Scholar] [CrossRef] [PubMed]
- Gestel, J.V.; Vlamakis, H.; Kolter, R. Division of Labor in Biofilms: The Ecology of Cell Differentiation. In Microbial Biofilms, 2nd ed.; Ghannoum, M., Parsek, M., Whiteley, M., Mukherjee, P.K., Eds.; ASM Press: Washinigton, DC, USA, 2015; pp. 67–97. [Google Scholar] [CrossRef]
- Niklas, K.J.; Newman, S.A. The Origins of Multicellular Organisms. Evol. Dev. 2013, 15, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Rokas, A. The Origins of Multicellularity and the Early History of the Genetic Toolkit For Animal Development. Annu. Rev. Genet. 2008, 42, 235–251. [Google Scholar] [CrossRef]
- Tameshige, T.; Hirakawa, Y.; Torii, K.U.; Uchida, N. Cell Walls as a Stage for Intercellular Communication Regulating Shoot Meristem Development. Front. Plant Sci. 2015, 6, 324. [Google Scholar] [CrossRef]
- Schumacher, J.A.; Hsieh, Y.W.; Chen, S.; Pirri, J.K.; Alkema, M.J.; Li, W.H.; Chang, C.; Chuang, C.F. Intercellular Calcium Signaling in a Gap Junction-coupled Cell Network Establishes Assymmetric Neuronal Fates in C. elegans. Development 2012, 139, 4191–4201. [Google Scholar] [CrossRef]
- Haas, P.; Gilmour, D. Chemokine Signaling Mediates Self-Organizing Tissue Migration in the Zebrafish Lateral Line. Dev. Cell 2006, 10, 673–680. [Google Scholar] [CrossRef]
- Ellison, D.; Mugler, A.; Brennan, M.D.; Lee, S.H.; Huebner, R.J.; Shamir, E.R.; Woo, L.A.; Kim, J.; Amar, P.; Nemenman, I.; et al. Cell–cell Communication Enhances the Capacity of Cell Ensembles to Sense Shallow Gradients During Morphogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, E679–E688. [Google Scholar] [CrossRef] [PubMed]
- Montero, J.A.; Nan, Y.G.; Macias, D.; Rodriguez-Leon, J.; Sanz-Ezquerro, J.J.; Merino, R.; Chimal-Monroy, J.; Nieto, M.A.; Hurle, J.M. Role of FGFs in the Control of Programmed Cell Death During Limb Development. Development 2001, 128, 2075–2084. [Google Scholar] [CrossRef]
- Lohmann, I.; McGinnis, N.; Bodmer, M.; McGinnis, W. The Drosophila Hox Gene Deformed Sculpts Head Morphology via Direct Regulation of the Apoptosis Activator reaper. Cell 2002, 110, 457–466. [Google Scholar] [CrossRef]
- Hart, Y.; Reich-Zeliger, S.; Antebi, Y.E.; Zaretsky, I.; Mayo, A.E.; Alon, U.; Friedman, N. Paradoxical Signaling by a Secreted Molecule Leads to Homeostasis of Cell Levels. Cell 2014, 158, 1022–1032. [Google Scholar] [CrossRef]
- Zhou, X.; Franklin, R.A.; Adler, M.; Jacox, J.B.; Bailis, W.; Shyer, J.A.; Flavell, R.A.; Mayo, A.; Alon, U.; Medzhitov, R. Circuit Design Features of a Stable Two-Cell System. Cell 2018, 172, 744–757.e17. [Google Scholar] [CrossRef]
- Wilson, S.E.; Netto, M.; Ambrósio, R. Corneal Cells: Chatty in Development, Homeostasis, Wound Healing, and Disease. Am. J. Ophthalmol. 2003, 136, 530–536. [Google Scholar] [CrossRef]
- ichi Nakahama, K. Cellular Communications in Bone Homeostasis and Repair. Cell. Mol. Life Sci. 2010, 67, 4001–4009. [Google Scholar] [CrossRef] [PubMed]
- Gurtner, G.C.; Werner, S.; Barrandon, Y.; Longaker, M.T. Wound Repair and Regeneration. Nature 2008, 453, 314–321. [Google Scholar] [CrossRef]
- Kobayashi, M.; Shigenaka, Y. The Mode of Action of Acetylcholine and 5-hydroxytryptamine at the Neuromuscular Junctions in a Molluscan Muscle (Radular Protractor). Comp. Biochem. Physiol. Part C Comp. Pharmacol. 1978, 60, 115–122. [Google Scholar] [CrossRef]
- Zimmerman, L.M.; Vogel, L.A.; Bowden, R.M. Understanding the Vertebrate Immune System: Insights from the Reptilian Perspective. J. Exp. Biol. 2010, 213, 661–671. [Google Scholar] [CrossRef]
- Shklarsh, A.; Ariel, G.; Schneidman, E.; Ben-Jacob, E. Smart Swarms of Bacteria-Inspired Agents with Performance Adaptable Interactions. PLoS Comput. Biol. 2011, 7, e1002177. [Google Scholar] [CrossRef]
- Payne, S.; Li, B.; Cao, Y.; Schaeffer, D.; Ryser, M.D.; You, L. Temporal Control of Self-organized Pattern Formation Without Morphogen Gradients in Bacteria. Mol. Syst. Biol. 2013, 9, 697. [Google Scholar] [CrossRef]
- Toda, S.; Blauch, L.R.; Tang, S.K.Y.; Morsut, L.; Lim, W.A. Programming Self-organizing Multicellular Structures with Synthetic Cell-cell Signaling. Science 2018, 361, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Tamsir, A.; Tabor, J.J.; Voigt, C.A. Robust Multicellular Computing Using Genetically Encoded NOR Gates and Chemical ‘Wires’. Nature 2010, 469, 212–215. [Google Scholar] [CrossRef]
- Zuroff, T.R.; Curtis, W.R. Developing Symbiotic Consortia for Lignocellulosic Biofuel Production. Appl. Microbiol. Biotechnol. 2012, 93, 1423–1435. [Google Scholar] [CrossRef]
- Hennig, S.; Rödel, G.; Ostermann, K. Artificial Cell-cell Communication as an Emerging Tool in Synthetic Biology Applications. J. Biol. Eng. 2015, 9, 1–2. [Google Scholar] [CrossRef]
- Eldar, A.; Elowitz, M.B. Functional Roles for Noise in Genetic Circuits. Nature 2010, 467, 167–173. [Google Scholar] [CrossRef]
- Smet, I.D.; Beeckman, T. Asymmetric Cell Division in Land Plants and Algae: The Driving Force for Differentiation. Nat. Rev. Mol. Cell Biol. 2011, 12, 177–188. [Google Scholar] [CrossRef]
- Naldi, A.; Carneiro, J.; Chaouiya, C.; Thieffry, D. Diversity and Plasticity of Th Cell Types Predicted from Regulatory Network Modelling. PLoS Comput. Biol. 2010, 6, e1000912. [Google Scholar] [CrossRef]
- Singh, A.M.; Chappell, J.; Trost, R.; Lin, L.; Wang, T.; Tang, J.; Matlock, B.K.; Weller, K.P.; Wu, H.; Zhao, S.; et al. Cell-Cycle Control of Developmentally Regulated Transcription Factors Accounts for Heterogeneity in Human Pluripotent Cells. Stem Cell Rep. 2014, 2, 398. [Google Scholar] [CrossRef]
- Revell, C.; Blumenfeld, R.; Chalut, K.J. Force-based Three-Dimensional Model Predicts Mechanical Drivers of Cell Sorting. Proc. R. Soc. B Biol. Sci. 2019, 286, 20182495. [Google Scholar] [CrossRef]
- Rudge, T.J.; Federici, F.; Steiner, P.J.; Kan, A.; Haseloff, J. Cell Polarity-Driven Instability Generates Self-Organized, Fractal Patterning of Cell Layers. ACS Synth. Biol. 2013, 2, 705–714. [Google Scholar] [CrossRef]
- Boyer, M.; Wisniewski-Dyé, F. Cell-cell Signalling in Bacteria: Not Simply a Matter of Quorum. FEMS Microbiol. Ecol. 2009, 70, 1–19. [Google Scholar] [CrossRef]
- Mitri, S.; Clarke, E.; Foster, K.R. Resource Limitation Drives Spatial Organization in Microbial Groups. ISME J. 2015, 10, 1471–1482. [Google Scholar] [CrossRef]
- Youk, H.; Lim, W.A. Secreting and Sensing the Same Molecule Allows Cells to Achieve Versatile Social Behaviors. Science 2014, 343, 1242782. [Google Scholar] [CrossRef]
- Maire, T.; Youk, H. Molecular-Level Tuning of Cellular Autonomy Controls the Collective Behaviors of Cell Populations. Cell Syst. 2015, 1, 349–360. [Google Scholar] [CrossRef]
- Li, F.; Long, T.; Lu, Y.; Ouyang, Q.; Tang, C. The Yeast Cell-cycle Network is Robustly Designed. Proc. Natl. Acad. Sci. USA 2004, 101, 4781–4786. [Google Scholar] [CrossRef]
- Albert, R.; Othmer, H.G. The Topology of the Regulatory Interactions Predicts the Expression Pattern of the Segment Polarity Genes in Drosophila melanogaster. J. Theor. Biol. 2003, 223, 1–18. [Google Scholar] [CrossRef]
- Davidich, M.I.; Bornholdt, S. Boolean Network Model Predicts Cell Cycle Sequence of Fission Yeast. PLoS ONE 2008, 3, e1672. [Google Scholar] [CrossRef]
- Kauffman, S. Homeostasis and Differentiation in Random Genetic Control Networks. Nature 1969, 224, 177–178. [Google Scholar] [CrossRef] [PubMed]
- Berg, H.C. Random Walks in Biology, New and Expanded ed.; Princeton University Press: Princeton, NJ, USA, 1993. [Google Scholar]
- Kang, S.; Kahan, S.; McDermott, J.; Flann, N.; Shmulevich, I. Biocellion: Accelerating Computer Simulation of Multicellular Biological System Models. Bioinformatics 2014, 30, 3101–3108. [Google Scholar] [CrossRef] [PubMed]
- Daniels, B.C.; Kim, H.; Moore, D.; Zhou, S.; Smith, H.B.; Karas, B.; Kauffman, S.A.; Walker, S.I. Criticality Distinguishes the Ensemble of Biological Regulatory Networks. Phys. Rev. Lett. 2018, 121, 138102. [Google Scholar] [CrossRef] [PubMed]
- Balleza, E.; Alvarez-Buylla, E.R.; Chaos, A.; Kauffman, S.; Shmulevich, I.; Aldana, M. Critical Dynamics in Genetic Regulatory Networks: Examples from Four Kingdoms. PLoS ONE 2008, 3, e2456. [Google Scholar] [CrossRef]
- Heller, E.; Fuchs, E. Tissue Patterning and Cellular Mechanics. J. Cell Biol. 2015, 211, 219–231. [Google Scholar] [CrossRef]
- Martinez-Sanchez, M.E.; Mendoza, L.; Villarreal, C.; Alvarez-Buylla, E.R. A Minimal Regulatory Network of Extrinsic and Intrinsic Factors Recovers Observed Patterns of CD4+ T Cell Differentiation and Plasticity. PLoS Comput. Biol. 2015, 11, e1004324. [Google Scholar] [CrossRef]
- Zhou, J.X.; Samal, A.; d’Hérouël, A.F.; Price, N.D.; Huang, S. Relative Stability of Network States in Boolean Network Models of Gene Regulation in Development. Biosystems 2016, 142–143, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, N.; Mendoza, L.; Álvarez-Buylla, E.R. A Computational Model of the Endothelial to Mesenchymal Transition. Front. Genet. 2020, 11, 40. [Google Scholar] [CrossRef] [PubMed]
- Newman, S.A. Cell Differentiation: What Have We Learned in 50 Years? J. Theor. Biol. 2020, 485, 110031. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Errede, B.; Behar, M.; Mather, W.; Nayak, S.; Hasty, J.; Dohlman, H.G.; Elston, T.C. Yeast Dynamically Modify Their Environment to Achieve Better Mating Efficiency. Sci. Signal. 2011, 4, ra54. [Google Scholar] [CrossRef]
- Wartlick, O.; Mumcu, P.; Kicheva, A.; Bittig, T.; Seum, C.; Jülicher, F.; González-Gaitán, M. Dynamics of Dpp Signaling and Proliferation Control. Science 2011, 331, 1154–1159. [Google Scholar] [CrossRef]
- Joncker, N.T.; Fernandez, N.C.; Treiner, E.; Vivier, E.; Raulet, D.H. NK Cell Responsiveness Is Tuned Commensurate with the Number of Inhibitory Receptors for Self-MHC Class I: The Rheostat Model. J. Immunol. 2009, 182, 4572–4580. [Google Scholar] [CrossRef] [PubMed]
- Damiani, C.; Kauffman, S.; Villani, M.; Colacci, A.; Serra, R. Cell-cell Interaction and Diversity of Emergent Behaviours. IET Syst. Biol. 2011, 5, 137–144. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Echlin, M.; Aguilar, B.; Shmulevich, I. Characterizing the Impact of Communication on Cellular and Collective Behavior Using a Three-Dimensional Multiscale Cellular Model. Entropy 2023, 25, 319. https://doi.org/10.3390/e25020319
Echlin M, Aguilar B, Shmulevich I. Characterizing the Impact of Communication on Cellular and Collective Behavior Using a Three-Dimensional Multiscale Cellular Model. Entropy. 2023; 25(2):319. https://doi.org/10.3390/e25020319
Chicago/Turabian StyleEchlin, Moriah, Boris Aguilar, and Ilya Shmulevich. 2023. "Characterizing the Impact of Communication on Cellular and Collective Behavior Using a Three-Dimensional Multiscale Cellular Model" Entropy 25, no. 2: 319. https://doi.org/10.3390/e25020319
APA StyleEchlin, M., Aguilar, B., & Shmulevich, I. (2023). Characterizing the Impact of Communication on Cellular and Collective Behavior Using a Three-Dimensional Multiscale Cellular Model. Entropy, 25(2), 319. https://doi.org/10.3390/e25020319






