The Structure of Chaos: An Empirical Comparison of Fractal Physiology Complexity Indices Using NeuroKit2
Abstract
:1. Introduction
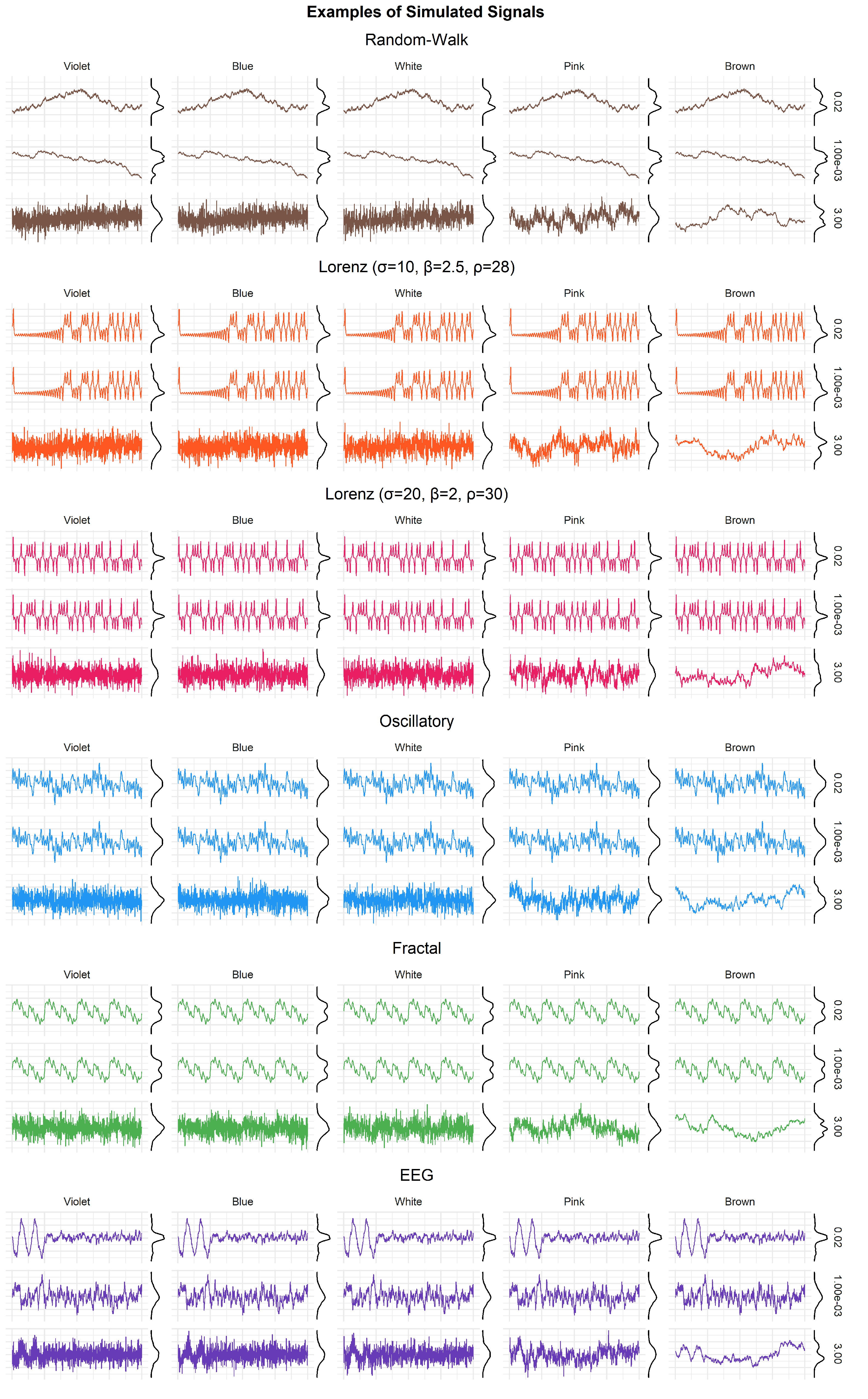
2. Methods
3. Results
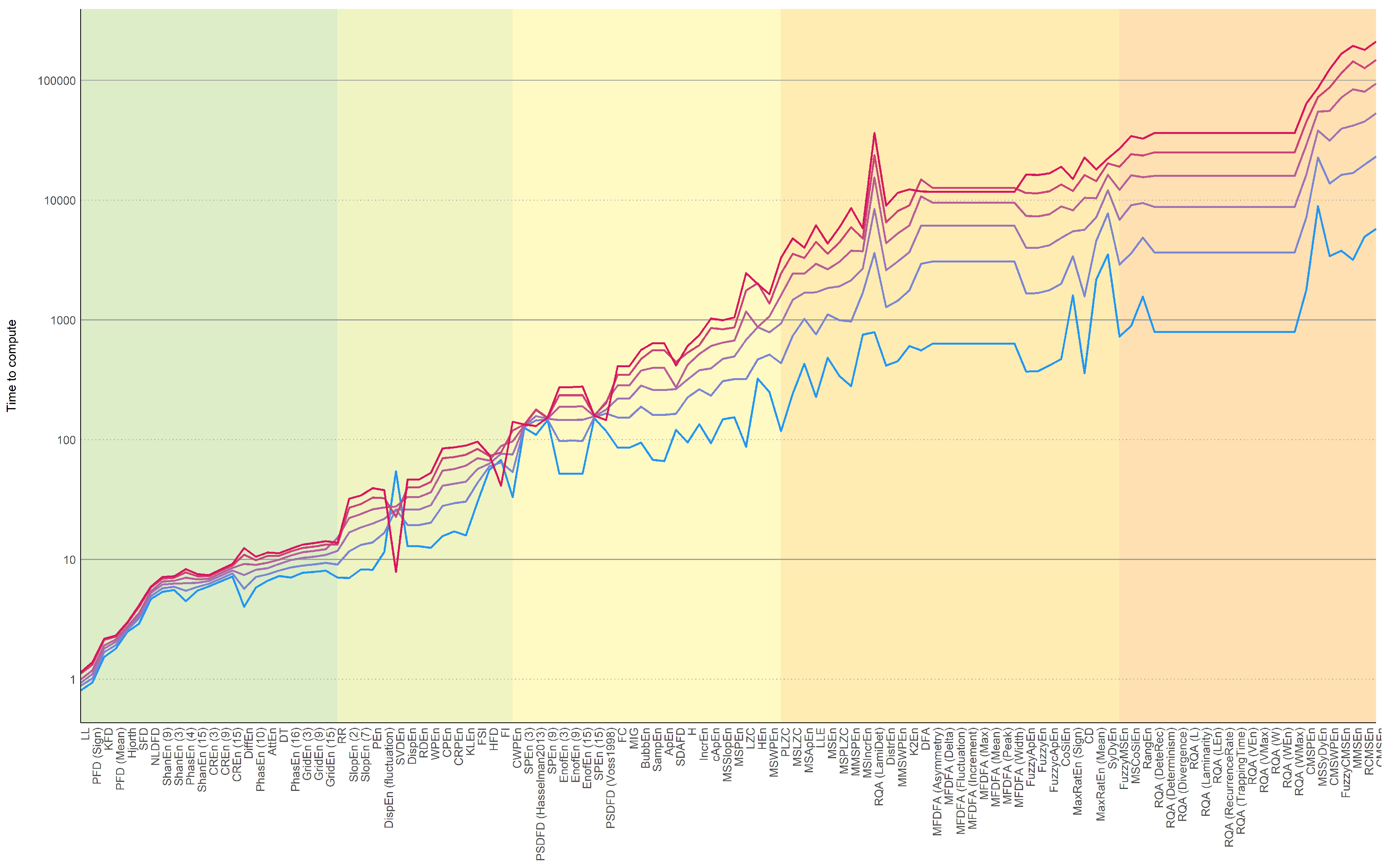
3.1. Computation Time
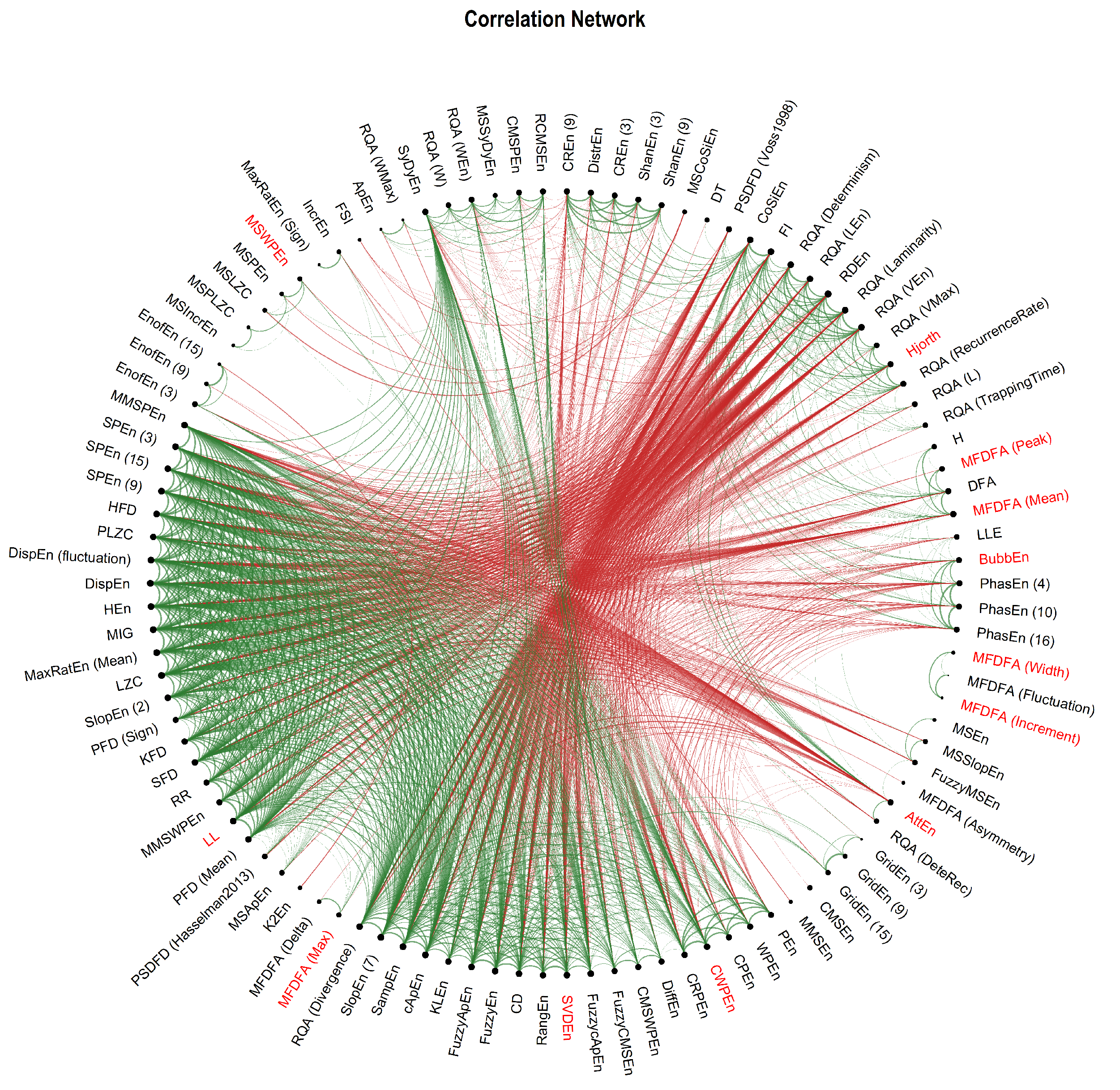
3.2. Correlation
3.3. Factor Analysis
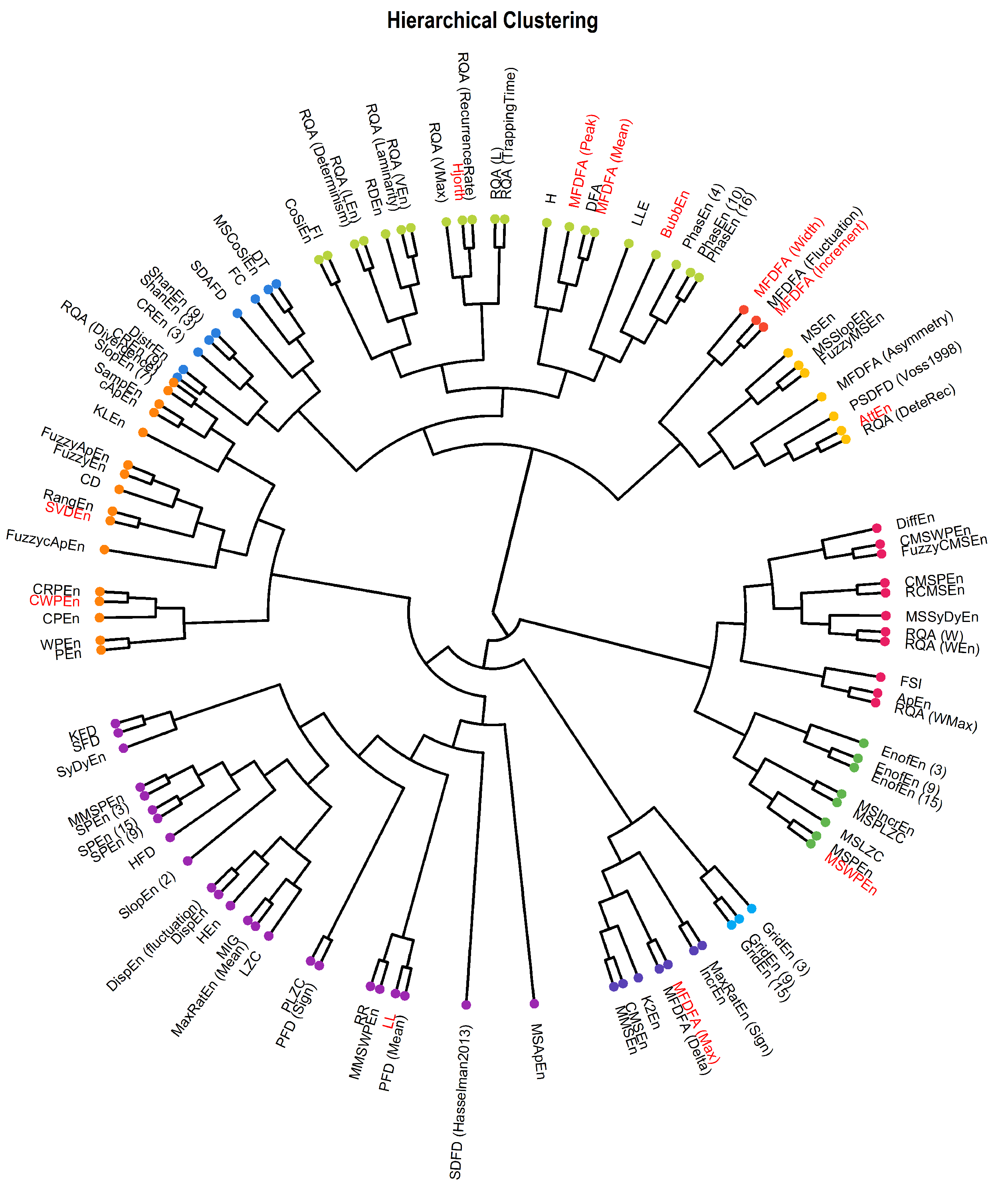
3.4. Hierarchical Clustering and Connectivity Network
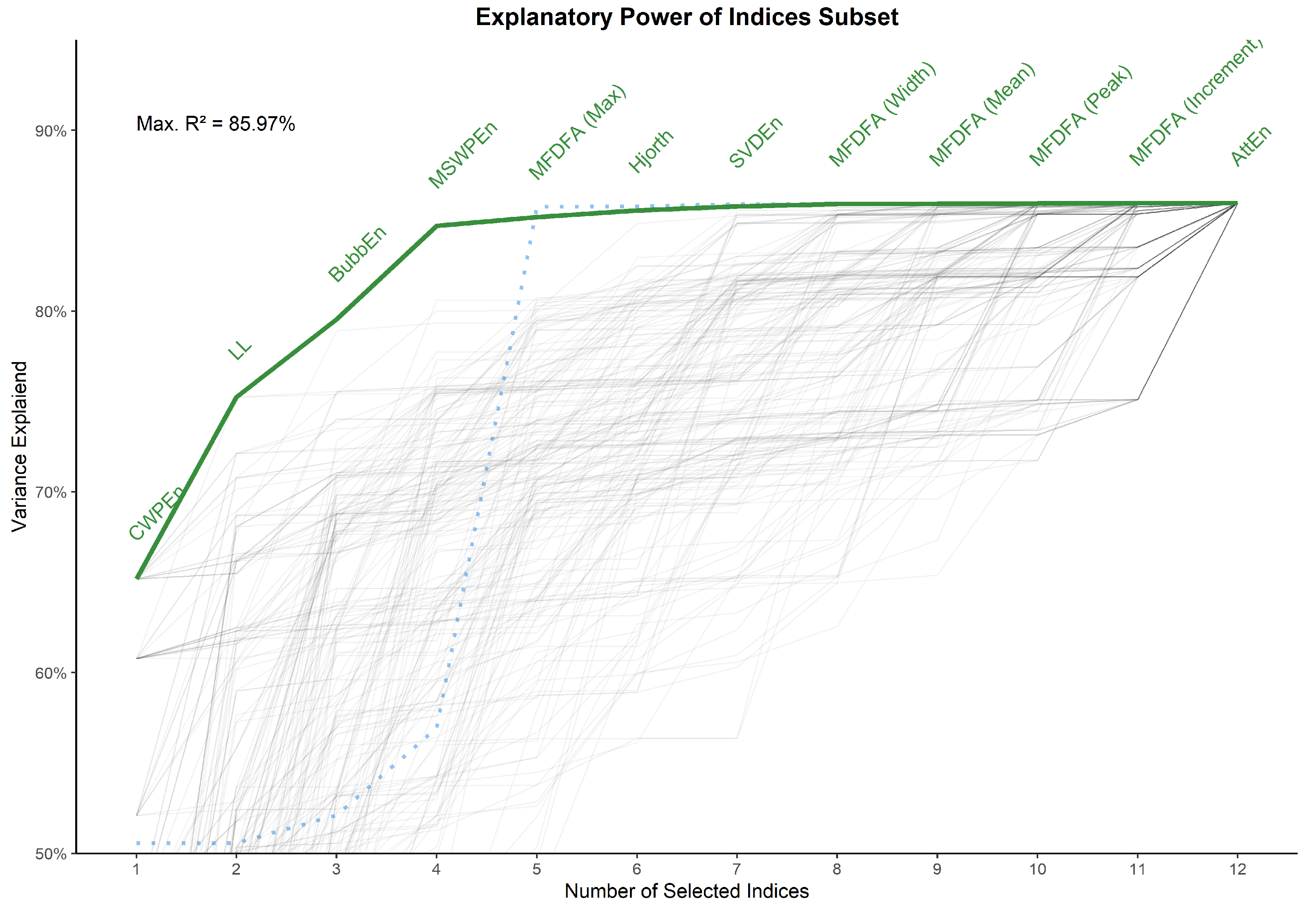
3.5. Indices Selection
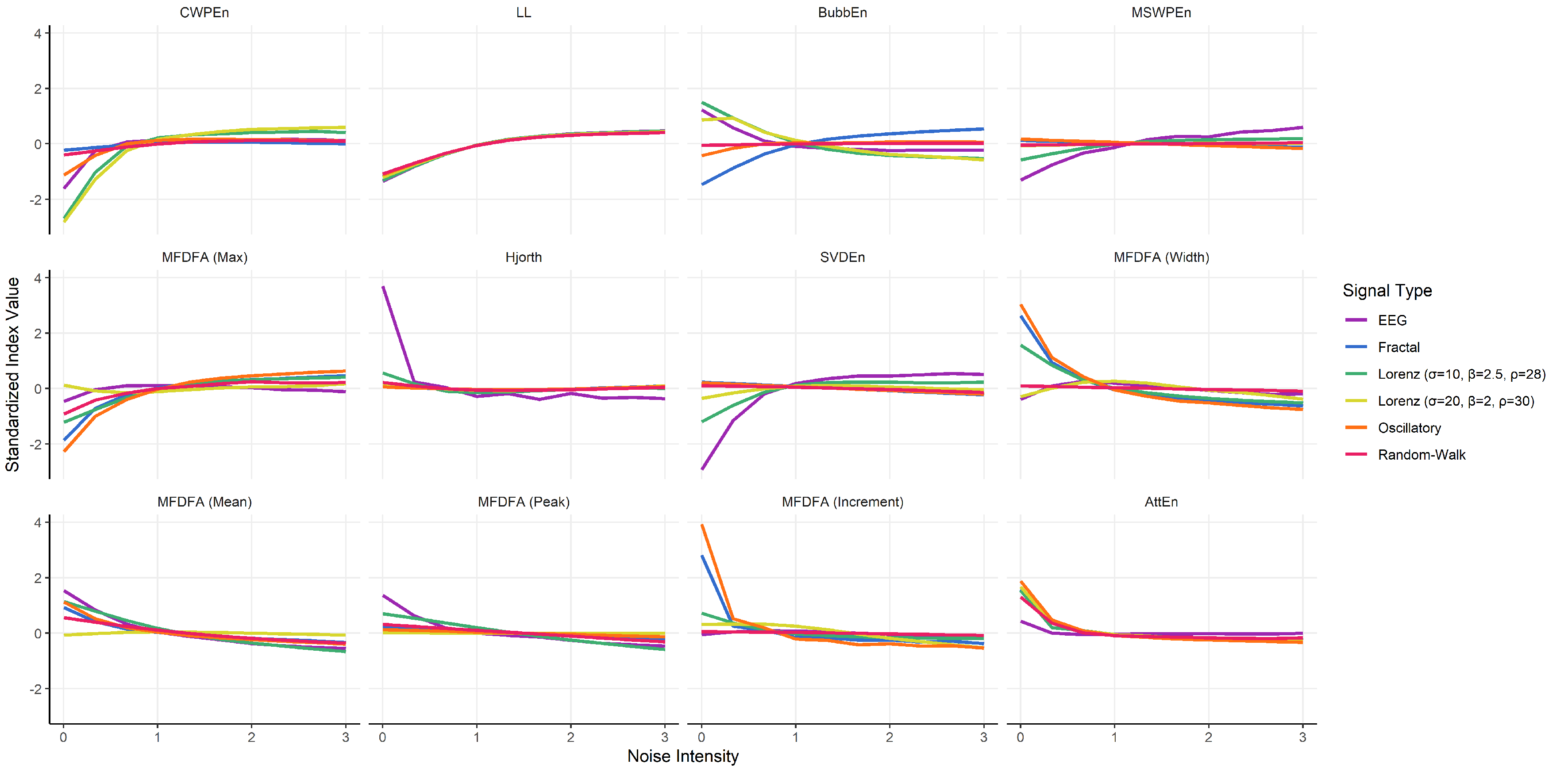
- CWPEn: The Conditional Weighted Permutation Entropy is based on the difference of weighted entropy between that obtained at an embedding dimension m and that obtained at [18].
- LL: The Line Length index stems out of a simplification of Katz’ fractal dimension (KFD) algorithm [19] and corresponds to the average of consecutive absolute differences. It is equivalent to NDLFD, the Fractal dimension via Normalized Length Density [20]. As it captures the amplitude 1-lag fluctuations, this index is likely sensitive to noise in the series.
- BubbEn: The Bubble Entropy is based on Permutation Entropy. It uses the Bubble sort algorithm and counts the number of swaps each vector undergoes in the embedding space instead of ranking their order [21].
- MSWPEn: The Multiscale Weighted Permutation Entropy is the entropy of weighted ordinal descriptors of the time-embedded signal computed at different scales obtained by a coarse-graining procedure [22].
- MFDFA (Max): The value of singularity spectrum D corresponding to the maximum value of singularity exponent H.
- Hjorth: Hjorth’s Complexity is defined as the ratio of the mean frequency of the first derivative of the signal to the mean frequency of the signal [23].
- SVDEn: The Singular Value Decomposition (SVD) Entropy quantifies the amount of eigenvectors needed for an adequate representation of the system [24].
- MFDFA (Width): The width of the multifractal singularity spectrum [25] obtained via Detrended Fluctuation Analysis (DFA).
- MFDFA (Mean): The mean of the maximum and minimum values of singularity exponent H, which quantifies the average fluctuations of the signal.
- MFDFA (Peak): The value of the singularity exponent H corresponding to peak of singularity dimension D. It is a measure of the self-affinity of the signal, and a high value is an indicator of high degree of correlation between the data points.
- MFDFA (Increment): The cumulative function of the squared increments of the generalized Hurst’s exponents between consecutive moment orders [26].
- AttEn: The Attention Entropy is based on the frequency distribution of the intervals between the local maxima and minima of the time series [27].
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bassingthwaighte, J.B.; Liebovitch, L.S.; West, B.J. Fractal Physiology; Springer: Berlin, Germany, 2013. [Google Scholar]
- Lau, Z.J.; Pham, T.; Annabel, S.; Makowski, D. Brain Entropy, Fractal Dimensions and Predictability: A Review of Complexity Measures for EEG in Healthy and Neuropsychiatric Populations. 2021. Available online: https://psyarxiv.com/f8k3x/ (accessed on 21 July 2022).
- Ehlers, C.L. Chaos and Complexity: Can It Help Us to Understand Mood and Behavior? Arch. Gen. Psychiatry 1995, 52, 960–964. [Google Scholar] [CrossRef]
- Goetz, S.J. Spatial Dynamics, Networks and Modelling? Edited by Aura Reggiani and Peter Nijkamp. Pap. Reg. Sci. 2007, 86, 523–524. [Google Scholar] [CrossRef]
- Yang, A.C.; Tsai, S.-J. Is Mental Illness Complex? From Behavior to Brain. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2013, 45, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Azami, H.; Rostaghi, M.; Abásolo, D.; Escudero, J. Refined Composite Multiscale Dispersion Entropy and Its Application to Biomedical Signals. IEEE Trans. Biomed. Eng. 2017, 64, 2872–2879. [Google Scholar] [PubMed]
- Manis, G.; Aktaruzzaman, M.; Sassi, R. Low Computational Cost for Sample Entropy. Entropy 2018, 20, 61. [Google Scholar] [CrossRef]
- Flood, M.W.; Grimm, B. EntropyHub: An Open-Source Toolkit for Entropic Time Series Analysis. PLoS ONE 2021, 16, e0259448. [Google Scholar] [CrossRef] [PubMed]
- Fulcher, B.D.; Jones, N.S. Hctsa: A Computational Framework for Automated Time-Series Phenotyping Using Massive Feature Extraction. Cell Syst. 2017, 5, 527–531. [Google Scholar] [CrossRef] [PubMed]
- Makowski, D.; Pham, T.; Lau, Z.J.; Brammer, J.C.; Lespinasse, F.; Pham, H.; Schölzel, C.; Chen, S.H.A. NeuroKit2: A Python Toolbox for Neurophysiological Signal Processing. Behav. Res. Methods 2021, 53, 1689–1696. [Google Scholar] [CrossRef]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array Programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- McKinney, W. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; Volume 445, pp. 51–56. [Google Scholar]
- Girault, J.-M.; Menigot, S. Palindromic Vectors, Symmetropy and Symmentropy as Symmetry Descriptors of Binary Data. Entrpy 2022, 24, 82. [Google Scholar] [CrossRef]
- Makowski, D.; Ben-Shachar, M.; Patil, I.; Lüdecke, D. Methods and Algorithms for Correlation Analysis in R. J. Open Source Softw. 2020, 5, 2306. [Google Scholar] [CrossRef]
- Lüdecke, D.; Patil, I.; Ben-Shachar, M.S.; Wiernik, B.M.; Waggoner, P.; Makowski, D. see: An R Package for Visualizing Statistical Models. J. Open Source Softw. 2021, 6, 3393. [Google Scholar] [CrossRef]
- Lüdecke, D.; Ben-Shachar, M.; Patil, I.; Makowski, D. Extracting, Computing and Exploring the Parameters of Statistical Models Using R. J. Open Source Softw. 2020, 5, 2445. [Google Scholar] [CrossRef]
- Makowski, D.; Lüdecke, D.; Ben-Shachar, M.S.; Patil, I. modelbased: Estimation of Model-Based Predictions, Contrasts and Means. 2022. Available online: https://CRAN.R-project.org/package=modelbased (accessed on 21 July 2022).
- Unakafov, A.M.; Keller, K. Conditional Entropy of Ordinal Patterns. Phys. D Nonlinear Phenom. 2014, 269, 94–102. [Google Scholar] [CrossRef]
- Esteller, R.; Echauz, J.; Tcheng, T.; Litt, B.; Pless, B. Line Length: An Efficient Feature for Seizure Onset Detection. In Proceedings of the 2001 Conference 23rd Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Istanbul, Turkey, 25–28 October 2001; IEEE: Piscataway, NJ, USA, 2001; Volume 2, pp. 1707–1710. [Google Scholar]
- Kalauzi, A.; Bojić, T.; Rakić, L. Extracting Complexity Waveforms from One-Dimensional Signals. Nonlinear Biomed. Phys. 2009, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Manis, G.; Aktaruzzaman, M.; Sassi, R. Bubble Entropy: An Entropy Almost Free of Parameters. IEEE Trans. Biomed. Eng. 2017, 64, 2711–2718. [Google Scholar] [CrossRef]
- Fadlallah, B.; Chen, B.; Keil, A.; Príncipe, J. Weighted-Permutation Entropy: A Complexity Measure for Time Series Incorporating Amplitude Information. Phys. Rev. E 2013, 87, 022911. [Google Scholar] [CrossRef] [PubMed]
- Hjorth, B. EEG Analysis Based on Time Domain Properties. Electroencephalogr. Clin. Neurophysiol. 1970, 29, 306–310. [Google Scholar] [CrossRef]
- Roberts, S.J.; Penny, W.; Rezek, I. Temporal and Spatial Complexity Measures for Electroencephalogram Based Brain-Computer Interfacing. Med. Biol. Eng. Comput. 1999, 37, 93–98. [Google Scholar] [CrossRef]
- Kantelhardt, J.W.; Zschiegner, S.A.; Koscielny-Bunde, E.; Havlin, S.; Bunde, A.; Stanley, H.E. Multifractal Detrended Fluctuation Analysis of Nonstationary Time Series. Phys. A Stat. Mech. Appl. 2002, 316, 87–114. [Google Scholar] [CrossRef]
- Faini, A.; Parati, G.; Castiglioni, P. Multiscale Assessment of the Degree of Multifractality for Physiological Time Series. Philos. Trans. R. Soc. A 2021, 379, 20200254. [Google Scholar] [CrossRef]
- Yang, J.; Choudhary, G.I.; Rahardja, S.; Franti, P. Classification of Interbeat Interval Time-Series Using Attention Entropy. IEEE Trans. Affect. Comput. 2020. [Google Scholar] [CrossRef]
- Namdari, A.; Li, Z. A Review of Entropy Measures for Uncertainty Quantification of Stochastic Processes. Adv. Mech. Eng. 2019, 11, 168781401985735. [Google Scholar] [CrossRef]
- Xiong, W.; Faes, L.; Ivanov, P.C.H. Entropy Measures, Entropy Estimators, and Their Performance in Quantifying Complex Dynamics: Effects of Artifacts, Nonstationarity, and Long-Range Correlations. Phys. Rev. E 2017, 95, 062114. [Google Scholar] [CrossRef]
- Schölzel, C. Nonlinear Measures for Dynamical Systems; Zenodo: Geneva, Switzerland, 2019. [Google Scholar] [CrossRef]
- Vallat, R. AntroPy: Entropy and Complexity of (EEG) Time-Series in Python; GitHub: San Francisco, CA, USA, 2022; Available online: https://github.com/raphaelvallat/antropy (accessed on 21 July 2022).


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Makowski, D.; Te, A.S.; Pham, T.; Lau, Z.J.; Chen, S.H.A. The Structure of Chaos: An Empirical Comparison of Fractal Physiology Complexity Indices Using NeuroKit2. Entropy 2022, 24, 1036. https://doi.org/10.3390/e24081036
Makowski D, Te AS, Pham T, Lau ZJ, Chen SHA. The Structure of Chaos: An Empirical Comparison of Fractal Physiology Complexity Indices Using NeuroKit2. Entropy. 2022; 24(8):1036. https://doi.org/10.3390/e24081036
Chicago/Turabian StyleMakowski, Dominique, An Shu Te, Tam Pham, Zen Juen Lau, and S. H. Annabel Chen. 2022. "The Structure of Chaos: An Empirical Comparison of Fractal Physiology Complexity Indices Using NeuroKit2" Entropy 24, no. 8: 1036. https://doi.org/10.3390/e24081036
APA StyleMakowski, D., Te, A. S., Pham, T., Lau, Z. J., & Chen, S. H. A. (2022). The Structure of Chaos: An Empirical Comparison of Fractal Physiology Complexity Indices Using NeuroKit2. Entropy, 24(8), 1036. https://doi.org/10.3390/e24081036






