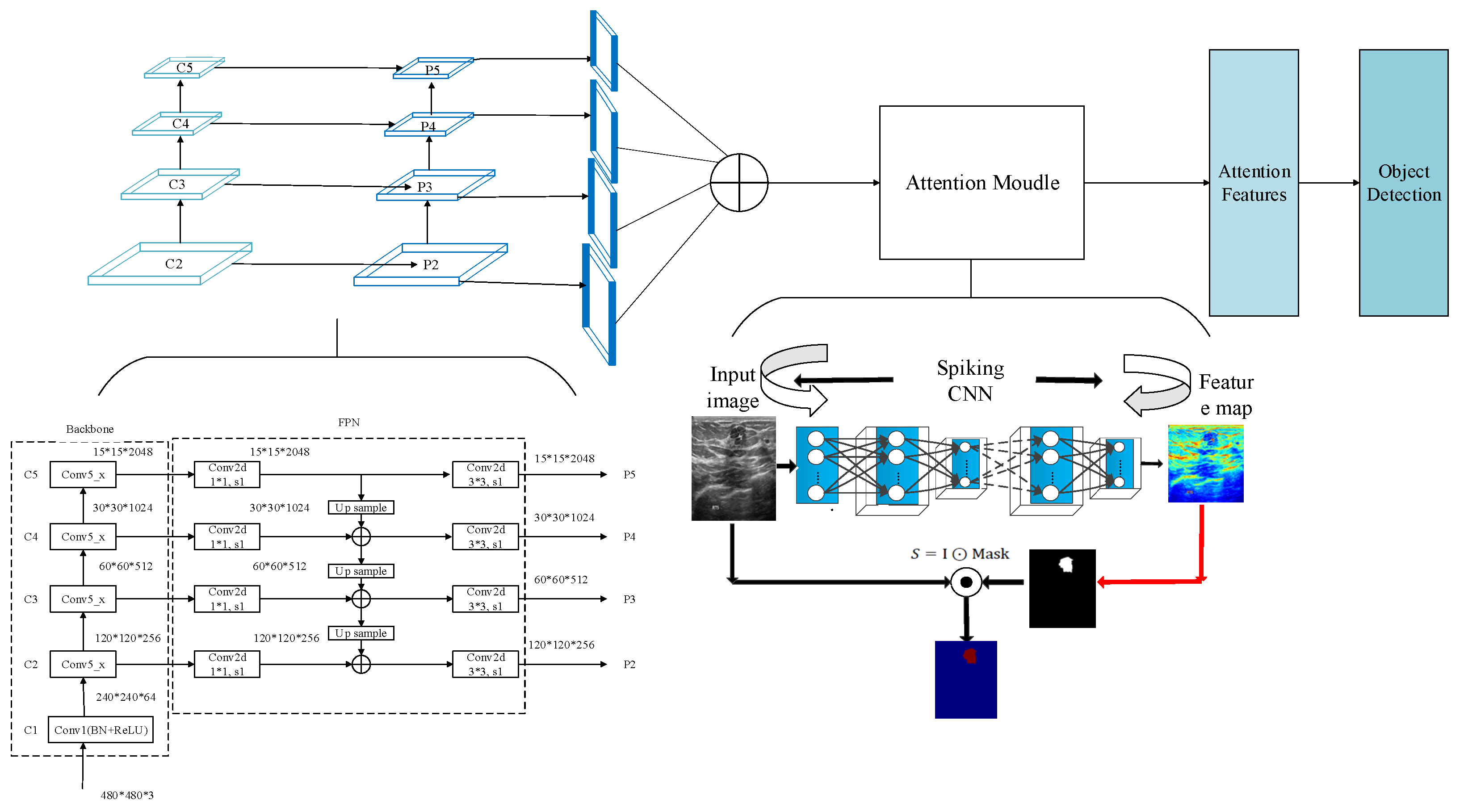
In this section, a multi-scale saliency fusion model and a transformation method from DNN to SNN are proposed. In the multi-scale saliency fusion model, as shown in
Figure 1, a feature pyramid network is used to obtain multi-scale features, and the attention module fuses the spatial and channel attention mechanisms.
3.1. Spiking Neural Networks
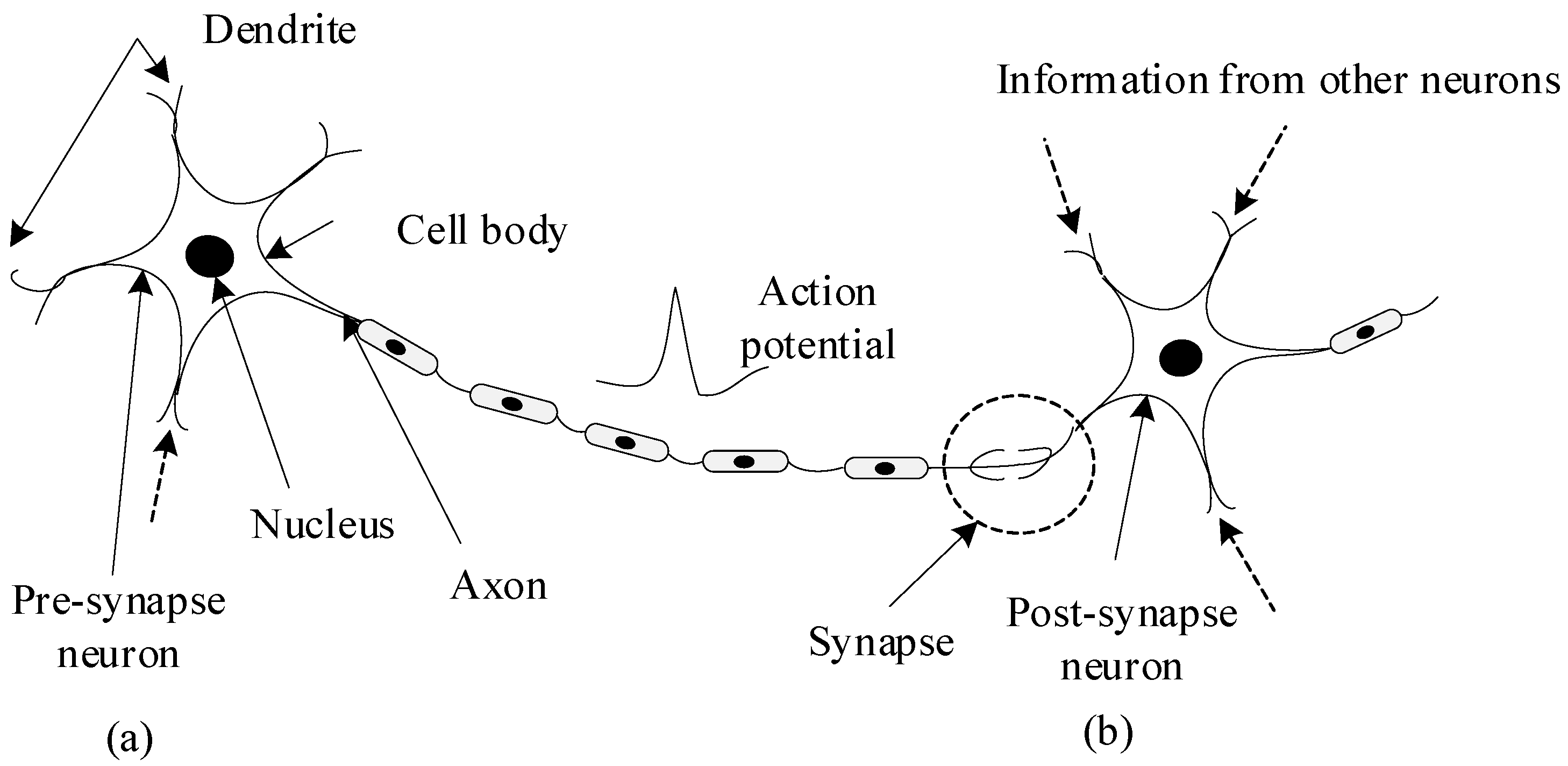
In this section, the introduction of SNN and the method of converting DNN to SNN are given. The SNNs are composed of spiking neurons interconnected by synapses. Spiking neurons simulate the information transmission mechanism of biological neurons, as shown in
Figure 2. This mimics the process that the ion channel on the cell membrane is opened by neurons receiving stimulation, and then the charged ions inside and outside the cell membrane flow to generate an action potential. When the action potential reaches a certain threshold, an action potential is generated. The action potential is transmitted along the axon to the nerve terminal. Finally, it is transmitted to the postsynaptic neuron through the synapse.
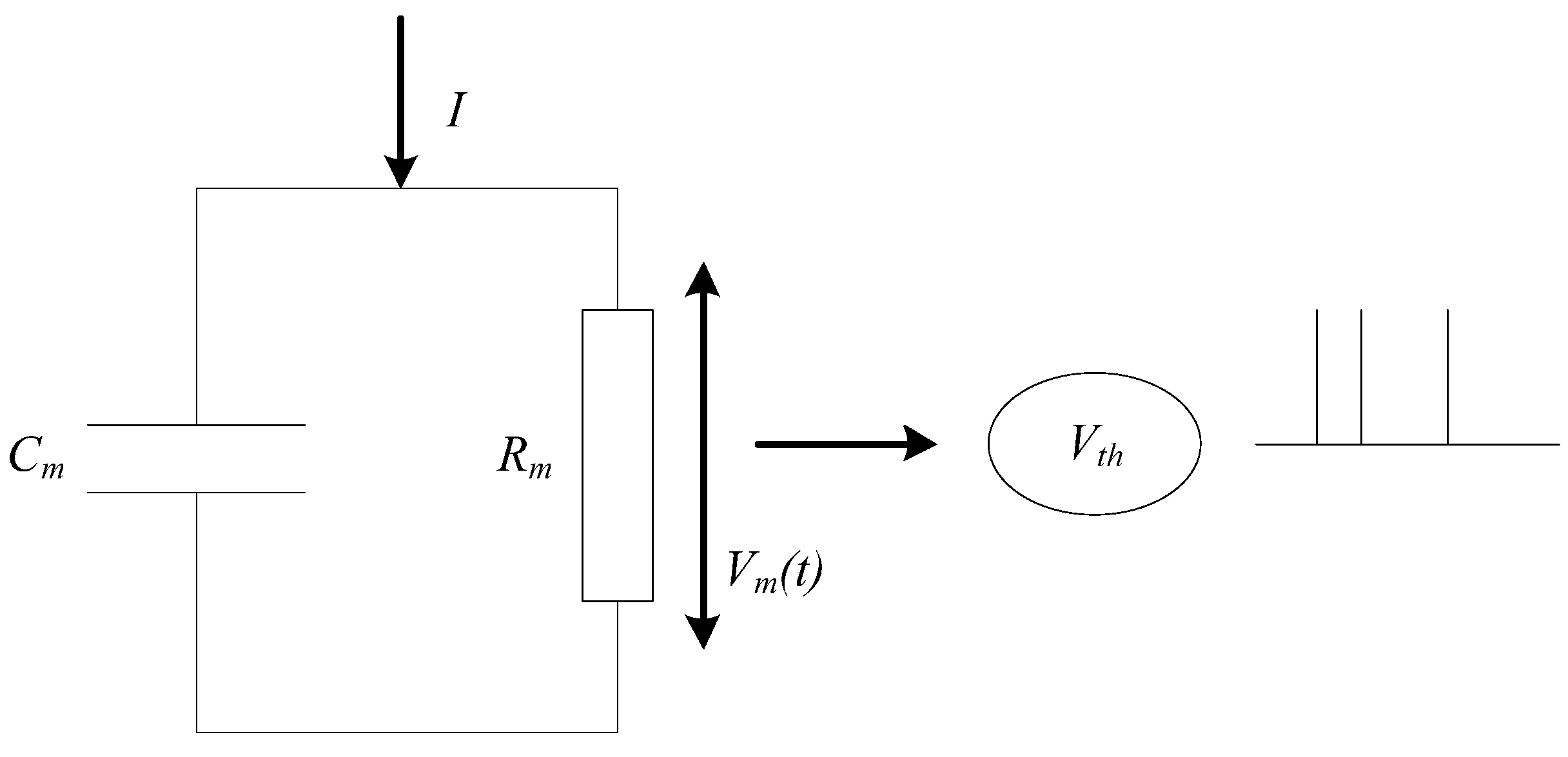
Considering the complexity of network scale and model, a simple leaky integrate-and-fire (LIF) [
25] neuron model is used for SNN in this paper. The basic circuit of the LIF model consists of a capacitor and a resistor in parallel. As shown in
Figure 3, the driving current can be divided into two parts. It can be calculated as follows:
where
is the membrane capacitance,
is the voltage of the membrane,
is the resistance of membrane, and
is the total current of membrane. Here,
is the time constant of leakage current, which is calculated as follows:
When the neuron receives a constant current stimulation and the cell membrane is at a resting potential of 0 mv, that is,
, the membrane potential can be calculated as follows:
where
is the firing time of the previous spike. If the value of
is less than the firing threshold
, no spike is generated; on the contrary, if the value of
reaches the threshold
, an output spike is generated at
. Therefore, the threshold of spike firing can be calculated as follows:
The internal spike time interval i.e.,
can be calculated as follows:
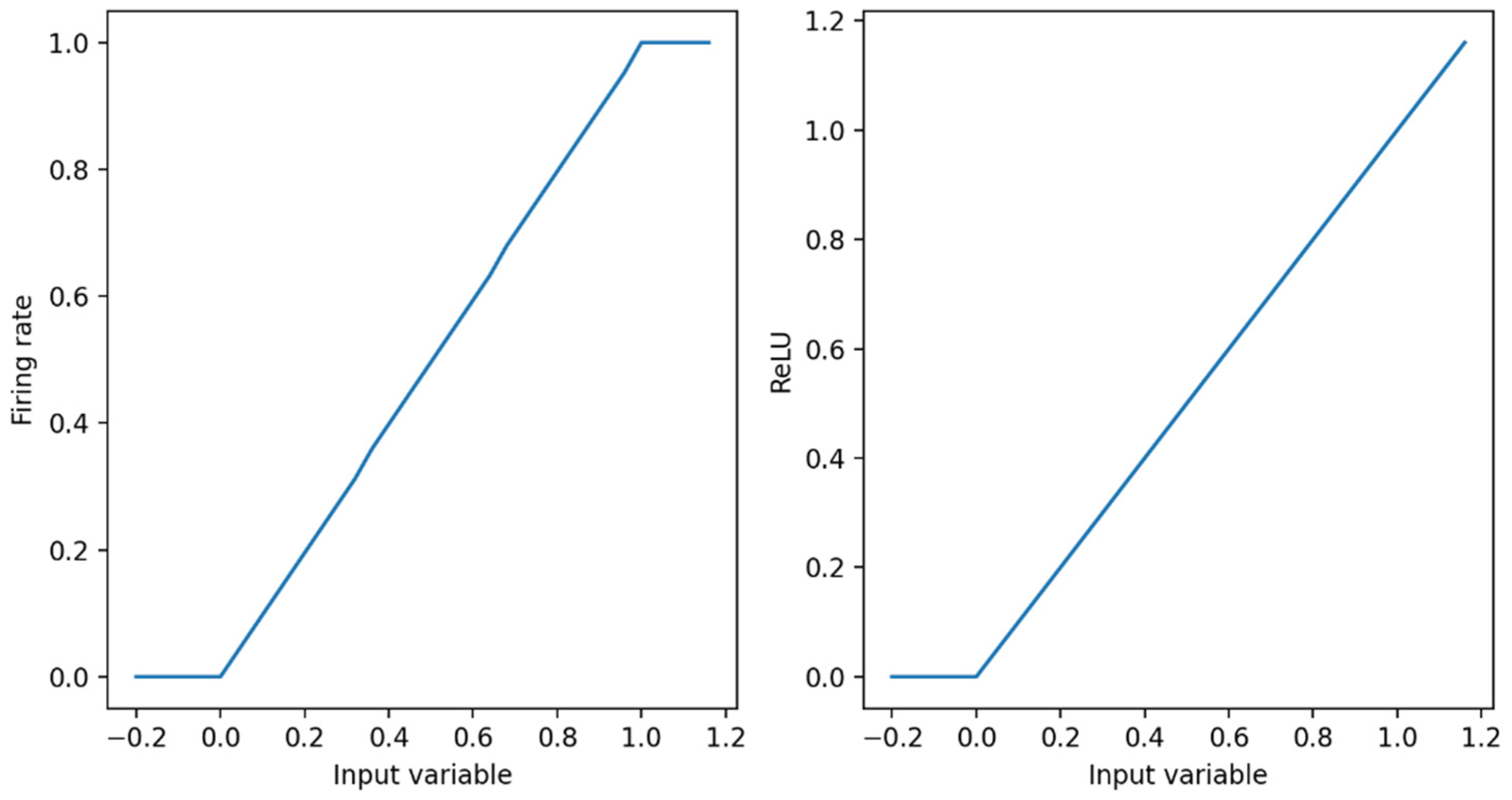
The ReLU activation function in DNN is very close to the curve of the spiking neuron model, as shown in
Figure 4. Therefore, the DNN can be converted into the SNN. We are able to prove this view theoretically. In this paper, the relationship between the firing frequency f of the first layer of the neural network and the activation in the corresponding ANN are discussed [
26].
Suppose the input is constant as
. The process of changes in membrane potential
V with time in SNN can be calculated as follows:
where
is the output spikes. The average firing rate in T time steps can be obtained by summation of membrane potential. It can be calculated as follows:
Then, move all items containing
to the left, and divide both sides by T at the same time, as follows:
Therefore, in the case of an infinite simulation time step, the following is true:
In the training process, DNN uses batch normalization to normalize the output value to a zero mean value to accelerate the training and convergence. It can be calculated as follows:
where
x is input value,
and
are mean and variance, respectively, and
and
are obtained in the training process.
After training, these transformations can be integrated into the weight vector to maintain the performance of batch normalization. However, there is no need to repeat the normalization calculation for each sample. Therefore, this work refers to the method proposed by [
27] to calculate the normalization. It can be calculated as follows:
This method does not need to transform the batch normalization layer after transforming the weight of the previous layer. Furthermore, when the batch normalization parameter is integrated into other weights, the loss is reduced.
3.2. Multi-Scale Saliency Fusion Model
An image pyramid network uses the same image to construct pyramid features through different scales [
28]. Compared with single-scale object detection, the advantage of an image pyramid is that it is possible to obtain different scale feature maps by adjusting the resolution of the image, and then to detect different scale objects. Because the image resolution is different, the size of the object and the semantic information of its features are also different. Pyramid features make up for the loss of semantic information in the process of down-sampling, so its detection precision can be improved to a certain extent.
Although the image pyramid network has a certain improvement effect on the detection precision, its disadvantage is that the large datasets occupy a lot of memory and consume a lot of time, so it has been gradually replaced by the feature pyramid network in the development process of object detection. The feature pyramid network (FPN) can achieve both speed and precision, and greatly improves the performance of object detection by improving multi-scale features with strong semantics. However, before the feature fusion in the FPN stage, there are semantic differences between the features of different network layers. The features of different network layers independently pass through the 1 × 1 convolutional layer, the purpose of which is to reduce the channels of the feature vector. However, there is a huge semantic gap between features of different scales. Due to the inconsistency of semantic information, fusing these features directly will reduce the expressive ability of multi-scale features. Therefore, this paper uses the FPN to obtain multi-scale features in the network and improves the detection precision of the network by fusing the high-resolution and the semantic information. The structure is shown in
Figure 5.
In the process of constructing pyramid feature mapping, the output features of the second stage to the last residuals in the fifth stage of the backbone network are reduced by a 1 × 1 convolution operation to obtain different scale features as {C2, C3, C4, C5}. Then, they are connected by top-down and horizontal connections to form pyramid features {P2, P3, P4, P5}. The convolution operation of 1 × 1 is to reduce the number of convolution kernels, that is, to reduce the number of channels of the feature maps without changing the size of the feature maps.
The human visual system often does not understand and process all information. Instead, it focuses attention on some significant or interesting information, which helps to filter out unimportant information and improve the efficiency of information processing. To make rational use of the limited visual information processing resources, humans select and focus on specific parts of the visual area. This visual processing mechanism is called the saliency mechanism [
29,
30,
31]. In detection tasks, extracting the detailed information of a specific area is the key to improving detection efficiency. The saliency mechanism can select the focus position in the input information of an image, which makes the detection network pay more attention to the more significant feature information in the input data so that the features extracted by the network are more distinguishable. In this paper, the saliency module is integrated after the feature pyramid module. Through the saliency mechanism, the number of false detections caused by background information can be reduced, thereby improving the detection precision of the network.
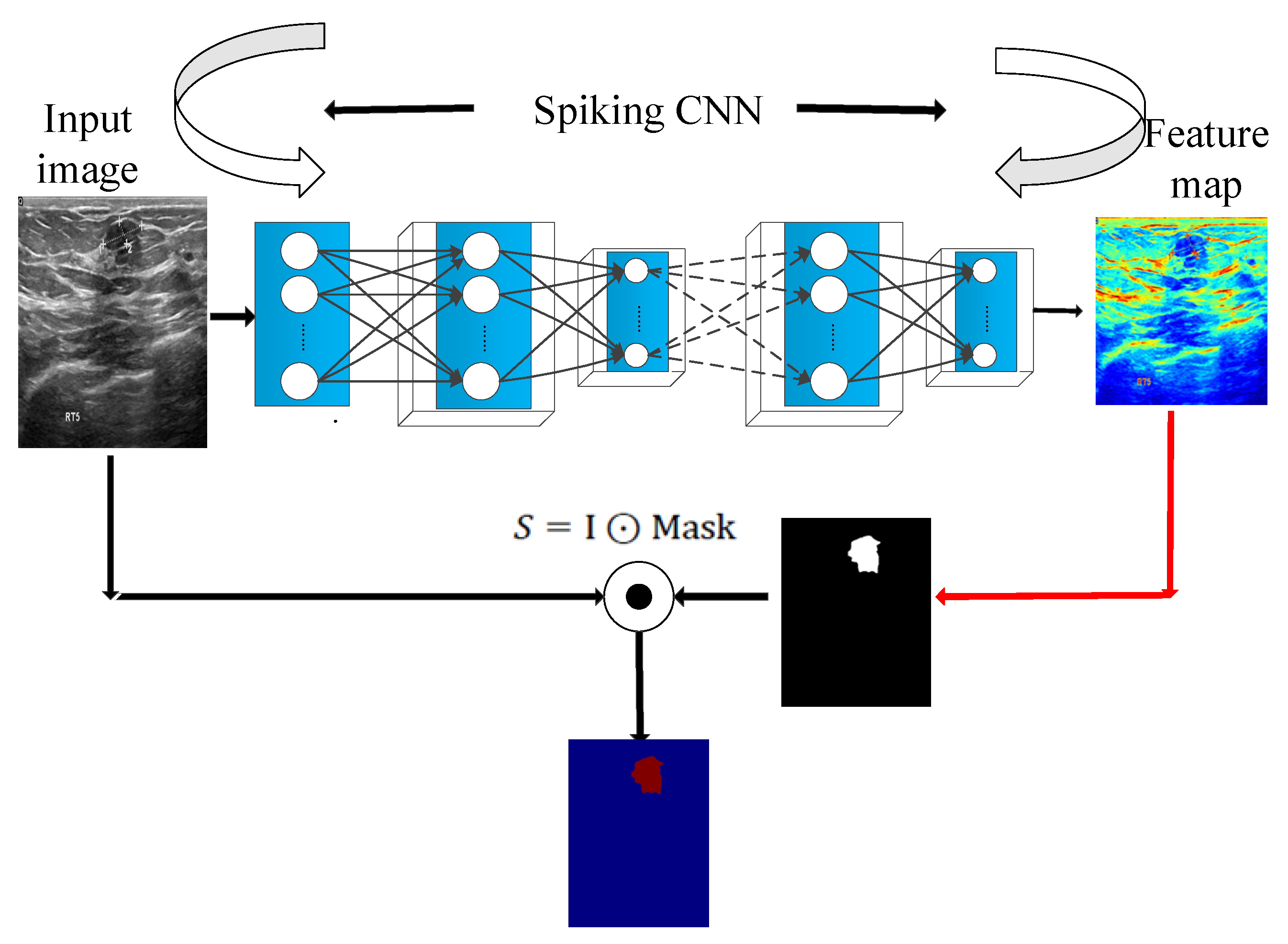
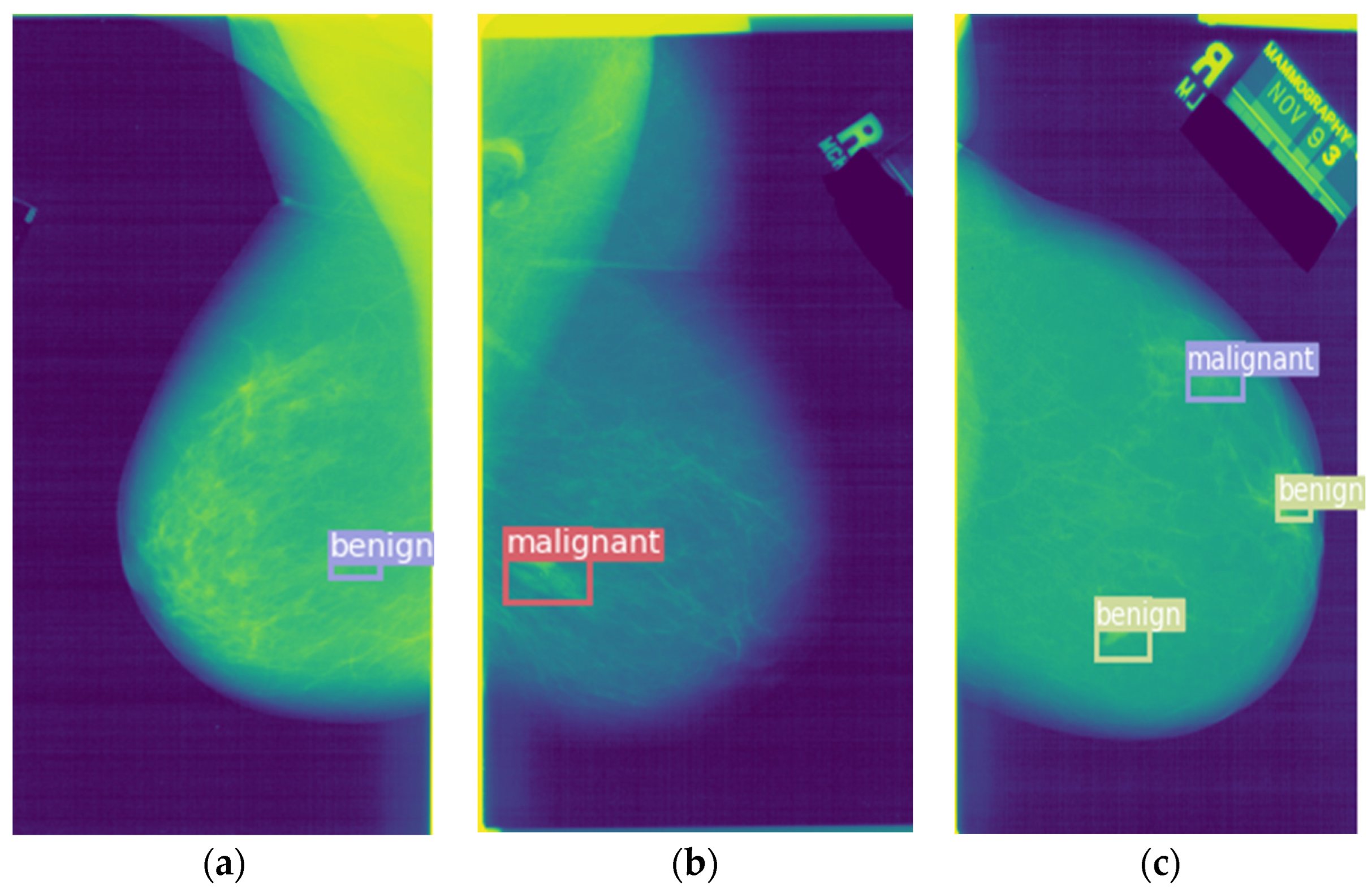
In the saliency module is shown in
Figure 6, a malignant tumor image is taken as an example. The spiking CNN is employed to extract the features. Then, the two-dimensional feature maps generated by the spike convolution layer are summed and the mask is calculated to obtain the saliency feature map.