Prebiotic Aggregates (Tissues) Emerging from Reaction–Diffusion: Formation Time, Configuration Entropy and Optimal Spatial Dimension
Abstract
:1. Introduction
- Replication: Reactants interact to produce a compound , mainly from another chemical compound, a substrate . Metabolism is assumed implicitly in the replication process. This replication process retains a sense of order, which will be measured in terms of configuration entropy.
- Heredity: on a large scale of perturbations, there is a viable continuity of equivalent traits (patterns) promoting adaptation.
2. Schnakenberg’s Model and Homogeneous Solution Instability
3. Necessary Conditions for Generating Proto-Tissues Structures
4. Unstable Manifold: Tissue Formation
- Equation (5) is necessary, but not sufficient, to satisfy Equation (8).
- New structures are favored for smaller values of the set , i.e., simple patterns. Tangentially, it is noted that simple patterns progress in the early stage of development of an embryo [11].
- The generalization of Equation (8) to small spatial dimensions of one or two is direct (formally, or , respectively). In the next sections, results are obtained for any spatial dimension including fractional dimensions.
- The generation of structures requires a minimal spatial size (see Equation (11)), as follows:where the parameter characterizes the cell size.
5. Number of Equivalent Structures and Heredity
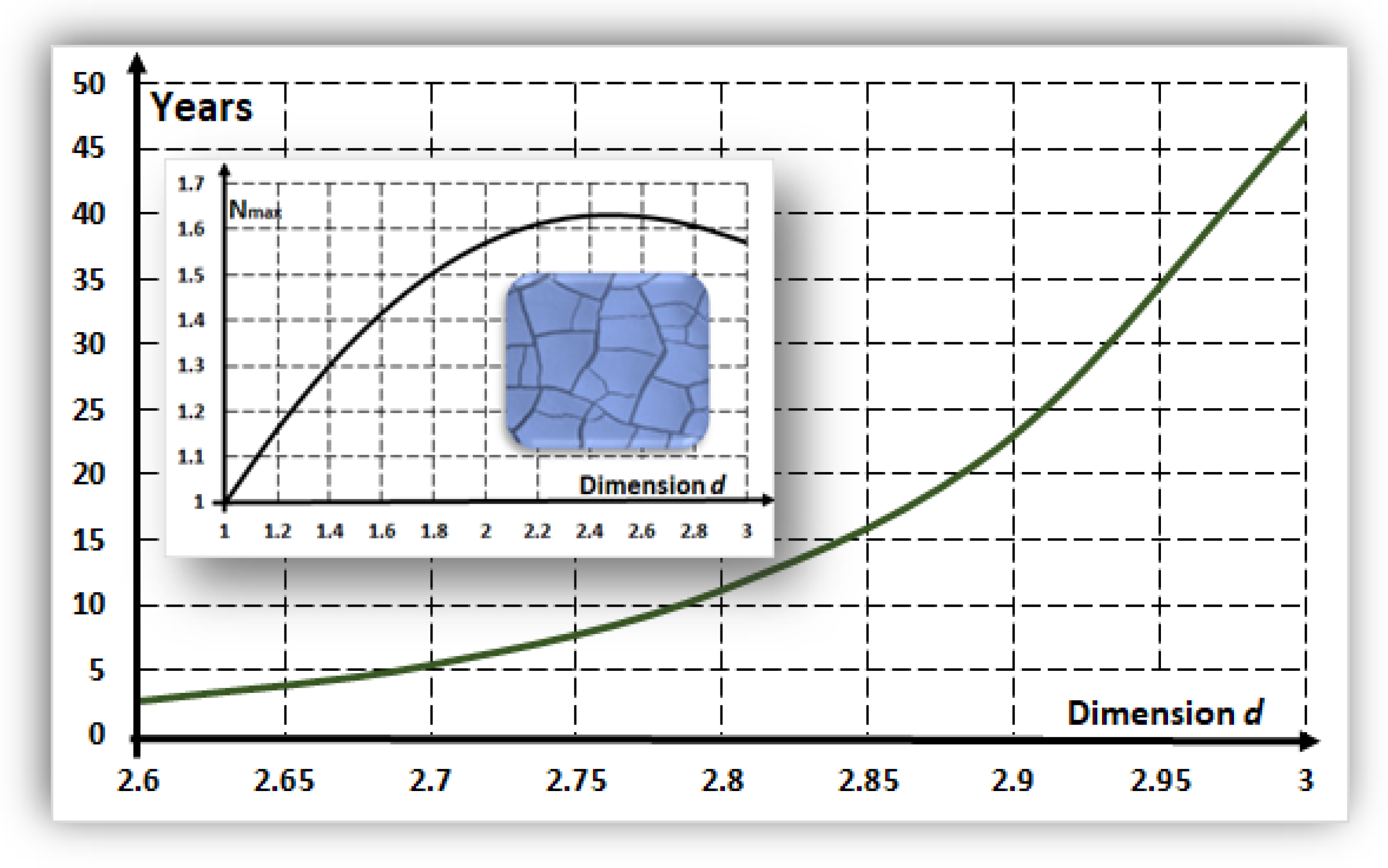
6. Time-Formation for Proto-Tissues: Role of Dimension
- (a)
- (b)
- A protocell of size m (i.e., 10 μm) and a membrane with thickness m.
- (c)
- As an estimation, is assumed to be approximately equal to the number of cells in the proto-tissue.
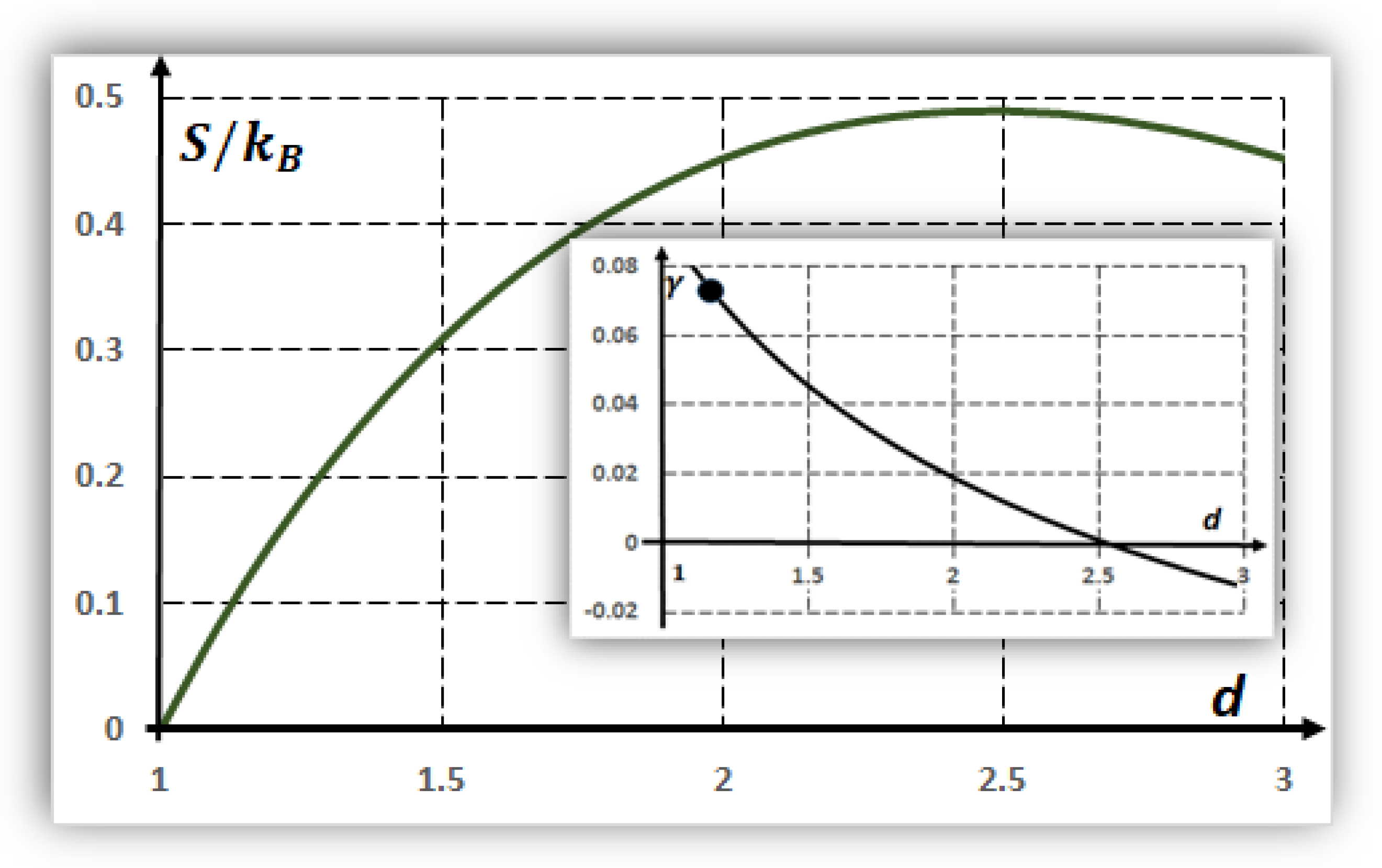
7. Environmental Fluctuations and Optimal Dimension: Configuration Entropy
- (a)
- If the spatial dimension is , then and weak fluctuations, , exist around this fractional dimension. These results indicate stable refuges against fluctuations. This point is fully complementary with a maximal diversity of structures when (Section 6).
- (b)
- In the same way, for a spatial dimension of , i.e., a smooth slime sheet, , corresponding to environmental variations. Additionally, for , e.g., a bubble in a liquid medium, the thermal variations are .
- (c)
- As a geologic example, in the Atacama Desert of Chile, rock temperatures vary between approximately [53] 0 and 45 °C. If the average temperatures are in the order of 296 oK, then corresponds to a spatial dimension smaller than two for the formation of hypothetical proto-tissues in these extreme conditions.
8. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hazen, R.M. Life’s rocky start. Sci. Am. 2001, 284, 76–851. [Google Scholar] [CrossRef]
- Steel, M.; Penney, D. Common ancestry put to the test. Nature 2010, 465, 168–169. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, E. What Is Life; Cambridge Universite Press: Cambridge, UK, 1944. [Google Scholar]
- Benner, S.A. Defining life. Astrobiology 2010, 10, 1021–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Branscomb, E.; Russell, M.J. Turnstile and bifurcator: The disequilibrium converting engines that put metabolism on the road. Biochim. Biophys. Acta Bioenerg. 2013, 1827, 62–78. [Google Scholar] [CrossRef] [Green Version]
- Leyva, Y.; Marín, O.; García-Jacas, C.R. Constraining the prebiotic cell size limits in extremely hostile environments: A dynamics perspective. Astrobiology 2018, 18, 403–411. [Google Scholar] [CrossRef]
- Dorn, E.D.; Nealson, K.H.; Adami, C. Monomer abundance distribution patterns as a universal biosignature: Examples from terrestrial and digital life. J. Mol. Evol. 2011, 72, 283–295. [Google Scholar] [CrossRef] [Green Version]
- Barge, L.M.; Branscomb, E.; Brucato, J.R.; Cardoso, S.S.S.; Cartwright, J.H.E.; Danielache, S.O.; Galante, D.; Kee, T.P.; Miguel, Y.; Mojzsis, S.; et al. Thermodynamics, Disequilibrium, Evolution: Far-from-equilibrium geological and chemical considerations for origin-of-life research. Orig. Life Evol. Biosph. 2017, 47, 39–56. [Google Scholar] [CrossRef] [Green Version]
- Turing, A.M. The chemical basis of morphogenesis. Philos. Trans. R. Soc. Lond. B 1952, 237, 37–72. [Google Scholar]
- Maynard-Smith, J. Shaping Life: Gene, Embryo and Evolution; Weidenfield and Nicolson: London, UK, 1998. [Google Scholar]
- Maynard-Smith, J.; Szathmáry, E. The Origins of Life: From the Birth of Life to the Origin of Language; Oxford Press: Oxford, UK, 1999. [Google Scholar]
- Huang, K. Statistical Mechanics; Wiley: Hoboken, NJ, USA, 1987. [Google Scholar]
- Landau, L.D.; Lifschitz, E.M. Statistical Mechanics; Elsevier: Amsterdam, The Netherlands, 1981. [Google Scholar]
- Toda, M.; Kubo, R.; Saito, N. Statistical Physics I; Springer: Berlin, Germany, 1982. [Google Scholar]
- Pathria, R.K. Statistical Mechanics; Elsevier: Amsterdam, The Netherlands, 2009. [Google Scholar]
- Schnakenberg, J. Simple chemical reaction systems with limit cycle behavior. J. Theor. Biol. 1979, 81, 389–400. [Google Scholar] [CrossRef]
- Hanczyc, M.M. Metabolism and motility in prebiotic structures. Philos. Trans. R. Soc. B 2011, 366, 2885–2893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haken, F. Synergetics, Introduction and Advanced Topics; Springer: Berlin, Germany, 2004. [Google Scholar]
- Murray, J.D. Mathematical Biology I: An Introduction; Springer: Berlin, Germany, 2002. [Google Scholar]
- Murray, J.D. Mathematical Biology II: An Introduction; Springer: Berlin, Germany, 2003. [Google Scholar]
- Clerc, M.G.; Escaff, D.; Kenkre, V.M. Patterns and localized structures in population dynamics. Phys. Rev. E 2005, 72, 056217. [Google Scholar] [CrossRef] [Green Version]
- Cantrell, R.S.; Cosner, C. Spatial Ecology via Reaction-Diffusion Equations; Wiley: Hoboken, NJ, USA, 2003. [Google Scholar]
- Vespignani, A. Modeling dynamical process in complex technical systems. Nat. Phys. 2012, 8, 32–39. [Google Scholar] [CrossRef]
- Boccara, N. Modeling Complex Systems, 2nd ed.; Springer: Berlin, Germany, 2010. [Google Scholar]
- Lakshmanan, M.; Rajasekar, S. Nonlinear Dynamics: Integrability, Chaos and Patterns; Springer: Berlin, Germany, 2003. [Google Scholar]
- Benguria, R.D.; Depassier, M.C. Speed of fronts of generalized reaction-diffusion equations. Phys. Rev. E 1998, 57, 6493. [Google Scholar] [CrossRef] [Green Version]
- Deloubrière, O.; Hilhorst, H.J.; Taüber, U.C. Multispecies pair annihilation reactions. Phys. Rev. Lett. 2002, 89, 250601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Descalzi, O.; Hayase, Y.; Brand, H.R. Oscillating localized structures in reaction-diffusion systems. Int. J. Bifurc. Chaos 2004, 14, 4097–4104. [Google Scholar] [CrossRef]
- Hilhorst, H.J.; Washenberger, M.J.; Taüber, U.C. Symmetries and species segregation in diffusion-limited pair annihilation. J. Stat. Mech. 2004, 2004, P10002. [Google Scholar] [CrossRef] [Green Version]
- Roman, C.; Davydovych, V. Non-Linear Reaction-Diffusion Systems; Springer: Berlin, Germany, 2017. [Google Scholar]
- Valverde, S.; Ohse, S.; Turalska, M.; West, B.J.; Garcia-Ojalvo, J. Structural determinants of criticality in biological networks. Front. Physiol. 2015, 6, 127. [Google Scholar] [CrossRef] [Green Version]
- Al Noufaey, K.S. Semi-analytical solutions of the Schnakenberg model of a reaction-diffusion cell with feedback. Results Phys. 2018, 9, 609–614. [Google Scholar] [CrossRef]
- Flores, J.C. Competitive exclusion and axiomatic set-theory: De Morgan’s laws, ecological virtual process, symmetries and frozen diversity. Acta Biotheor. 2016, 64, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Li, M.; Booth, R.; Mann, S. Predatory behavior in synthetic protocell communities. Nat. Chem. 2017, 9, 110–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandelbrot, B. Fractals and Chaos; Springer: Berlin, Germany, 2004. [Google Scholar]
- Falconer, K. Fractal Geometry: Mathematical Foundations and Applications; John Wiley & Sons: Hoboken, NJ, USA, 2003. [Google Scholar]
- Gordon, N. Introducing Fractal Geometry; Icon: Duxford, UK, 2000; ISBN 978-1-84046-123-7. [Google Scholar]
- Jones-Smith, K.; Mathur, H. Fractal Analysis: Revisiting Pollock’s drip paintings. Nature 2006, 444, E9–E10. [Google Scholar] [CrossRef] [PubMed]
- Ben-Avraham, D.; Havlin, S. Diffusion and Reactions in Fractals and Disordered Systems; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Falconer, K. Fractals, a Very Short Introduction; Oxford University Press: Oxford, UK, 2013. [Google Scholar]
- Jin, Y.; Wu, Y.; Li, H.; Zhao, M.; Pan, J. Definition of fractal topography to essential understanding of scale-invariance. Sci. Rep. 2017, 7, 46672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landau, L.D.; Lifshitz, E.M. Theory of Elasticity, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2007. [Google Scholar]
- Gray, N.H. Symmetry in a natural fracture pattern: The origin of columnar joint networks. Comput. Math. Appl. 1986, 12, 531–545. [Google Scholar] [CrossRef] [Green Version]
- Goehring, L.; Mahadevan, L.; Morris, S.W. Nonequilibrium scale selection mechanism for columnar jointing. Proc. Natl. Acad. Sci. USA 2009, 106, 387–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goehring, L.; Morris, S.W. Cracking mud, freezing dirt and breaking rocks. Phys. Today 2014, 67, 39–44. [Google Scholar] [CrossRef]
- NASA. Possible Signs of Ancient Drying in Martian Rock; Jet Propulsion Laboratory: Pasadena, CA, USA, 2017. Available online: https://www.nasa.gov/image-feature/jpl/pia21263/possible-signs-of-ancient-drying-in-martian-rock (accessed on 24 November 2021).
- Flores, J.C. Mean-field crack networks on desiccated films and their applications: Girl with a pearl earring. Soft Matter 2017, 13, 1352–1356. [Google Scholar] [CrossRef] [Green Version]
- Flores, J.C. Entropy signature for crack networks in old painting: Saturation prospectus. Entropy 2018, 20, 772. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Lowensohn, J.; Burton, J.C. Universal scaling of polygonal desiccation crack patterns. Phys. Rev. E 2019, 99, 012802. [Google Scholar] [CrossRef] [Green Version]
- Rich, A. On the problem of evolution and biochemical information transfer. In Horizons; Kasha, M., Pullmann, B., Eds.; Academic Press: New York, NY, USA, 1962. [Google Scholar]
- Hazen, R.M.; Sverjensky, D.A. Mineral Surfaces, Geochemical Complexity and the Origins of Life; Cold Spring Harbor Laboratory Press: Suffolk County, NY, USA, 2010. [Google Scholar]
- Flores, J.C. Dimensional ensemble and (topological) fracton thermodynamics: The slow route to equilibrium. Sci. Rep. 2019, 9, 12793. [Google Scholar] [CrossRef]
- McKay, C.P.; Molaro, J.L.; Marinova, M.M. High-frequency rock temperatures data from hyper-arid desert environments in the Atacama and the Antarctic dry Valley and implications for rock weathering. Geomorphology 2009, 110, 182–187. [Google Scholar] [CrossRef]
- Dyson, F. Origin of Life; Cambridge University Press: Cambridge, UK, 1986. [Google Scholar]
- Hyman, T.; Brangwynne, C. In retrospect: The origin of life. Nature 2012, 491, 524–525. [Google Scholar] [CrossRef]
- Brack, A. The Molecular Origins of Life; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar]
- Shapiro, R. A simpler origin for life. Sci. Am. 2007, 296, 46–53. [Google Scholar] [CrossRef]
- Javaux, E.J. Challenges in evidencing the earliest traces of life. Nature 2019, 572, 451–460. [Google Scholar] [CrossRef]
- Demongeot, J.; Glade, N.; Moreira, A.; Vial, L. RNA Relics and Origin of Life. Int. J. Mol. Sci. 2009, 10, 3420–3441. [Google Scholar] [CrossRef]
- Adamski, P.; Eleveld, M.; Sood, A.; Kun, Á.; Szilágyi, A.; Czárán, T.; Szathmáry, E.; Otto, S. From self-replication to replicator systems en route to de novo life. Nat. Rev. Chem. 2020, 4, 386–403. [Google Scholar] [CrossRef]
- Jordan, S.F.; Rammu, H.; Zheludev, I.; Hartley, A.; Maréchal, A.; Lane, N. Promotion of protocell self-assembly from mixed amphiphiles at the origin of life. Nat. Ecol. Evol. 2019, 3, 1705–1714. [Google Scholar] [CrossRef] [PubMed]
- Michalski, J.R.; Onstott, T.C.; Mojzsis, S.J.; Mustard, J.; Chan, Q.H.S.; Niles, P.B.; Johnson, S.S. The Martian subsurface as a potential window into the origin of life. Nat. Geosci. 2018, 11, 21–26. [Google Scholar] [CrossRef]
- Totani, T. Emergence of life in an inflationary universe. Sci. Rep. 2020, 10, 1671. [Google Scholar] [CrossRef] [PubMed]
- Greaves, J.S.; Richards, A.M.S.; Bains, W.; Rimmer, P.B.; Sagawa, H.; Clements, D.L.; Seager, S.; Petkowski, J.J.; Sousa-Silva, C.; Ranjan, S.; et al. Phosphine gas in the cloud decks of Venus. Nat. Astron. 2021, 5, 655–664. [Google Scholar] [CrossRef]
- Shapiro, R. A Replicator was not involved in the origin of life. Life 2000, 49, 173–176. [Google Scholar]
- Parisi, G. Complex Systems: A Physicist’s Viewpoint. arXiv 2013, arXiv:cond-mat/0205297v1. [Google Scholar] [CrossRef] [Green Version]
- Damer, B.; Deamer, D. The hot spring hypothesis for an origin of life. Astrobiology 2020, 20, 429–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maturana, H.R.; Varela, F.J. Autopoiesis and Cognition: The Realization of the Living; D. Reidel Publishing Company: Dordrecht, The Netherlands, 1972. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flores, J.C. Prebiotic Aggregates (Tissues) Emerging from Reaction–Diffusion: Formation Time, Configuration Entropy and Optimal Spatial Dimension. Entropy 2022, 24, 124. https://doi.org/10.3390/e24010124
Flores JC. Prebiotic Aggregates (Tissues) Emerging from Reaction–Diffusion: Formation Time, Configuration Entropy and Optimal Spatial Dimension. Entropy. 2022; 24(1):124. https://doi.org/10.3390/e24010124
Chicago/Turabian StyleFlores, Juan Cesar. 2022. "Prebiotic Aggregates (Tissues) Emerging from Reaction–Diffusion: Formation Time, Configuration Entropy and Optimal Spatial Dimension" Entropy 24, no. 1: 124. https://doi.org/10.3390/e24010124
APA StyleFlores, J. C. (2022). Prebiotic Aggregates (Tissues) Emerging from Reaction–Diffusion: Formation Time, Configuration Entropy and Optimal Spatial Dimension. Entropy, 24(1), 124. https://doi.org/10.3390/e24010124




