An Interpretation Architecture for Deep Learning Models with the Application of COVID-19 Diagnosis
Abstract
1. Introduction
1.1. Related Work
1.1.1. Deep Learning Approaches for COVID-19 Analysis
1.1.2. Interpretation of Deep Learning Models
1.2. Our Work and Contributions
- (1)
- We propose an interpretation architecture of deep learning models, and apply the architecture to COVID-19 diagnosis. Our interpretation architecture has good generality and can act as the interpretation solution for any CNN-based deep learning classification methods in other research areas.
- (2)
- Our architecture makes a comprehensive interpretation of deep models from different perspectives, including the training trends, diagnostic performances, learned features, feature extractors, neuron patterns in hidden layers, the areas of interest for supporting diagnostic decision, and etc. To our best knowledge, very little study has made a comprehensive interpretation about the deep learning-based diagnosis model. Furthermore, all of our interpretation results can be presented intuitively through visualization.
- (3)
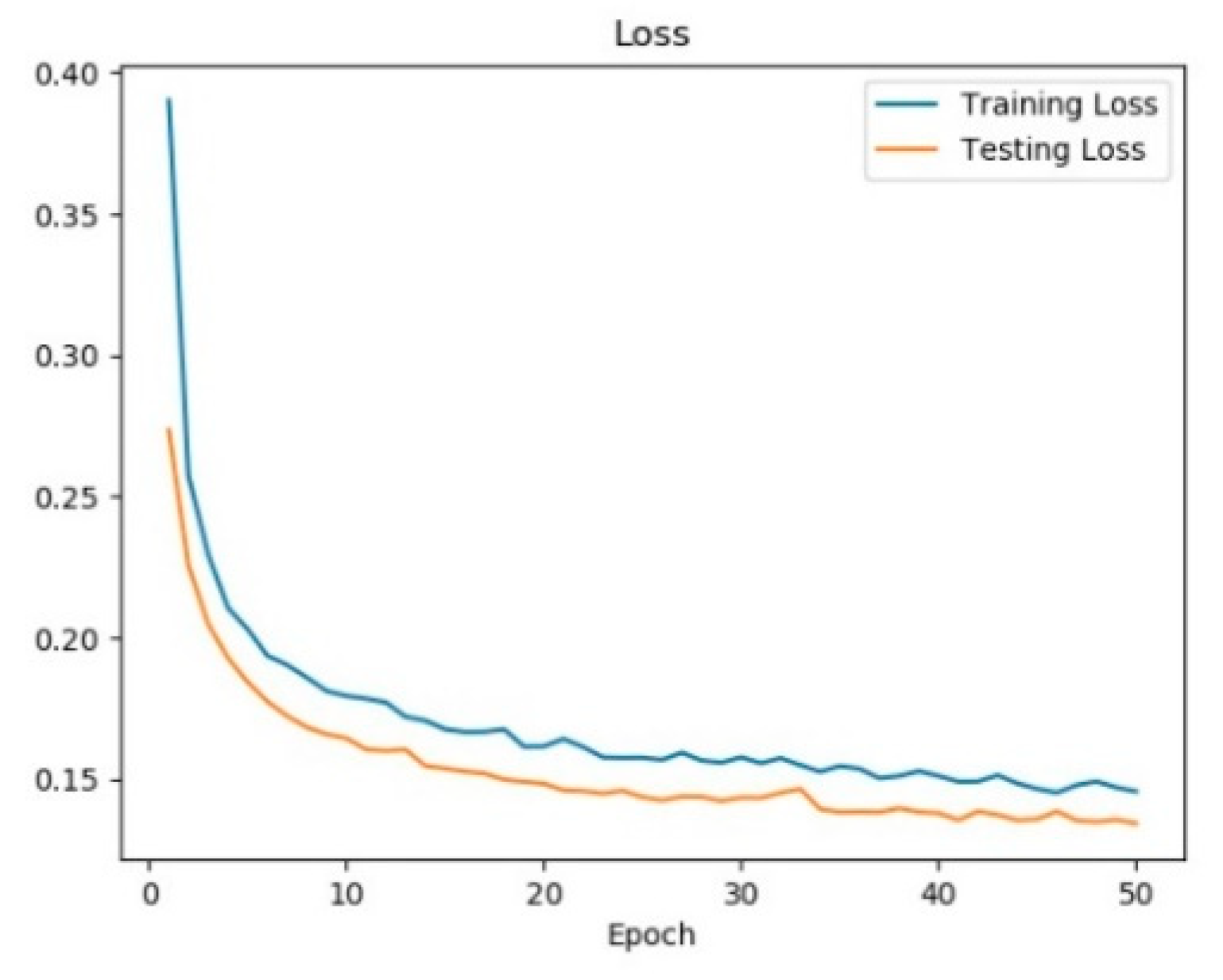
- Unlike most of the previous studies that focus on analyzing the testing phase of a CNN, in our architecture we propose to interpret the deep learning model from both the training phase and the testing phase to provide a general understanding of the CNN. The understanding of the training dynamic is quite necessary, through which researchers can observe the evolution of the diagnostic model, know whether the training is on the right track and whether the model converges, find latent mistakes, and etc.
- (4)
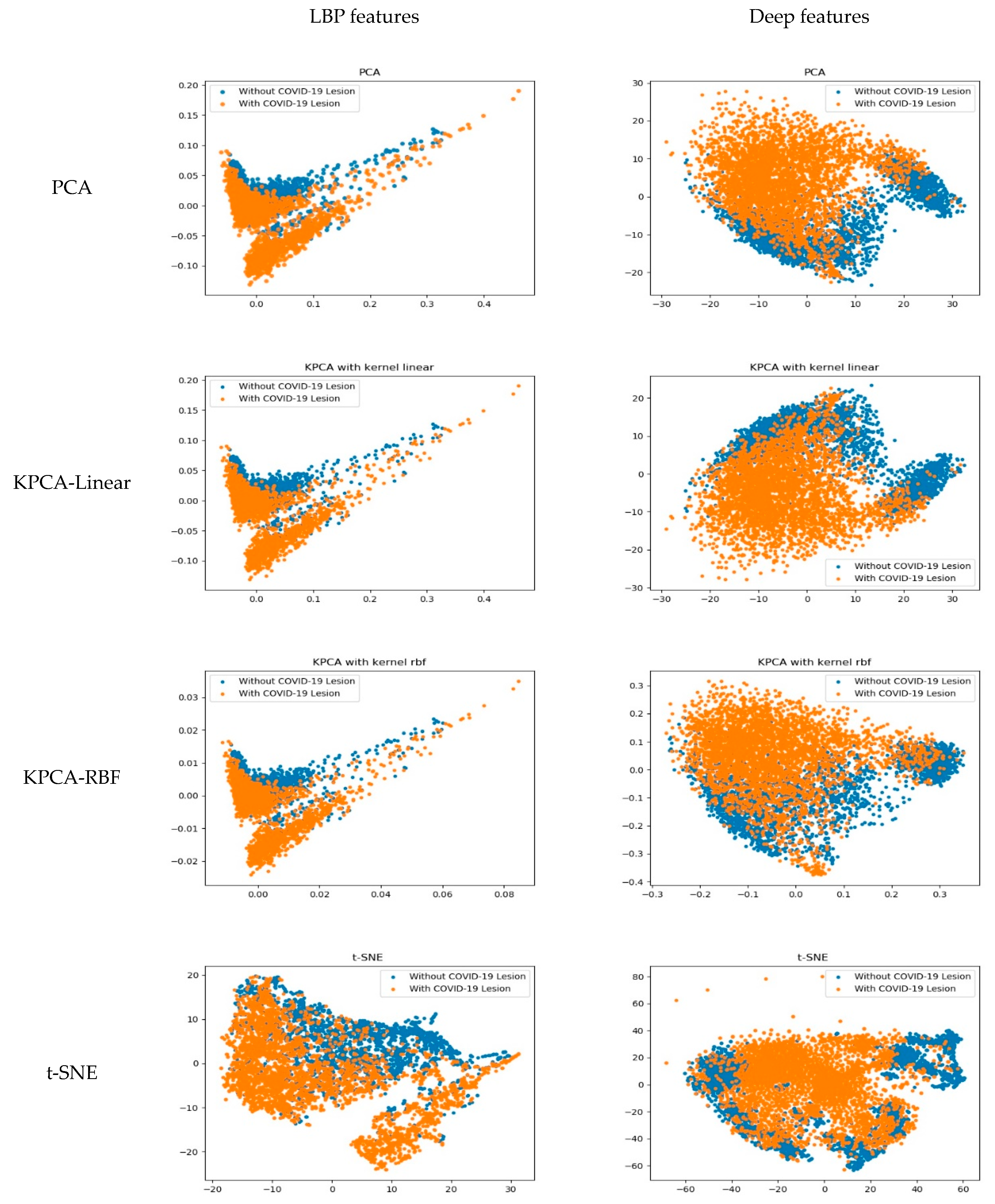
- To analyze the diagnostic performance of the deep learning-based COVID-19 diagnostic model, we design the comparison between the deep learning model and traditional classification method. Furthermore, we novelly make an explanation about the difference of diagnosis accuracy through visualizing the learned features of both methods. To our best knowledge, very little study has made a performance comparison together with the visual explanation of the difference in performance.
- (5)
- In our architecture, we provide a new possible pattern for CAD in the clinical field, through providing the supports for diagnostic decision. In the CAD of clinical COVID-19 diagnosis, our visualized attention maps for supporting diagnostic decisions can promisingly act as an important reference. For a new scanned CT image, our deep learning diagnostic model can output the diagnostic decision and the attention map for supporting the decision simultaneously. Clinician can first observe the diagnosis result, and then refer to the visualized attention map to make sure whether the diagnostic model focuses on the right regions, and whether the diagnosis result of the deep learning model for this image is trustable.
- (6)
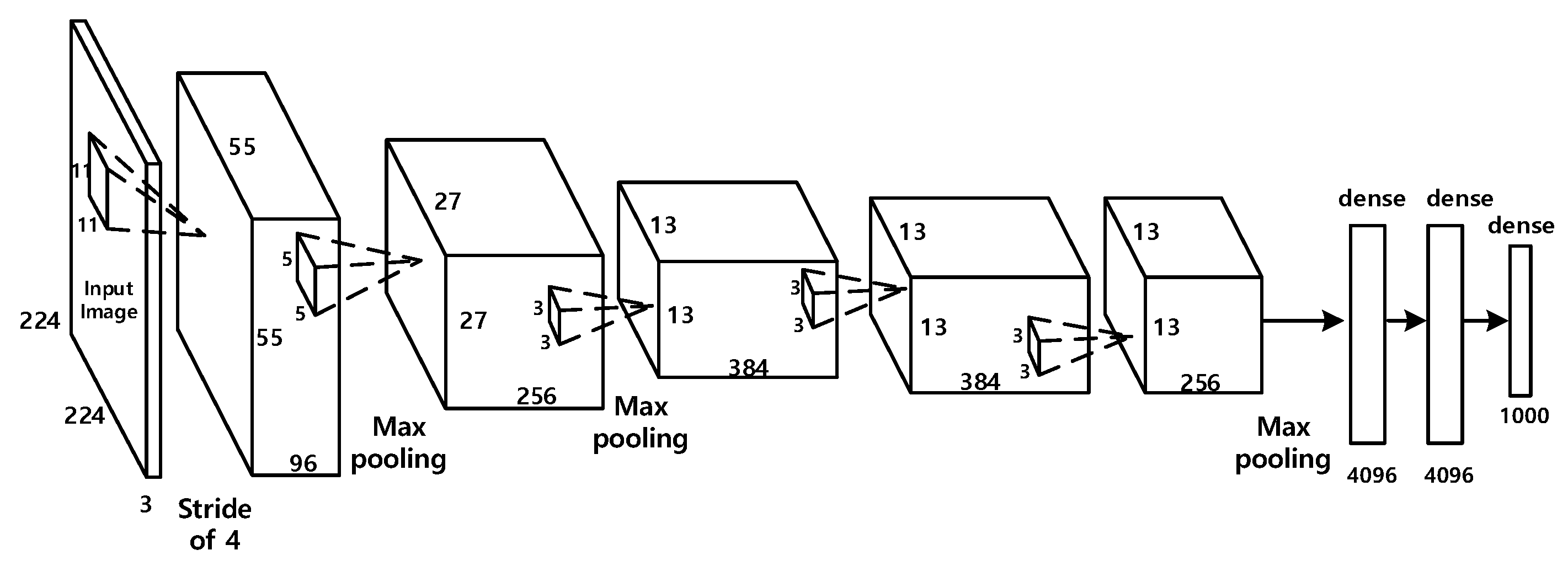
- Besides the interpretation architecture, we construct a deep learning model for the diagnosis of COVID-19. We choose the Alexnet framework as the deep learning model. Then we modify the original Alexnet to adapt it to the binary classification of COVID-19. The modified Alexnet model is trained using transfer learning strategy.
- (7)
- Overall, our proposed interpretation architecture provides an intuitive analysis of the deep learning models, which can possibly help to build better trust and take a step forward in the application of CAD methods into the clinical COVID-19 diagnosis field. First, through the visualization of the training trends, the comparison and explanation of classification performance, and the visualization of the CNN layers, researchers can gain a general judgement about the robustness and accuracy of the deep model. Then, for each new CT image, the clinician can obtain the diagnostic result from the deep model, and then make sure whether the diagnostic result for this image is trustable by referring to the visualized attention maps as supports. The clinical COVID-19 diagnosis can benefit from the high efficiency and accuracy of the CAD systems, and obtain high reliability through our interpretation architecture at the same time.
2. Materials and the Diagnostic Model
2.1. Materials
2.2. The Deep Learning-Based Diagnostic Model
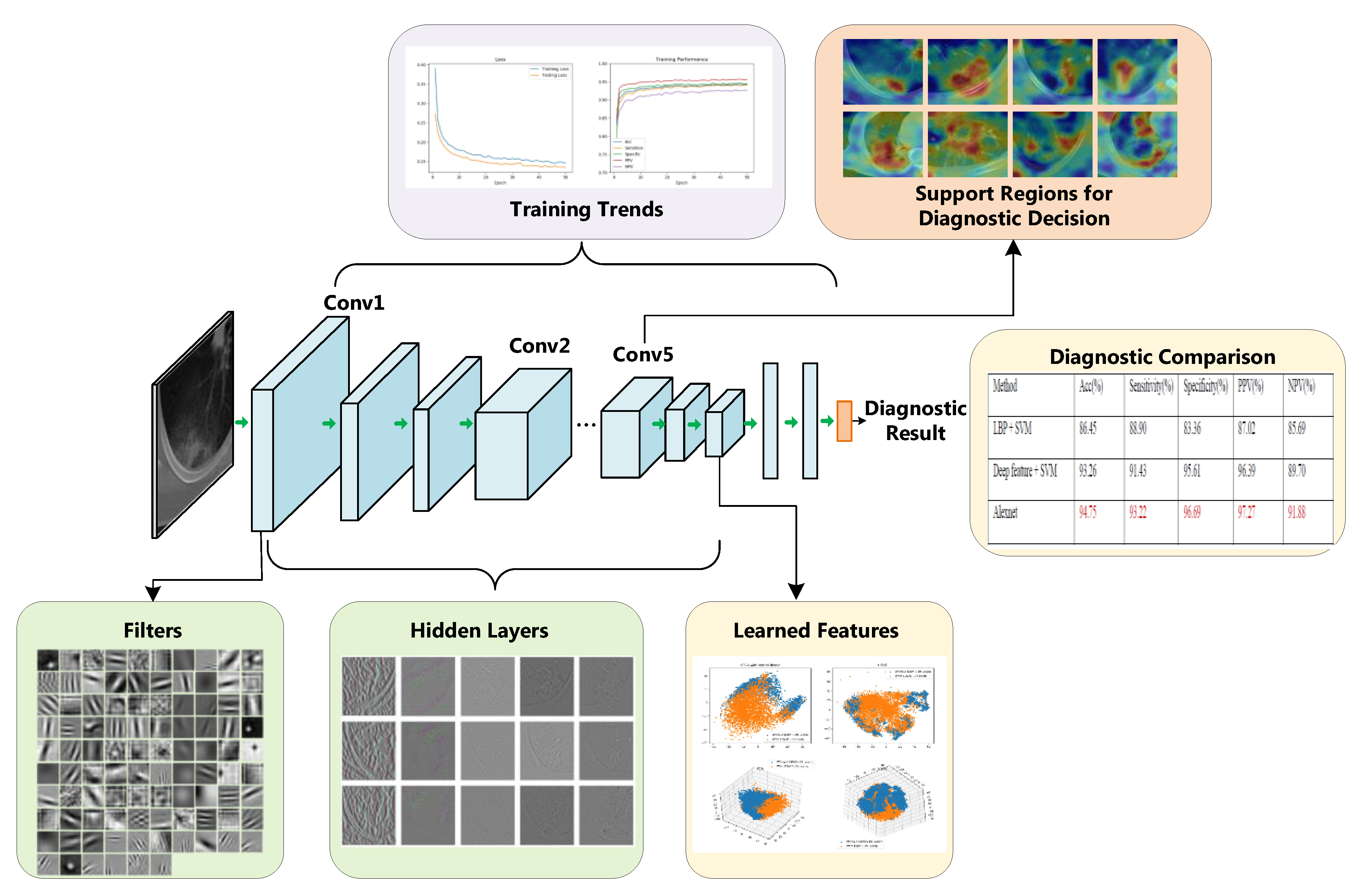
3. The Interpretation Architecture and Results
3.1. Training Dynamic
3.2. Diagnostic Performance
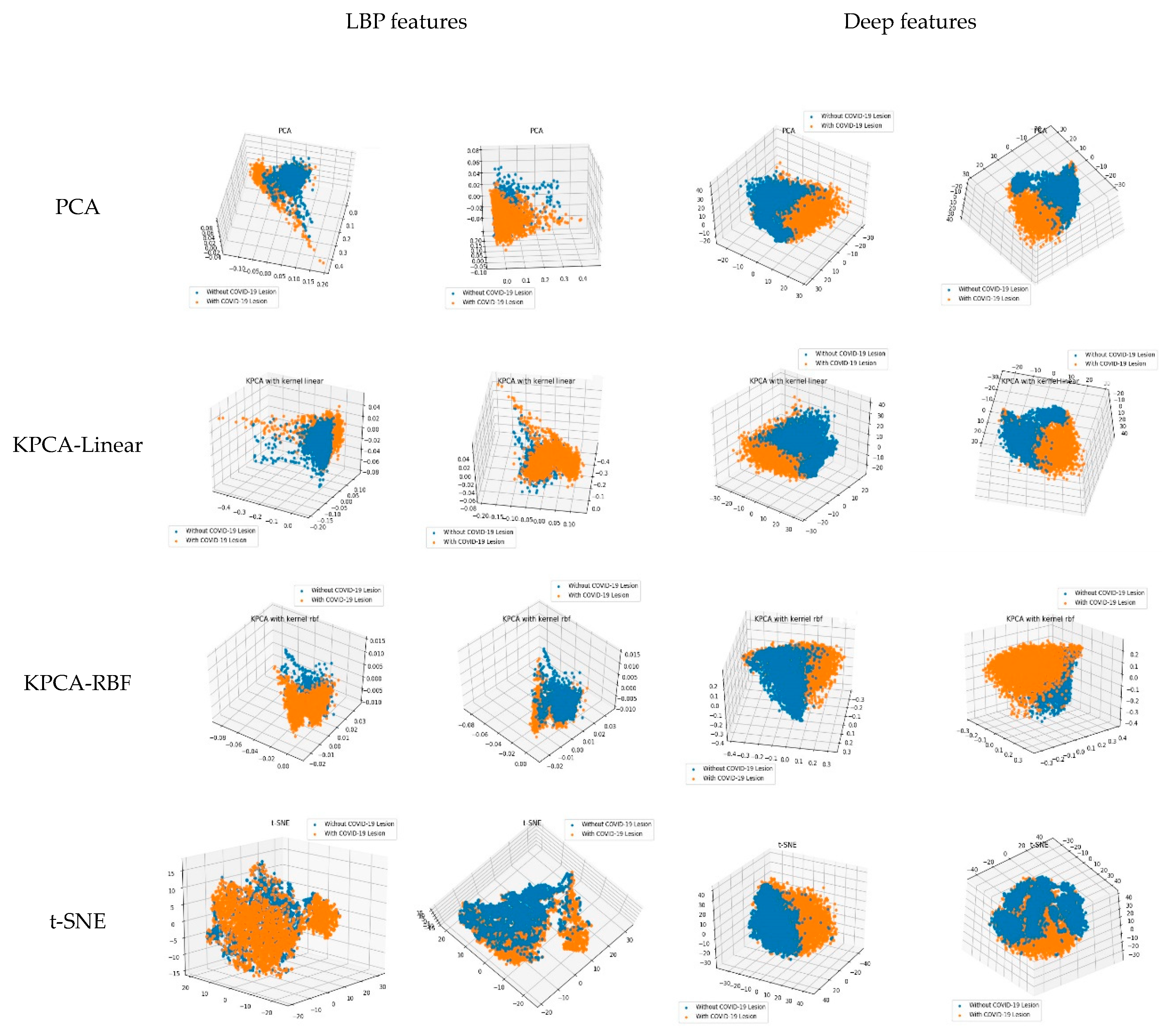
3.3. The Learned Features
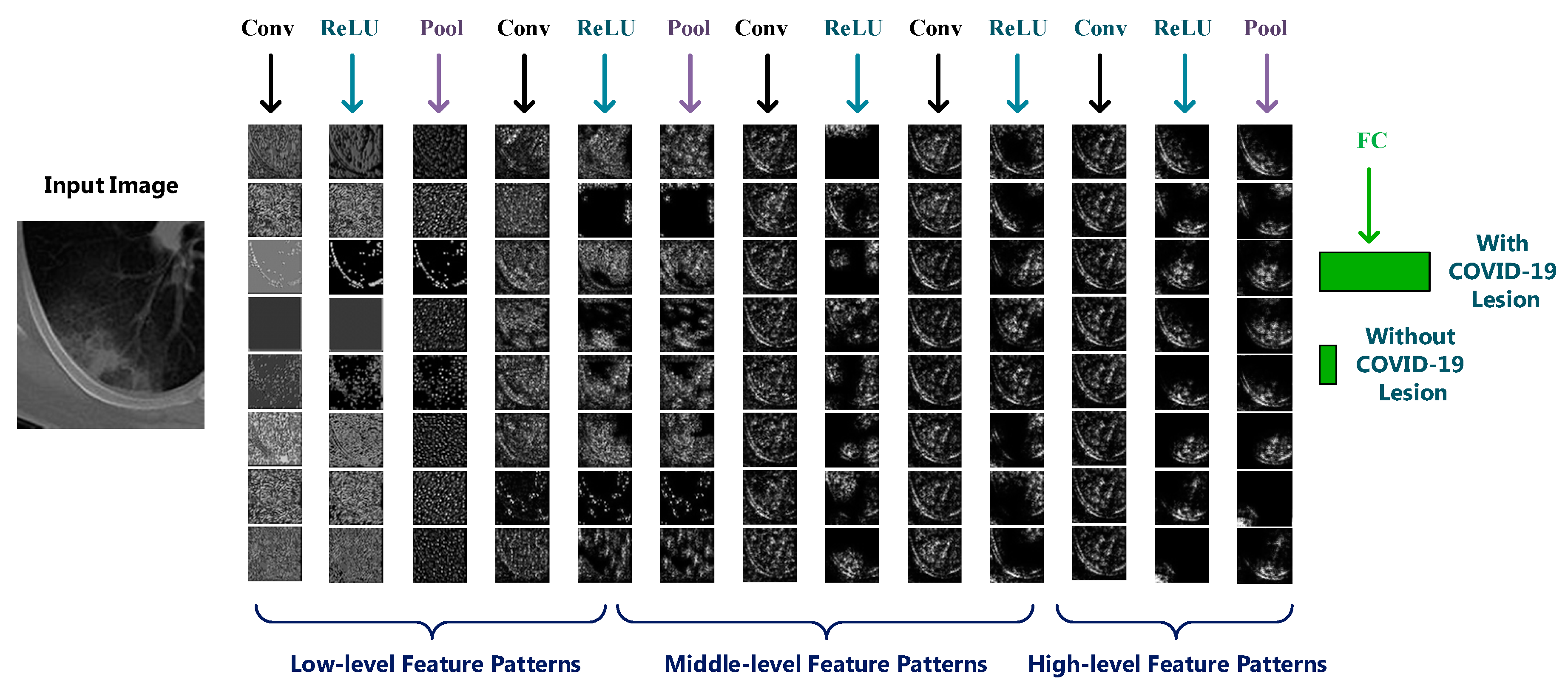
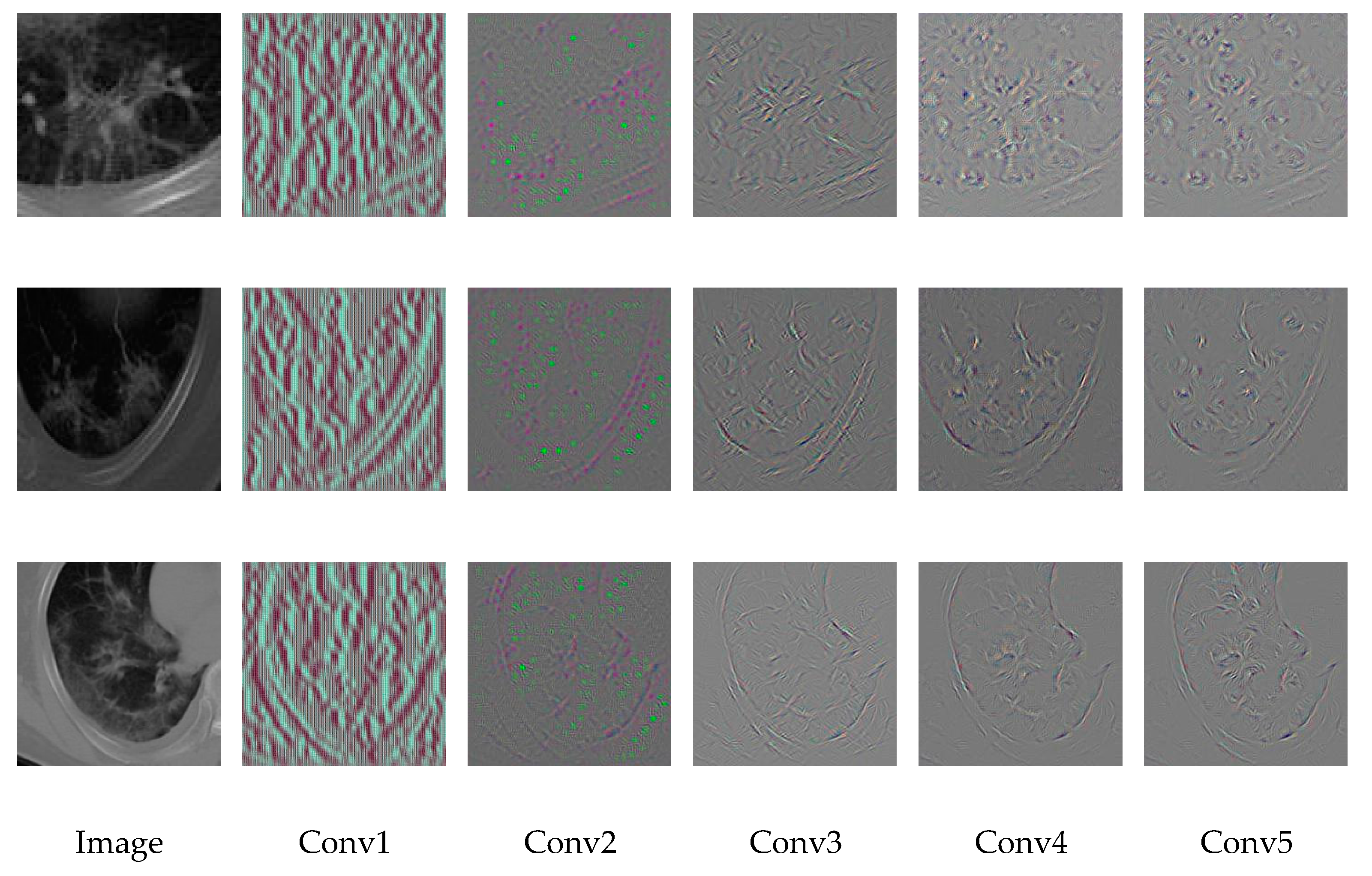
3.4. Visualization of CNN Layers
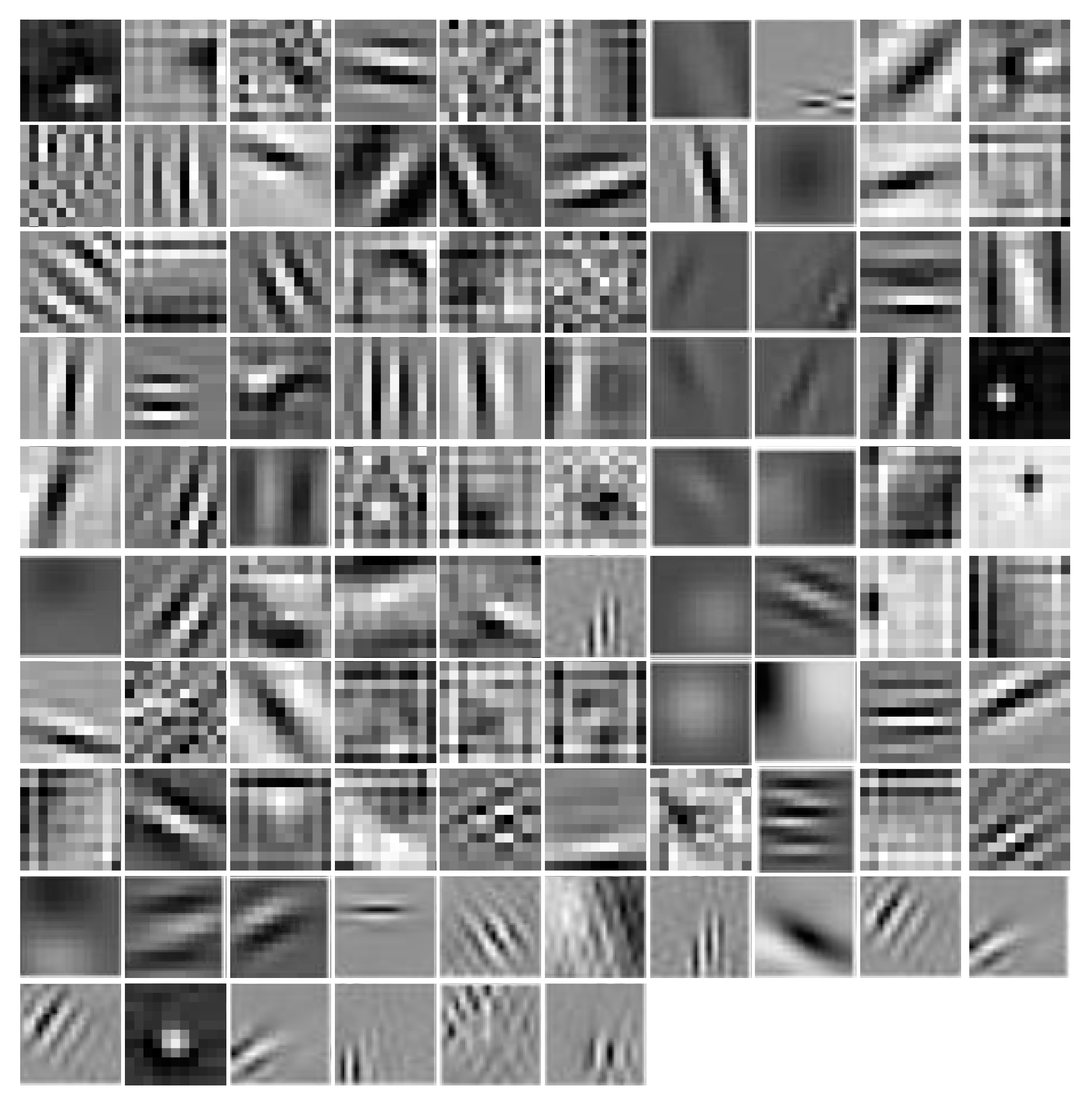
3.4.1. First Layer Filters
3.4.2. Hidden Layers Visualization
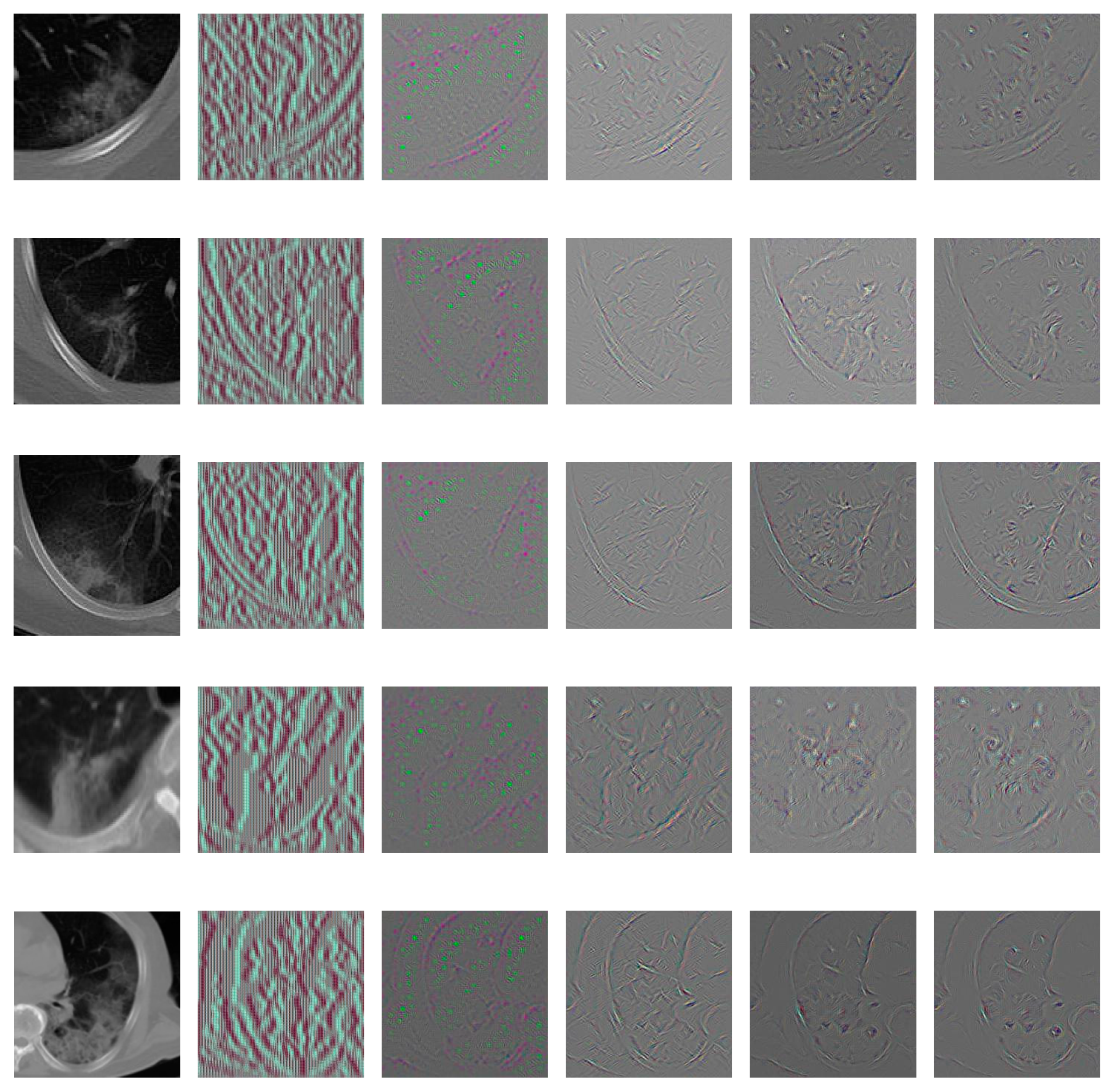
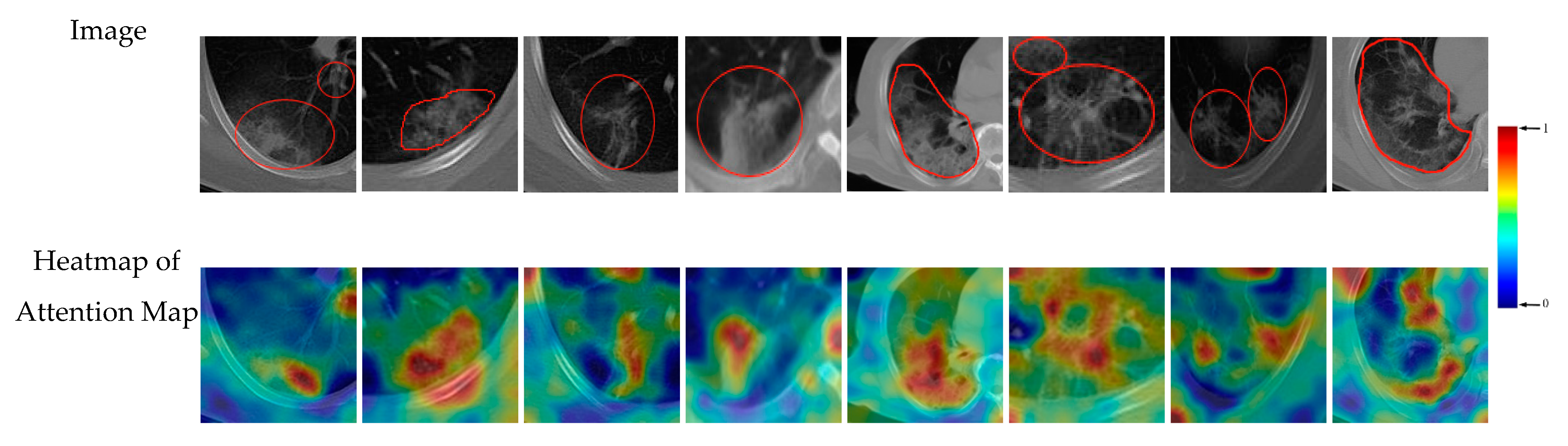
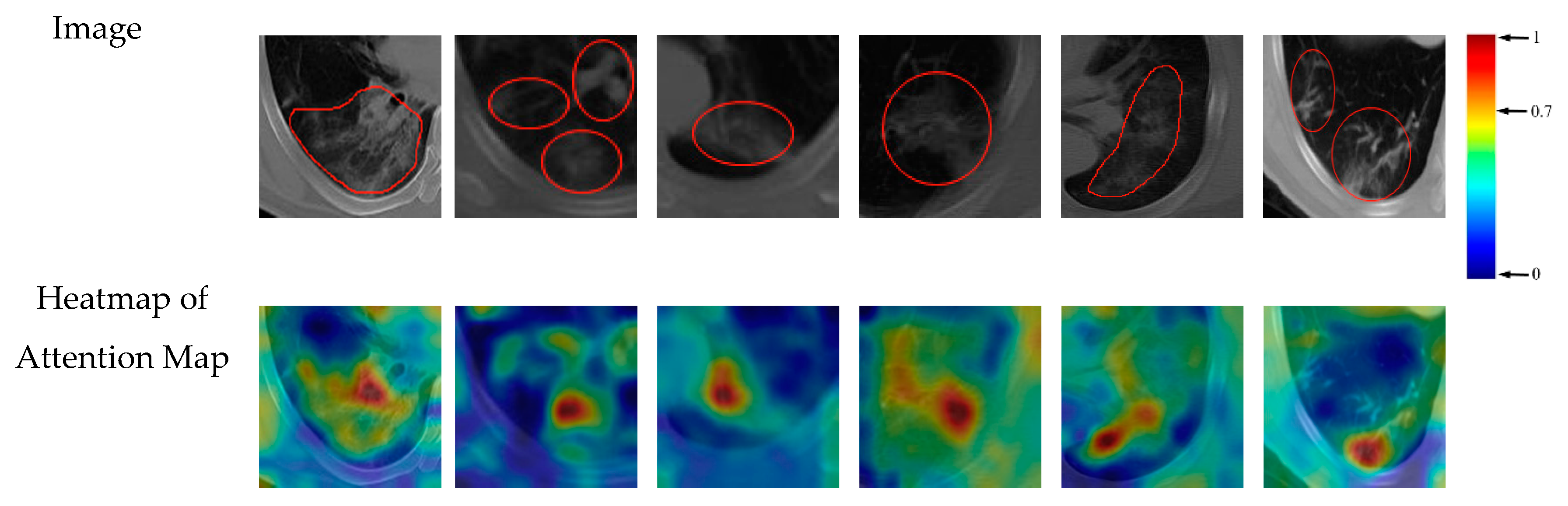
3.5. Support Regions for Diagnostic Decision
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- World Health Organization. Novel Coronavirus (COVID-19) Situation. Available online: https://who.sprinklr.com (accessed on 11 January 2021).
- Priesemann, V.; Brinkmann, M.; Ciesek, S.; Cuschieri, S.; Czypionka, T.; Giordano, G.; Gurdasani, D.; Hanson, C.; Hens, N.; Iftekhar, E.; et al. Calling for pan-European commitment for rapid and sustained reduction in SARS-CoV-2 infections. Lancet 2021, 397, 92–93. [Google Scholar] [CrossRef]
- Perc, M.; Miksi, N.G.; Slavinec, M.; Stožer, A. Forecasting COVID-19. Front. Phys. 2020, 8, 1–5. [Google Scholar] [CrossRef]
- Xie, X.; Zhong, Z.; Zhao, W.; Zheng, C.; Wang, F.; Liu, J. Chest CT for typical 2019-nCoV pneumonia: Relationship to negative RT-PCR testing. Radiology 2020, 296, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Khatami, F.; Saatchi, M.; Zadeh, S.S.T.; Aghamir, Z.S.; Shabestari, A.N.; Reis, L.O.; Aghamir, S.M. A meta-analysis of accuracy and sensitivity of chest CT and RT-PCR in COVID-19 diagnosis. Sci. Rep. 2020, 10, 22402. [Google Scholar] [CrossRef]
- Mahdavi, S.; Kheirieh, A.; Daliri, S.; Kalantar, M.H.; Valikhani, M.; Khosravi, A.; Enayatrad, M.; Emamian, M.H.; Chaman, R.; Rohani-Rasaf, M. More Reliability of Suspicious Symptoms plus Chest CT-Scan than RT_PCR Test for the Diagnosis of COVID-19 in an 18-days-old Neonate. IDCases 2020, 21, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Huang, C. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical characteristics of 138 hospitalized patients with 2019 novel Coronavirus Infected pneumonia in wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Kim, H.; Hong, H.; Yoon, S.H. Diagnostic Performance of CT and Reverse Transcriptase-Polymerase Chain Reaction for Coro navirus Disease 2019: A Meta-Analysis. Radiology 2020, 296, 145–155. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Luo, L.; Luo, Z.; Lyu, J.X.; Ng, M.Y.; Shen, X.P.; Wen, Z. Diagnostic performance between CT and initial real-time RT-PCR for clinically suspected 2019 coronavirus disease (COVID-19) patients outside Wuhan, China. Respir. Med. 2020, 168, 105980. [Google Scholar] [CrossRef]
- Apostolopoulos, I.D.; Mpesiana, T.A. Covid-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020, 43, 635–640. [Google Scholar] [CrossRef]
- Jamshidi, M.B.; Lalbakhsh, A.; Talla, J.; Peroutka, Z.; Hadjilooei, F.; Lalbakhsh, P.; Jamshidi, M.; La Spada, L.; Mirmozafari, M.; Dehghani, M.; et al. Artificial Intelligence and COVID-19: Deep Learning Approaches for Diagnosis and Treatment. IEEE Access 2020, 8, 109581–109595. [Google Scholar] [CrossRef]
- Hu, S.; Gao, Y.; Niu, Z.; Jiang, Y.; Li, L.; Xiao, X.; Wang, M.; Fang, E.F.; Menpes-Smith, W.; Xia, J.; et al. Weakly Supervised Deep Learning for COVID-19 Infection Detection and Classification from CT Images. IEEE Access 2020, 8, 118869–118883. [Google Scholar] [CrossRef]
- Kang, H.; Xia, L.; Yan, F.; Wan, Z.; Shi, F.; Yuan, H.; Jiang, H.; Wu, D.; Sui, H.; Zhang, C.; et al. Diagnosis of Coronavirus Disease 2019 (COVID-19) with Structured Latent Multi-View Representation Learning. IEEE Trans. Med. Imaging 2020, 39, 2606–2614. [Google Scholar] [CrossRef]
- Mohammed, A.; Wang, C.; Zhao, M.; Ullah, M.; Naseem, R.; Wang, H.; Pedersen, M.; Alaya Cheikh, F. Semi-supervised Network for Detection of COVID-19 in Chest CT Scans. IEEE Access 2020, 8, 155987–156000. [Google Scholar] [CrossRef]
- Zebin, T.; Rezvy, S. COVID-19 detection and disease progression visualization: Deep learning on chest X-rays for classification and coarse localization. Appl. Intell. 2020, 50, 1010–1021. [Google Scholar] [CrossRef]
- Rawat, W.; Wang, Z. Deep Convolutional Neural Networks for Image Classification: A Comprehensive Review. Neural Comput. 2017, 29, 2352–2449. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cao, Y.; Zha, Z.; Zhang, J.; Xiong, Z. Deep Degradation Prior for Low-Quality Image Classification. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 11046–11055. [Google Scholar]
- Hamida, A.B.; Benoit, A.; Lambert, P.; Amar, C.B. 3-D Deep Learning Approach for Remote Sensing Image Classification. IEEE Trans. Getsci. Remote Sens. 2018, 56, 4420–4434. [Google Scholar] [CrossRef]
- Peterson, J.C.; Battleday, R.M.; Griffiths, T.L.; Russakovsky, O. Human uncertainty makes classification more robust. In Proceedings of the International Conference on Computer Vision, Seoul, Korea, 27 October–2 November 2019; pp. 9617–9626. [Google Scholar]
- Agustsson, E.; Uijlings, J.R.; Ferrari, V. Interactive Full Image Segmentation by Considering All Regions Jointly. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 11622–11631. [Google Scholar]
- Ahn, J.; Cho, S.; Kwak, S. Weakly Supervised Learning of Instance Segmentation with Inter-pixel Relations. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 2209–2218. [Google Scholar]
- Wang, D.; Su, J.; Yu, H. Feature Extraction and Analysis of Natural Language Processing for Deep Learning English Language. IEEE Access 2020, 8, 46335–46345. [Google Scholar] [CrossRef]
- Young, T.; Hazarika, D.; Poria, S.; Cambria, E. Recent Trends in Deep Learning Based Natural Language Processing. arXiv 2018, arXiv:1708.02709v5. Available online: https://arxiv.org/abs/1708.02709v5 (accessed on 2 January 2021).
- Farooq, M.; Hafeez, A. Covid-resnet: A Deep Learning Framework for Screening of Covid19 from Radiographs. arXiv 2020, arXiv:2003.14395. Available online: https://arxiv.org/abs/2003.14395 (accessed on 2 January 2021).
- Hemdan, E.E.; Shouman, M.A.; Karar, M.E. Covidx-net: A Framework of Deep Learning Classifiers to Diagnose Covid-19 in X-ray Images. arXiv 2020, arXiv:2003.11055. Available online: https://arxiv.org/abs/2003.11055 (accessed on 2 January 2021).
- He, X.; Yang, X.; Zhang, S.; Zhao, J.; Xie, P. Sample-Efficient Deep Learning for COVID-19 Diagnosis Based on CT Scans. MedRxiv 2020. [Google Scholar] [CrossRef]
- Abdani, S.R.; Zulkifley, M.A.; Zulkifley, N.H. A Lightweight Deep Learning Model for COVID-19 Detection. In Proceedings of the IEEE Symposium on Industrial Electronics & Applications, TBD, Malaysia, 17–18 July 2020; pp. 1–5. [Google Scholar]
- Masood, A.; Sengur, A. Deep Learning Approaches for COVID-19 Detection Based on Chest X-ray Images. Expert Syst. Appl. 2021, 164, 1–11. [Google Scholar]
- Shi, F.; Wang, J.; Shi, J.; Wu, Z.; Wang, Q.; Tang, Z.; He, K.; Shi, Y.; Shen, D. Review of Artificial Intelligence Techniques in Imaging Data Acquisition, Segmentation and Diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2020, 14, 4–5. [Google Scholar] [CrossRef]
- Guiot, J.; Vaidyanathan, A.; Deprez, L.; Zerka, F.; Danthine, D.; Frix, A.-N.; Thys, M.; Henket, M.; Canivet, G.; Mathieu, S.; et al. Development and Validation of an Automated Radiomic CT Signature for Detecting COVID-19. Diagnostics 2021, 11, 41. [Google Scholar] [CrossRef]
- Alom, M.; Rahman, M.; Nasrin, M.; Taha, T.; Asari, V. COVID_MTNet: COVID-19 Detection with Multi-Task Deep Learning Approaches. arXiv 2020, arXiv:2004.03747. Available online: https://arxiv.org/abs/2004.03747 (accessed on 2 January 2021).
- Zhi, Q.; Austin, B.; Glass, L.; Xiao, C.; Sun, J. FLANNEL: Focal Loss Based Neural Network Ensemble for COVID-19 Detection. J. Am. Med. Inform. Assoc. 2020, 1–9. [Google Scholar] [CrossRef]
- Shan, F.; Gao, Y.; Wang, J.; Shi, W.; Shi, N.; Han, M.; Xue, Z.; Shi, Y. Lung Infection Quantification of COVID-19 in CT Images with Deep Learning. arXiv 2020, arXiv:2003.04655. Available online: https://arxiv.org/abs/2003.04655 (accessed on 2 January 2021).
- Zhang, P.; Zhong, Y.; Deng, Y.; Tang, X.; Li, X. CoSinGAN: Learning COVID-19 Infection Segmentation from a Single Radiologi-cal Image. Diagnostics 2020, 10, 901. [Google Scholar] [CrossRef]
- Li, L.; Qin, L.; Xu, Z.; Yin, Y.; Wang, X.; Kong, B.; Bai, J.; Lu, Y.; Fang, Z.; Song, Q.; et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology 2020, 296, 2. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Wang, B.; Xu, H.; Luo, C.; Wei, L.; Zhao, W.; Hou, X.; Ma, W.; Xu, Z.; Zheng, Z.; et al. AI-Assisted CT Imaging Analysis for COVID-19 Screening: Building and Deploying a Medical AI System in Four Weeks. MedRxiv 2020. [Google Scholar] [CrossRef]
- Huang, L.; Han, R.; Ai, T.; Yu, P.; Kang, H.; Tao, Q.; Xia, L. Serial quantitative chest CT assessment of COVID-19: Deep-Learning Approach. Radiol. Cardiothorac. Imaging 2020, 2, e200075. [Google Scholar] [CrossRef]
- Gao, Y.; Xu, Z.; Feng, J.; Jin, C.; Han, X.; Wu, H.; Shi, H. Longitudinal assessment of COVID-19 using a deep learning–based quantitative CT pipeline: Illustration of two cases. Radiol. Cardiothorac. Imaging 2020, 2, e200082. [Google Scholar]
- Lipton, Z. The Mythos of Model Interpretability. Commun. ACM 2016, 61. [Google Scholar] [CrossRef]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning Deep Features for Discriminative Localization. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2921–2929. [Google Scholar]
- Selvaraju, R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-based Localization. In Proceedings of the International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Tiulpin, A.; Thevenot, J.; Rahtu, E.; Lehenkari, P.; Saarakkala, S. Automatic Knee Osteoarthritis Diagnosis from Plain Radiographs: A Deep Learning-Based Approach. Sci. Rep. 2018, 8, 1727. [Google Scholar] [CrossRef] [PubMed]
- Panwar, H.; Gupta, P.; Siddiqui, M.; Morales-Menendez, R.; Bhardwaj, P.; Singh, V. A Deep Learning and Grad-CAM based Color Visualization Approach for Fast Detection of COVID-19 Cases using Chest X-ray and CT-Scan Images. Chaos Solitons Fractals 2020, 140, 110190. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.; Singh, S.; Guestrin, C. Why Should I Trust You? Explaining the Predictions of Any Classifier. In Proceedings of the ACM SIGKDD, San Francisco, CA, USA, 13–17 August 2016. [Google Scholar]
- Xu, Y.; Jia, Z.; Wang, L.; Ai, Y.; Zhang, F.; Lai, M.; Chang, E. Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinform. 2017, 18, 281. [Google Scholar] [CrossRef] [PubMed]
- Rieke, J.; Eitel, F.; Weygandt, M.; Haynes, J.; Ritter, K. Visualizing Convolutional Networks for MRI-based Diagnosis of Alzheimer’s Disease. arXiv 2018, arXiv:1808.02874. Available online: https://arxiv.org/abs/1808.02874 (accessed on 2 January 2021).
- Krizhevsky, A.; Sutskever, I.; Hinton, G. ImageNet classification with deep convolutional neural networks. In Proceedings of the Conference and Workshop on Neural Information Processing Systems, Harrahs and Harveys, Lake Tahoe, NV, USA, 3–6 December 2012; pp. 1097–1105. [Google Scholar]
- Lu, S.; Lu, Z.; Zhang, Y. Pathological brain detection based on AlexNet and transfer learning. J. Comput. Sci. 2019, 30, 41–47. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.; Li, K.; Li, F. ImageNet: A large-scale hierarchical image database. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Qin, Z.; Yu, F.; Liu, C.; Chen, X. How convolutional neural networks see the world—A survey of convolutional neural network visualization methods. Math. Found. Comput. 2018, 1, 149–180. [Google Scholar] [CrossRef]
- Springenberg, J.; Dosovitskiy, A.; Brox, T.; Riedmiller, M. Striving for Simplicity: The All Convolutional Net. arXiv 2015, arXiv:1412.6806. Available online: https://arxiv.org/abs/1412.6806 (accessed on 2 January 2021).
- Ozbulak, U. PyTorch CNN Visualizations. Github, 2019. Available online: https://github.com/utkuozbulak/pytorch-cnn-visualizations (accessed on 3 February 2021).


| Method | ACC (%) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|
| LBP + SVM | 86.45 | 88.90 | 83.36 | 87.02 | 85.69 |
| Deep feature + SVM | 93.26 | 91.43 | 95.61 | 96.39 | 89.70 |
| Modified Alexnet | 94.75 | 93.22 | 96.69 | 97.27 | 91.88 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wan, Y.; Zhou, H.; Zhang, X. An Interpretation Architecture for Deep Learning Models with the Application of COVID-19 Diagnosis. Entropy 2021, 23, 204. https://doi.org/10.3390/e23020204
Wan Y, Zhou H, Zhang X. An Interpretation Architecture for Deep Learning Models with the Application of COVID-19 Diagnosis. Entropy. 2021; 23(2):204. https://doi.org/10.3390/e23020204
Chicago/Turabian StyleWan, Yuchai, Hongen Zhou, and Xun Zhang. 2021. "An Interpretation Architecture for Deep Learning Models with the Application of COVID-19 Diagnosis" Entropy 23, no. 2: 204. https://doi.org/10.3390/e23020204
APA StyleWan, Y., Zhou, H., & Zhang, X. (2021). An Interpretation Architecture for Deep Learning Models with the Application of COVID-19 Diagnosis. Entropy, 23(2), 204. https://doi.org/10.3390/e23020204







