Residue Cluster Classes: A Unified Protein Representation for Efficient Structural and Functional Classification
Abstract
1. Introduction
2. Materials and Methods
2.1. RCC Calculation Implementation
| Algorithm 1: RCC calculation |
| Input: Cartesian coordinates of protein atoms |
| Output: RCC coordinates |
| 1: Generate graph with the contacts of all residues. Two residues are in contact if they are within a distance threshold. |
| 2: Calculate maximal cliques using Tomita algorithm (see below) |
| 3: Calculate RCC from maximal cliques. |
2.1.1. Contact Map Calculation
| Algorithm 2: Contact map calculation |
| Input: Cartesian coordinates of protein atoms |
| Output: Contact graph |
| For each residue r in the protein: |
| Calculate its cube in the grid Gr using the hash function. |
| Get the list of residues L inside the cubes neighboring Gr. |
| For each residue s in L: |
| Calculate D distance between rc and sc. |
| Quick inclusion if D < d and continue. |
| Quick exclusion if minimum {D–rd, D–sd} > d and continue. |
| Perform contact check for each pair of atoms from r and s. |
2.1.2. Maximal Cliques Calculation
2.1.3. RCC Calculation
2.2. RCC Database
2.3. Model Training and Testing
3. Results
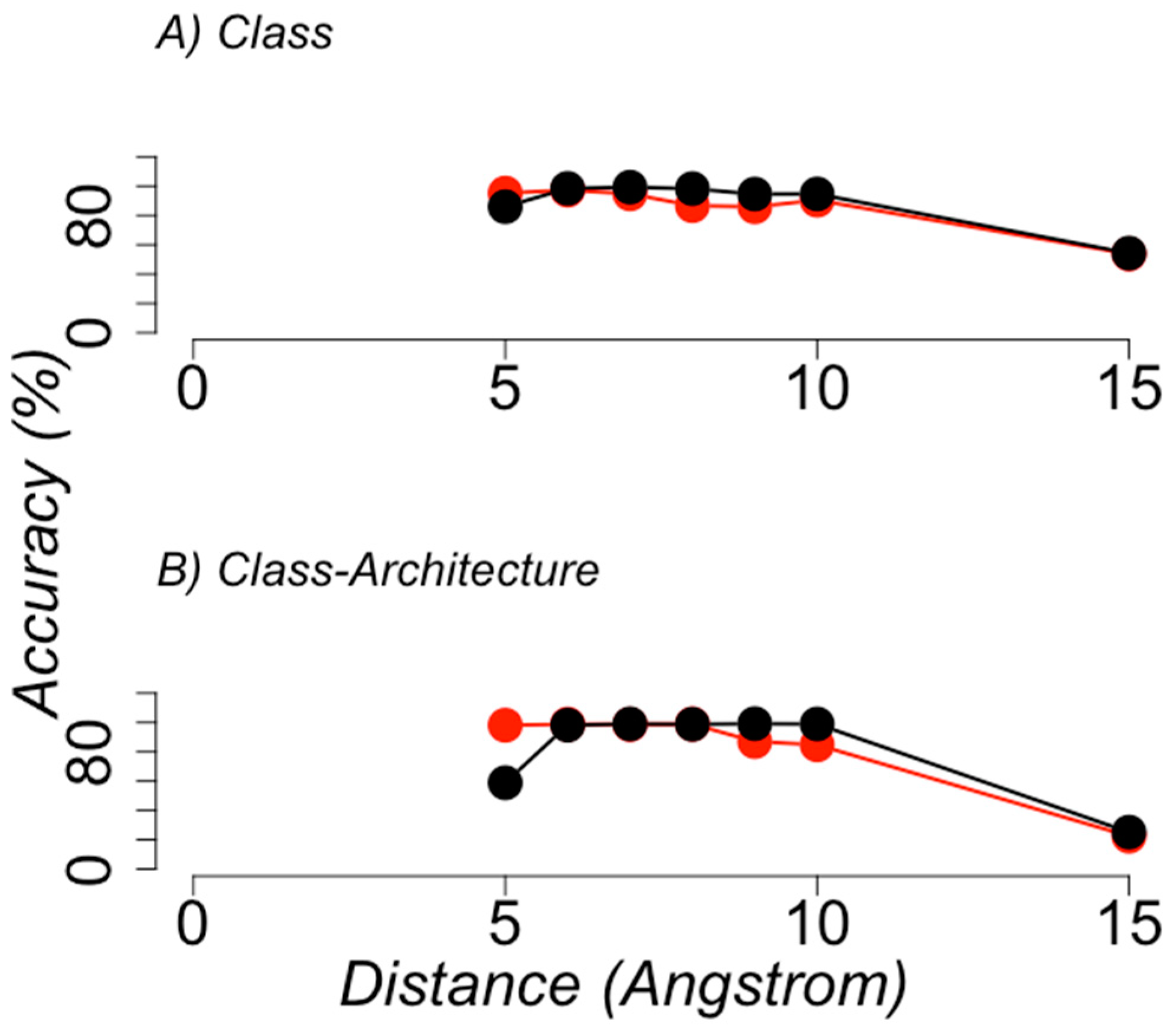
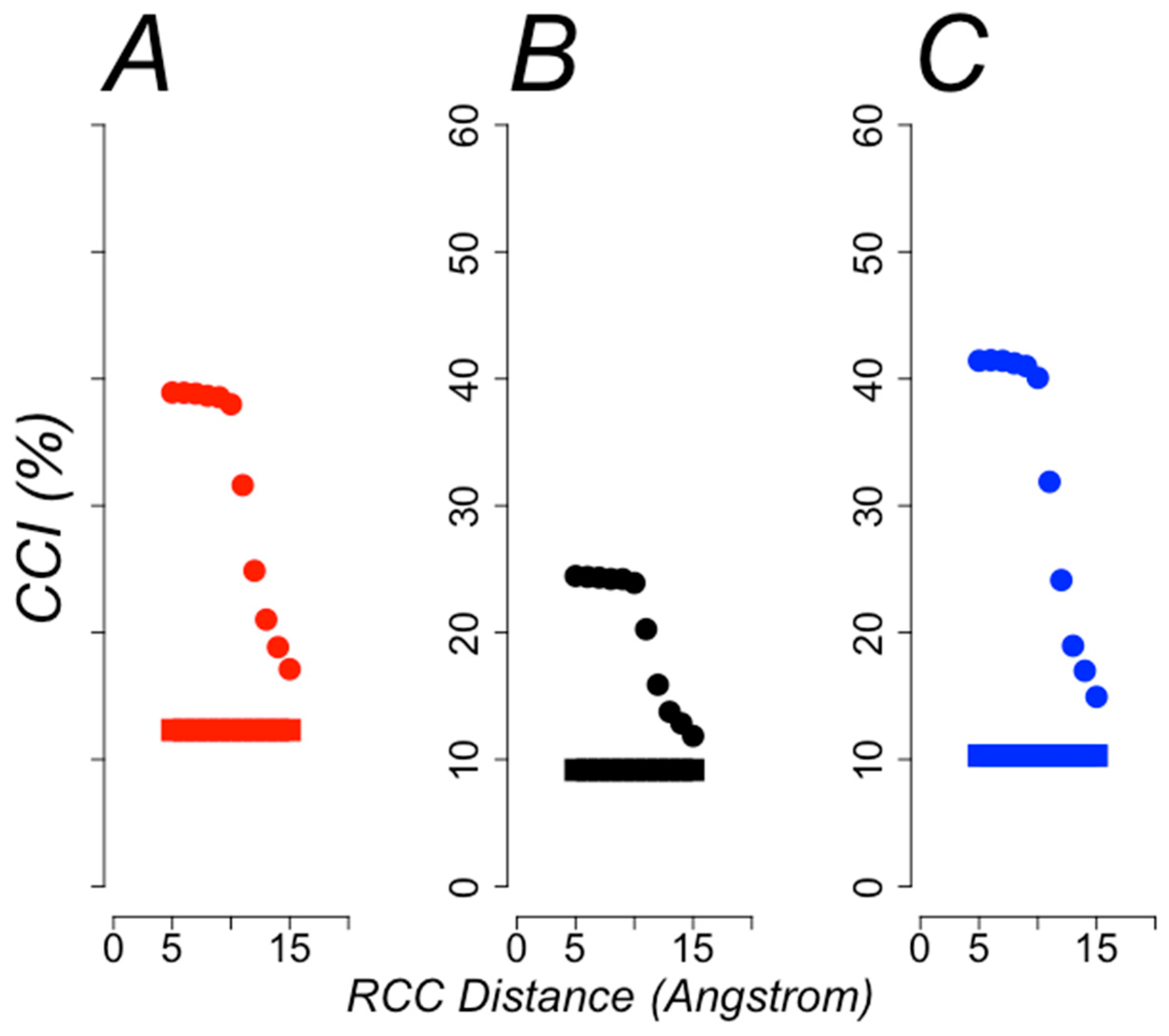
3.1. Protein Structural Classification
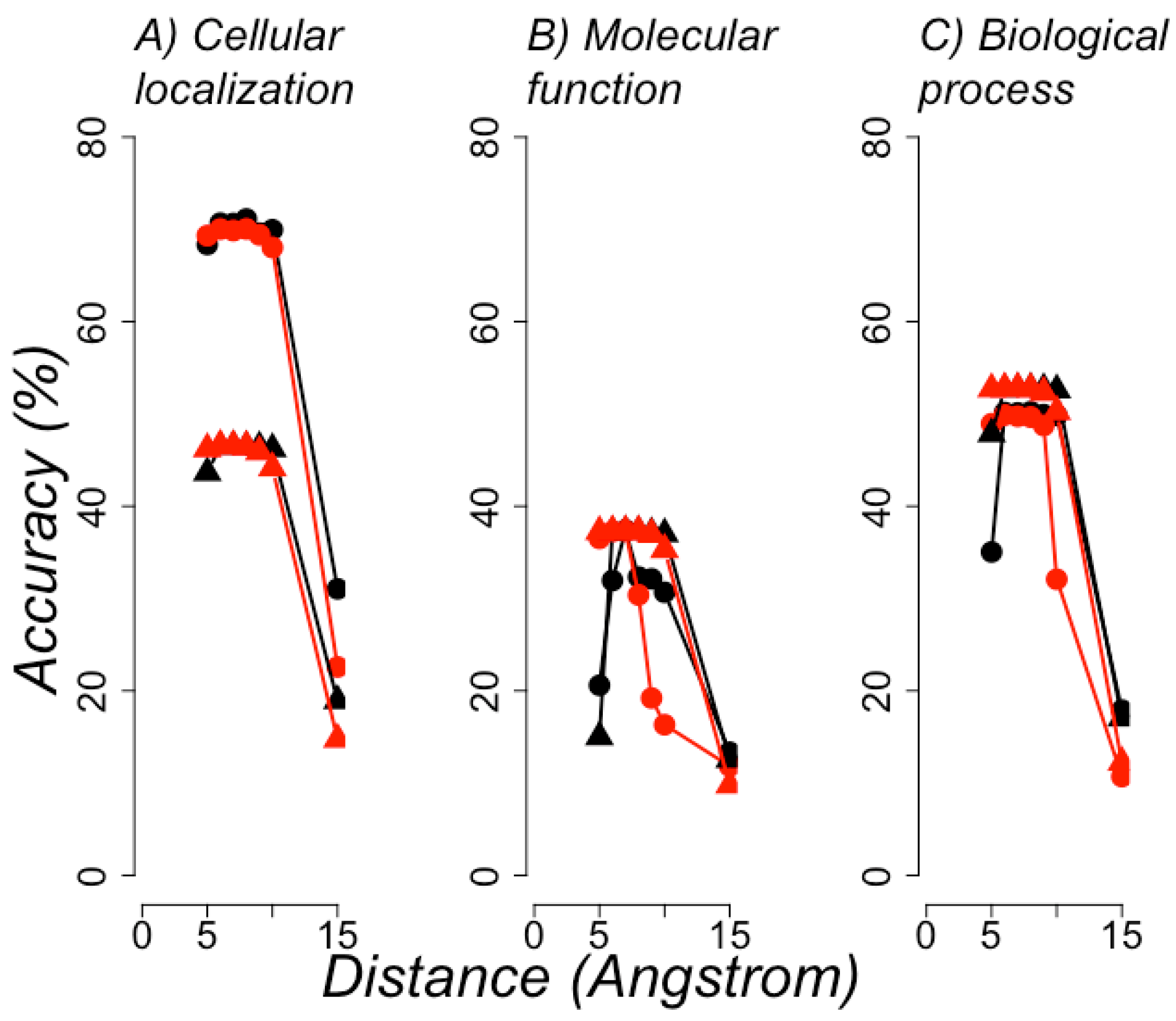
3.2. Protein Functional Classification
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Baker, D. Protein Structure Prediction and Structural Genomics. Science 2001, 294, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, R.; Archana, A.; Thangakani, A.M.; Jemimah, S.; Velmurugan, D.; Gromiha, M.M. PDBparam: Online Resource for Computing Structural Parameters of Proteins. Bioinform. Boil. Insights 2016, 10, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.M. The Proteomics Protocols Handbook; Humana Press, Inc.: Totowa, NJ, USA, 2005. [Google Scholar] [CrossRef]
- Zhang, Y.; Wen, J.; Yau, S.S.-T. Phylogenetic analysis of protein sequences based on a novel k-mer natural vector method. Genomics 2019, 111, 1298–1305. [Google Scholar] [CrossRef] [PubMed]
- Juan, D.; Pazos, F.; Valencia, A. Emerging methods in protein co-evolution. Nat. Rev. Genet. 2013, 14, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Sahraeian, S.M.; Luo, K.R.; Brenner, S.E. SIFTER search: A web server for accurate phylogeny-based protein function prediction. Nucleic Acids Res. 2015, 43, W141–W147. [Google Scholar] [CrossRef] [PubMed]
- AlQuraishi, M. AlphaFold at CASP13. Bioinformatics 2019, 35, 4862–4865. [Google Scholar] [CrossRef]
- Zhou, N.; Jiang, Y.; Bergquist, T.R.; Lee, A.J.; Kacsoh, B.Z.; Crocker, A.W.; Lewis, K.A.; Georghiou, G.; Nguyen, H.N.; Hamid, N.; et al. The CAFA challenge reports improved protein function prediction and new functional annotations for hundreds of genes through experimental screens. Genome Boil. 2019, 20, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Kulmanov, M.; Hoehndorf, R. DeepGOPlus: Improved protein function prediction from sequence. Bioinformatics 2019, 36, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, N.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2014, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Corral, R.C.; Chávez, E.; Del Rio, G. Machine Learnable Fold Space Representation based on Residue Cluster Classes. Comput. Boil. Chem. 2015, 59, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Vehlow, C.; Stehr, H.; Winkelmann, M.; Duarte, J.M.; Petzold, L.; Dinse, J.; Lappe, M. CMView: Interactive contact map visualization and analysis. Bioinformatics 2011, 27, 1573–1574. [Google Scholar] [CrossRef] [PubMed]
- Geng, C. DrawGridBox-PyMOLWiki. 2016. Available online: https://pymolwiki.org/index.php/DrawGridBox (accessed on 26 February 2020).
- Tomita, E.; Tanaka, A.; Takahashi, H. The worst-case time complexity for generating all maximal cliques and computational experiments. Theor. Comput. Sci. 2006, 363, 28–42. [Google Scholar] [CrossRef]
- Eppstein, D.; Löffler, M.; Strash, D. Listing All Maximal Cliques in Sparse Graphs in Near-Optimal Time. In Computer Vision; Springer Science and Business Media LLC: Berlin, Germany, 2010; Volume 6506, pp. 403–414. [Google Scholar]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Kotthoff, L.; Thornton, C.; Hoos, H.H.; Hutter, F.; Leyton-Brown, K. Auto-WEKA: Automatic Model Selection and Hyperparameter Optimization in WEKA. The NIPS ’17 Competition Build. Intell. Syst. 2019, 18, 81–95. [Google Scholar]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA data mining software. ACM SIGKDD Explor. Newsl. 2009, 11, 10–18. [Google Scholar] [CrossRef]
- Burley, S.K.; Berman, H.M.; Bhikadiya, C.; Bi, C.; Chen, L.; Di Costanzo, L.; Christie, C.; Dalenberg, K.; Duarte, J.M.; Dutta, S.; et al. RCSB Protein Data Bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2018, 47, D464–D474. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Oron, T.R.; Clark, W.T.; Bankapur, A.R.; D’Andrea, D.; Lepore, R.; Funk, C.S.; Kahanda, I.; Verspoor, K.M.; Ben-Hur, A.; et al. An expanded evaluation of protein function prediction methods shows an improvement in accuracy. Genome Boil. 2016, 17, 184. [Google Scholar] [CrossRef] [PubMed]
- Ching; Choudhary; Liao, W.-K.; Ross; Gropp, W. Efficient structured data access in parallel file systems. In Proceedings of the IEEE International Conference on Cluster Computing CLUSTR-03, Hong Kong, China, 1–4 December 2003; pp. 326–335. [Google Scholar] [CrossRef]
- Markov, I.L. Limits on fundamental limits to computation. Nature 2014, 512, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, C. Actor Model of Computation. 2010. Available online: http://arxiv.org/abs/1008.1459http://carlhewitt.info (accessed on 3 February 2020).
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Žídek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Improved protein structure prediction using potentials from deep learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef] [PubMed]
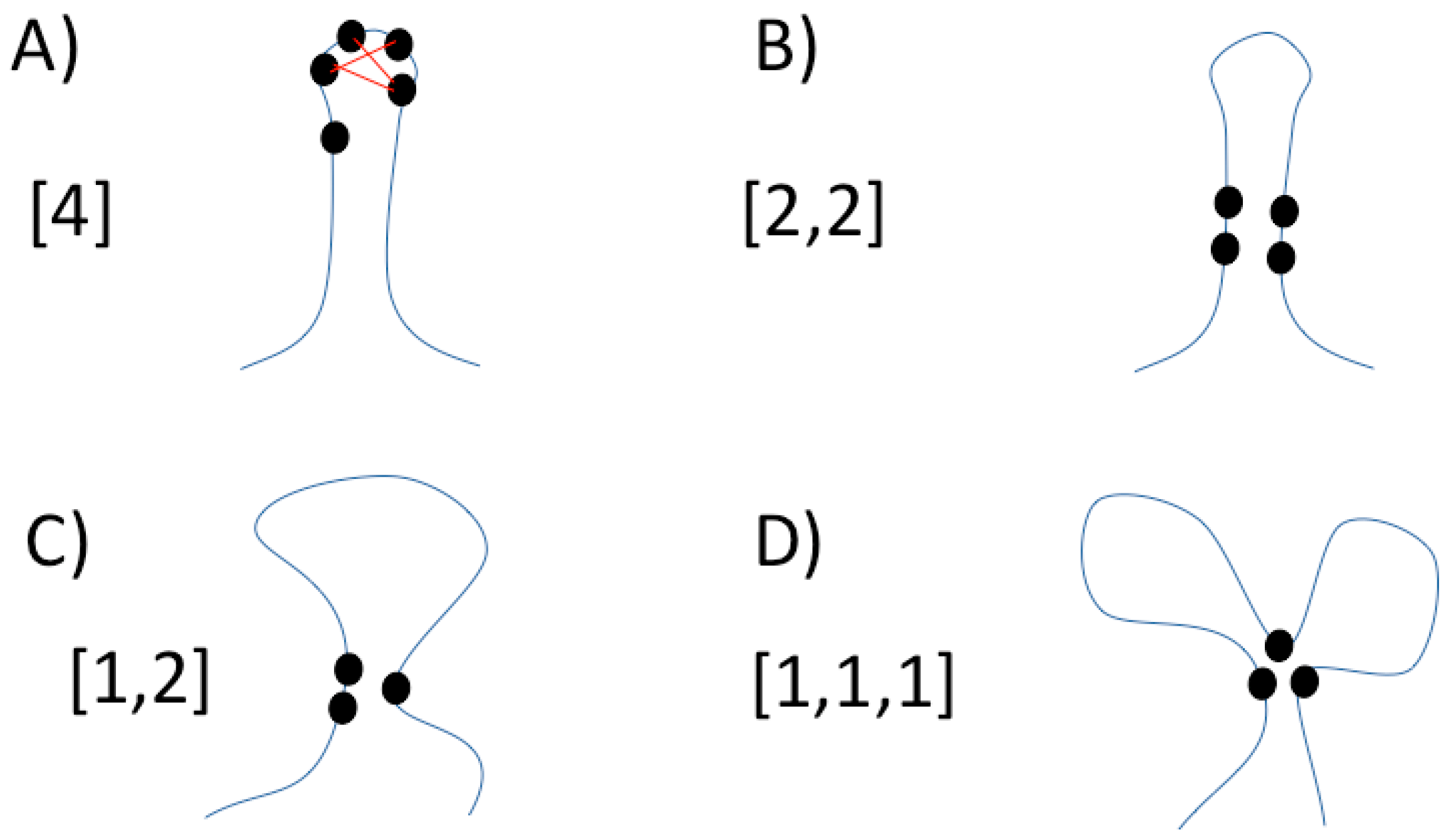
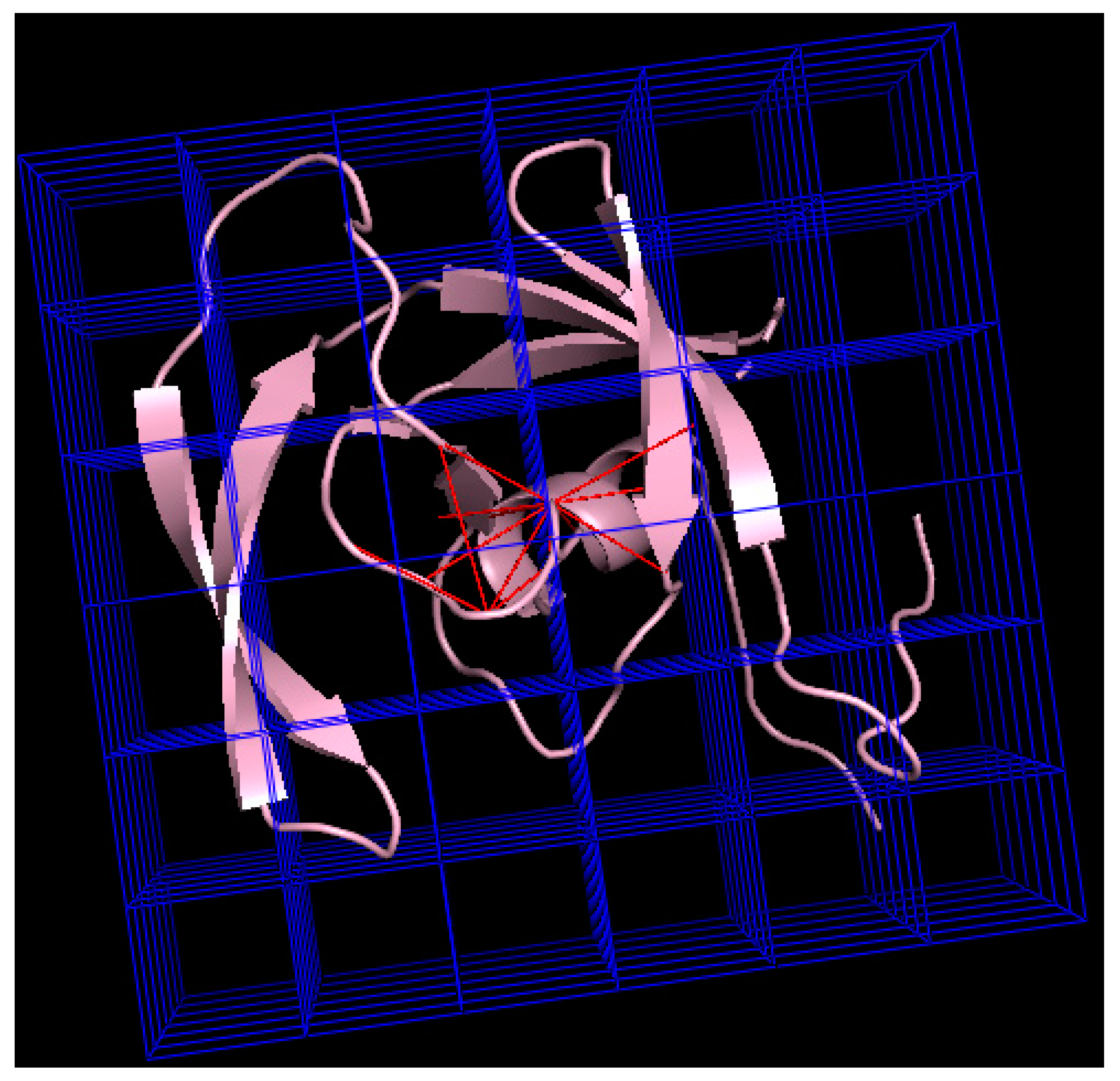
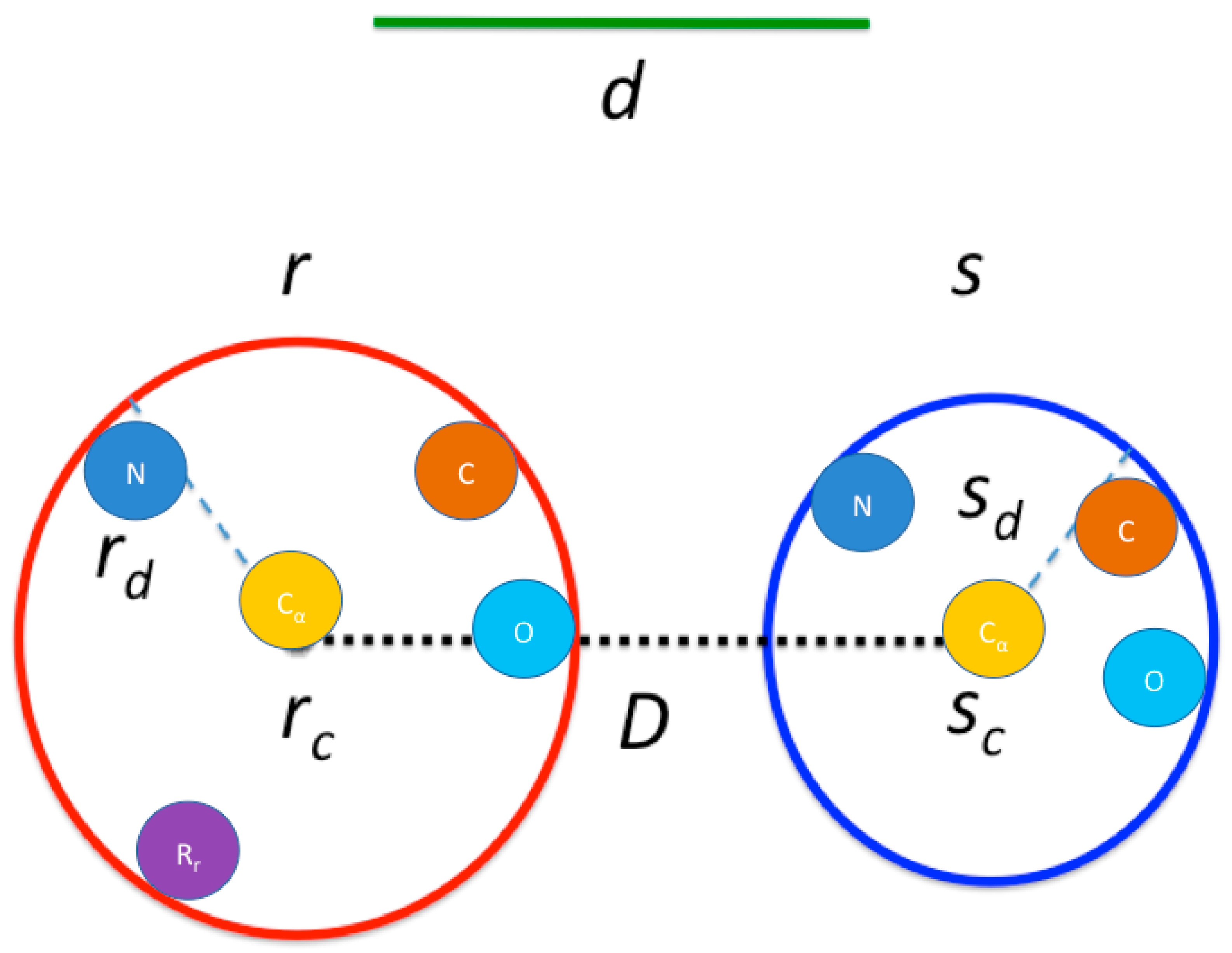
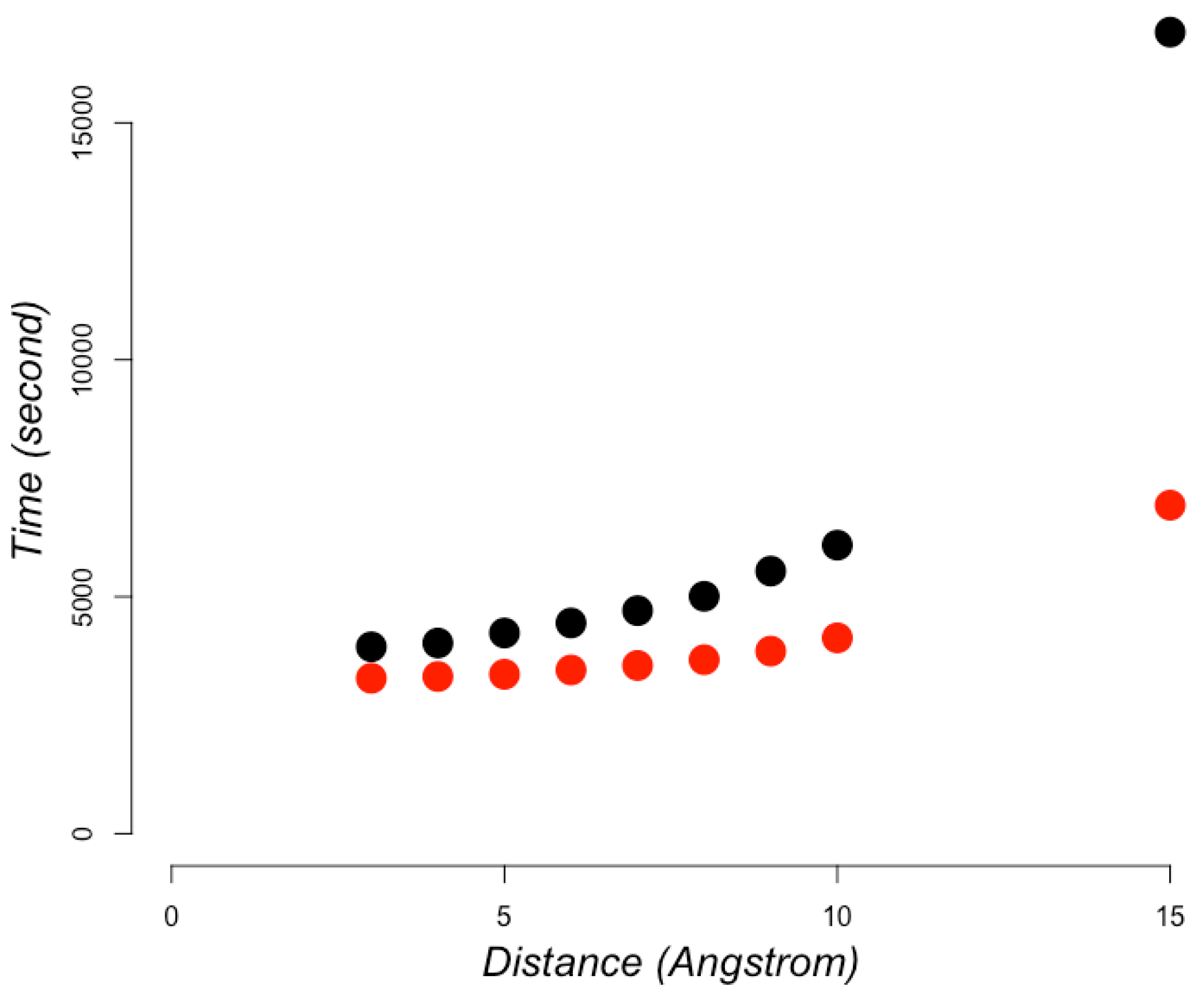
| PDB ID* | Length** | Reading*** (ms) | Graph Calculation (ms) | Maximal Cliques (ms) | Total Time (ms) |
|---|---|---|---|---|---|
| 1ORN (A) | 214 | 39 | 19 | 6 | 64 |
| 2HOX (A) | 425 | 110 | 37 | 6 | 153 |
| 3GVK (A) | 644 | 202 | 47 | 6 | 255 |
| 1F8N (A) | 818 | 329 | 67 | 6 | 402 |
| CATH Level | Mean Cross-Validation Accuracy * (Corral et al) | Mean Cross-Validation Accuracy ** (Current) |
|---|---|---|
| C | 0.96 | 0.98 |
| A | 0.88 | 0.89 |
| GO Function | CAFA2* Fmax | Fmax** | Fmax*** |
|---|---|---|---|
| C | 0.46 | 0.44 | 0.58 |
| F | 0.59 | 0.24 | 0.48 |
| P | 0.37 | 0.41 | 0.54 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fontove, F.; Del Rio, G. Residue Cluster Classes: A Unified Protein Representation for Efficient Structural and Functional Classification. Entropy 2020, 22, 472. https://doi.org/10.3390/e22040472
Fontove F, Del Rio G. Residue Cluster Classes: A Unified Protein Representation for Efficient Structural and Functional Classification. Entropy. 2020; 22(4):472. https://doi.org/10.3390/e22040472
Chicago/Turabian StyleFontove, Fernando, and Gabriel Del Rio. 2020. "Residue Cluster Classes: A Unified Protein Representation for Efficient Structural and Functional Classification" Entropy 22, no. 4: 472. https://doi.org/10.3390/e22040472
APA StyleFontove, F., & Del Rio, G. (2020). Residue Cluster Classes: A Unified Protein Representation for Efficient Structural and Functional Classification. Entropy, 22(4), 472. https://doi.org/10.3390/e22040472






