Abstract
This paper proposes a geometric estimator of dependency between a pair of multivariate random variables. The proposed estimator of dependency is based on a randomly permuted geometric graph (the minimal spanning tree) over the two multivariate samples. This estimator converges to a quantity that we call the geometric mutual information (GMI), which is equivalent to the Henze–Penrose divergence. between the joint distribution of the multivariate samples and the product of the marginals. The GMI has many of the same properties as standard MI but can be estimated from empirical data without density estimation; making it scalable to large datasets. The proposed empirical estimator of GMI is simple to implement, involving the construction of an minimal spanning tree (MST) spanning over both the original data and a randomly permuted version of this data. We establish asymptotic convergence of the estimator and convergence rates of the bias and variance for smooth multivariate density functions belonging to a Hölder class. We demonstrate the advantages of our proposed geometric dependency estimator in a series of experiments.
1. Introduction
Estimation of multivariate dependency has many applications in fields such as information theory, clustering, structure learning, data processing, feature selection, time series prediction, and reinforcement learning, see [1,2,3,4,5,6,7,8,9,10], respectively. It is difficult to accurately estimate the mutual information in high-dimensional settings, specially where the data is multivariate with an absolutely continuous density with respect to Lebesgue measure—the setting considered in this paper. An important and regular measure of dependency is the Shannon mutual information (MI), which has seen extensive use across many application domains. However, the estimation of mutual information can often be challenging. In this paper, we focus on a measure of MI that we call the Geometric MI (GMI). This MI measure is defined as the asymptotic large sample limit of a randomized minimal spanning tree (MST) statistic spanning the multivariate sample realizations. The GMI is related to a divergence measure called the Henze–Penrose divergence [11,12], and related to the multivariate runs test [13]. In [14,15], it was shown that this divergence measure can be used to specify a tighter bound for the Bayes error rate for testing if a random sample comes from one of two distributions the bound in [14,15] is tighter than previous divergence-type bounds such as the Bhattacharrya bound [16]. Furthermore, the authors of [17] proposed a non-parametric bound on multi-class classification Bayes error rate using a global MST graph.
Let and be random variables with unknown joint density and marginal densities and , respectively, and consider two hypotheses: , and are independent and , and are dependent,
The GMI is defined as the Henze–Penrose divergence between and which can be used as a dependency measure. In this paper, we prove that for large sample size n the randomized MST statistic spanning the original multivariate sample realizations and a randomly shuffled data set converges almost surely to the GMI measure. A direct implication of [14,15] is that the GMI provides a tighter bound on the Bayes misclassification rate for the optimal test of independence. In this paper, we propose an estimator based on a random permutation modification of the Friedman–Rafsky multivariate test statistic and show that under certain conditions the GMI estimator achieves the parametric mean square error (MSE) rate when the joint density is bounded and smooth. Importantly unlike other measures of MI, our proposed GMI estimator does not require explicit estimation of the joint and marginal densities.
Computational complexity is an important challenge in machine learning and data science. Most plug-in-based estimators, such as the kernel density estimator (KDE) or the K-nearest-neighbor (KNN) estimator with known convergence rate, require runtime complexity of , which is not suitable for large scale applications. Noshad et al. proposed a graph theoretic direct estimation method based on nearest-neighbor ratios (NNR) [18]. The NNR estimator is based on k-NN graph and computationally more tractable than other competing estimators with complexity . The construction of the minimal spanning tree lies at the heart of the GMI estimator proposed in this paper. Since the GMI estimator is based on the Euclidean MST the dual-tree algorithm by March et al. [19] can be applied. This algorithm is based on the construction of Borůvka [20] and implements the Euclidean MST in approximately time. In this paper, we experimentally show that for large sample size the proposed GMI estimator has faster runtime than the KDE plug-in method.
1.1. Related Work
Estimation of mutual information has a rich history. The most common estimators of MI are based on plug-in density estimation, e.g., using the histogram, kernel density or kNN density estimators [21,22]. Motivated by ensemble methods applied to divergence estimation [23,24], in [22] an ensemble method for combining multiple KDE bandwidths was proposed for estimating MI. Under certain smoothness conditions this ensemble MI estimator was shown to achieve parametric convergence rates.
Another class of estimators of multivariate dependency bypasses the difficult density estimation task. This class includes the statistically consistent estimators of Rényi- and KL mutual information which are motivated by the asymptotic limit of the length of the KNN graph, [25,26] when joint density is smooth. The estimator of [27] builds on KNN methods for Rényi entropy estimation. The authors of [26], showed that when MI is large the KNN and KDE approaches are ill-suited for estimating MI since the joint density may be insufficiently smooth when there are strong dependencies. To overcome this issue an assumption on the smoothness of the density is required, see [28,29], and [23,24]. For all these methods, the optimal parametric rate of MSE convergence is achieved when the densities are either d, or times differentiable [30]. In this paper, we assume that joint and marginal densities are smooth in the sense that they belong to Hölder continuous classes of densities , where the smoothness parameter and the Lipschitz constant .
A MI measure based on the Pearson chi-square divergence was considered in [31] that is computational efficient and numerically stable. The authors of [27,32] used nearest-neighbor graph and minimal spanning tree approaches, respectively, to estimate Rényi mutual information. In [22], a non-parametric mutual information estimator was proposed using a weighted ensemble method with parametric convergence rate. This estimator was based on plug-in density estimation, which is challenging in high dimension.
Our proposed dependency estimator differs from previous methods in the following ways. First, it estimates a different measure of mutual information, the GMI. Second, instead of using the KNN graph the estimator of GMI uses a randomized minimal spanning tree that spans the multivariate realizations. The proposed GMI estimator is motivated by the multivariate runs test of Friedman and Rafsky (FR) [33] which is a multivariate generalization of the univariate Smirnov maximum deviation test [34] and the Wald-Wolfowitz [35] runs test in one dimension. We also emphasize that the proposed GMI estimator does not require boundary correction, in contrast to other graph-based estimators, such as, the NNR estimator [18], scalable MI estimator [36], or cross match statistic [37].
1.2. Contribution
The contribution of this paper has three components
- (1)
- We propose a novel non-parametric multivariate dependency measure, referred to as geometric mutual information (GMI), which is based on graph-based divergence estimation. The geometric mutual information is constructed using a minimal spanning tree and is a function of the Friedman–Rafsky multivariate test statistic.
- (2)
- We establish properties of the proposed dependency measure analogous to those of Shannon mutual information, such as, convexity, concavity, chain rule, and a type of data-processing inequality.
- (3)
- We derive a bound on the MSE rate for the proposed geometric estimator. An advantage of the estimator is that it achieves the optimal MSE rate without the need for boundary correction, which is required for most plug-in estimators.
1.3. Organization
The rest of the paper is organized as follows. In Section 2, we define the geometric mutual information and establish some of its mathematical properties. In Section 2.2 and Section 2.3, we introduce a statistically consistent GMI estimator and derive a bound on its mean square error convergence rate. In Section 3 we verify the theory through experiments.
Throughout the paper, we denote statistical expectation by and the variance by abbreviation . Bold face type indicates random vectors. All densities are assumed to be absolutely continuous with respect to non-atomic Lebesgue measure.
2. The Geometric Mutual Information (GMI)
In this section, we first review the definition of the Henze–Penrose (HP) divergence measure defined by Berisha and Hero in [13,14]. The Henze–Penrose divergence between densities f and g with domain for parameter is defined as follows (see [13,14,15]):
where . This functional is an f-divergence [38], equivalently, as an Ali-Silvey distance [39], i.e., it satisfies the properties of non-negativity, monotonicity, and joint convexity [15]. The measure (1) takes values in and if and only if almost surely.
The mutual information measure is defined as follows. Let , , and be the marginal and joint distributions, respectively, of random vectors , where and are positive integers. Then by using (1), a Henze–Penrose generalization of the mutual information between and , is defined by
We will show below that has a geometric interpretation in terms of the large sample limit of a minimal spanning tree spanning n sample realizations of the merged labeled samples . Thus, we call the GMI between and . The GMI satisfies similar properties to other definitions of mutual information, such as Shannon and Rényi mutual information. Recalling (3) in [14], an alternative form of is given by
where
The function was defined in [13] and is called the geometric affinity between and . The next subsection of the paper is dedicated to the basic inequalities and properties of the proposed GMI measure (2).
2.1. Properties of the Geometric Mutual Information
In this subsection we establish basic inequalities and properties of the GMI, , given in (2). The following theorem shows that is a concave function in and a convex function in . The proof is given in Appendix A.1.
Theorem 1.
Denote by the GMI when and have joint density . Then the GMI satisfies
- (i)
- Concavity in : Let be conditional density of given and let and be densities on . Then for ,The inequality is strict unless either or are zero or .
- (ii)
- Convexity in : Let and be conditional densities of given and let be marginal density. Then for ,The inequality is strict unless either or are zero or .
The GMI, , satisfies properties analogous to the standard chain rule and the data-processing inequality [40]. For random variables , and with conditional density we define the conditional GMI
The next theorem establishes a relation between the joint and conditional GMI.
Theorem 2.
For given d-dimensional random vector with components and random variable Y,
where and the conditional GMI is defined in (7).
For Theorem 2 reduces to
Please note that when , the inequality (8) is trivial since . The proof of Theorem 2 is given in Appendix A.2. Theorem 2 is next applied to the case where and form a Markov chain. The proof of the following “leany” data-processing inequality (Proposition 1) is provided in Appendices section, Appendix A.3.
Proposition 1.
Suppose random vectors form a Markov chain denoted, , in the sense that . Then for
where
Furthermore, if both and together hold true, we have .
The inequality in (10) becomes interpretable as the standard data-processing inequality , when
since
2.2. The Friedman–Rafsky Estimator
Let a random sample from be available. Here we show that the GMI can be directly estimated without estimating the densities. The estimator is inspired by the MST construction of [33] that provides a consistent estimate of the Henze–Penrose divergence [14,15]. We denote by the i-th joint sample and by the sample set . Divide the sample set into two subsets and with the proportion and , where .
Denote by the set
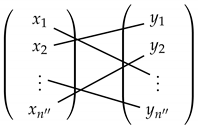

This means that for each given the first element the second element is replaced by a randomly selected . This results in a random shuffling of the binary relation relating in . The estimator of is derived based on the Friedman–Rafsky (FR) multivariate runs test statistic [33] on the concatenated data set, . The FR test statistic is defined as the number of edges in the MST spanning the merged data set that connect a point in to a point in . This test statistic is denoted by . Please note that since the MST is unique with probability one (under the assumption that all density functions are Lebesgue continuous) then all inter point distances between nodes are distinct. This estimator converges to almost surely as . The procedure is summarized in Algorithm 1.
| Algorithm 1: MST-based estimator of GMI |
| Input: Data set |
| 1: Find using arguments in Section 2.4 |
| 2: , |
| 3: Divide into two subsets and |
| 4: : shuffle first and second elements of pairs in |
| 5: |
| 6: Construct MST on |
| 7: edges connecting a node in to a node of |
| 8: |
| Output: , where |
Theorem 3 shows that the output in Algorithm 1 estimates the GMI with parameter . The proof is provided in Appendix A.4.
Theorem 3.
For given proportionality parameter , choose , such that and, as , we have and . Then
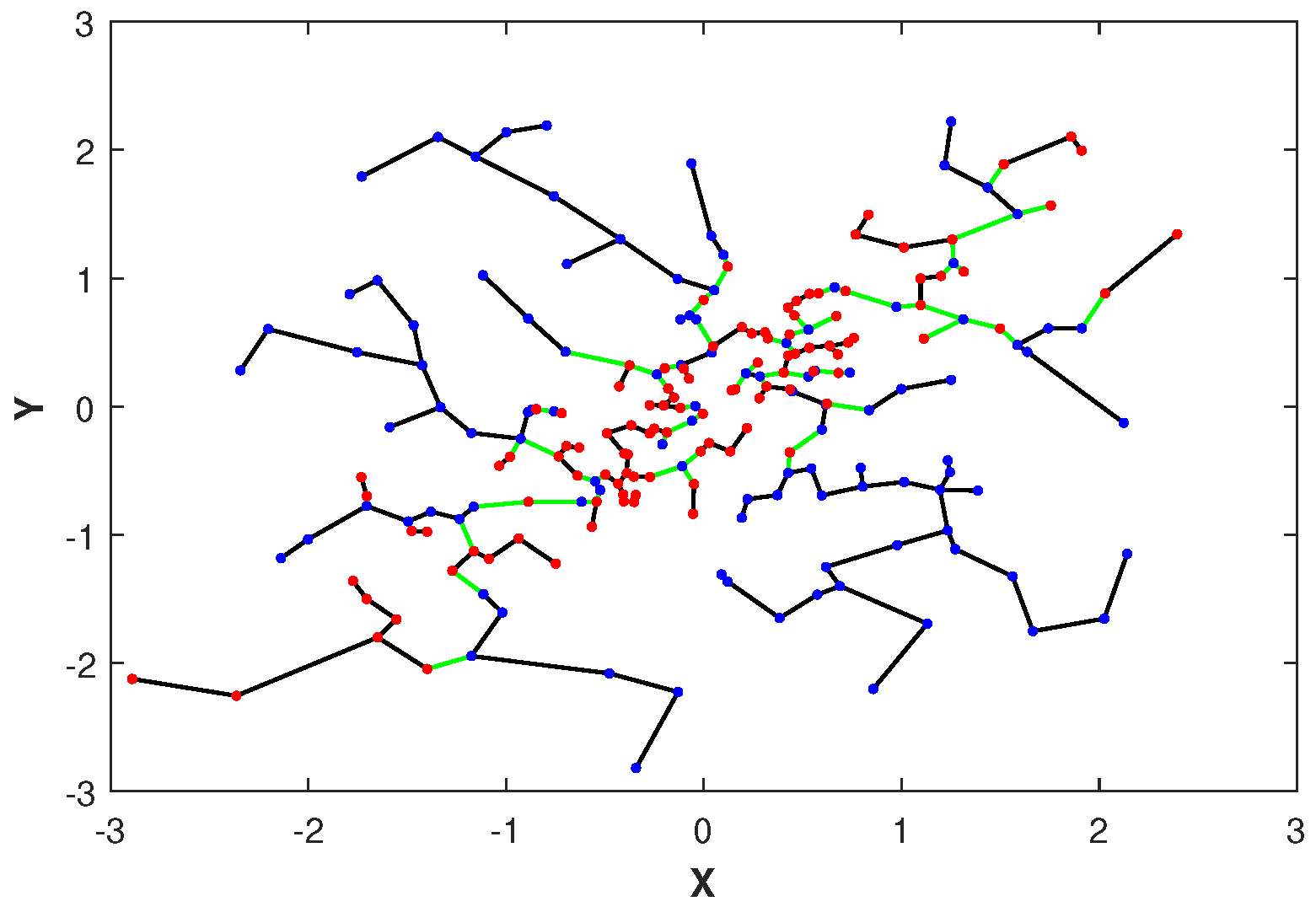
Please note that the asymptotic limit in (11) depends on the proportionality parameter . Later in Section 2.4, we discuss the choice of an optimal parameter . In Figure 1, we illustrate the MST constructed over merged independent () and highly dependent () data sets drawn from two-dimensional normal distributions with correlation coefficients . Notice that the edges of the MST connecting samples with different colors, corresponding to independent and dependent samples, respectively, are indicated in green. The total number of green edges is the FR test statistic .
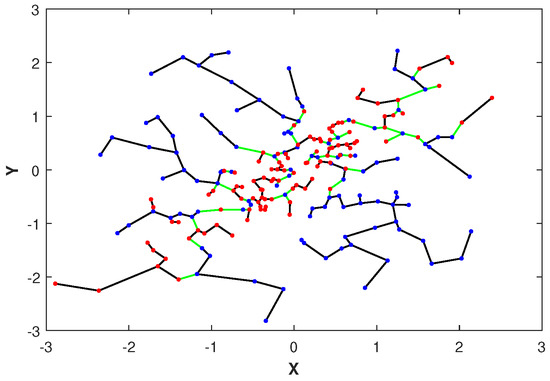
Figure 1.
The MST and FR statistic of spanning the merged set of normal points when and are independent (denoted in blue points) and when and are highly dependent (denoted in red points). The FR test statistic is the number of edges in the MST that connect samples from different color nodes (denoted in green) and it is used to estimate the GMI .
2.3. Convergence Rates
In this subsection we characterize the MSE convergence rates of the GMI estimator of Section 2.2 in the form of upper bounds on the bias and the variance. This MSE bound is given in terms of the sample size n, the dimension d, and the proportionality parameter . Deriving convergence rates for mutual information estimators has been of interest in information theory and machine learning [22,27]. The rates are typically derived in terms of a smoothness condition on the densities, such as the Hölder condition [41]. Here we assume , and have support sets , , and , respectively, and are smooth in the sense that they belong to Hölder continuous classes of densities K), [42,43]:
Definition 1.
(Hölder class): Let be a compact space. The Hölder class of functions , with Hölder parameters η and K, consists of functions g that satisfy
where is the Taylor polynomial (multinomial) of g of order k expanded about the point and is defined as the greatest integer strictly less than η.
To explore the optimal choice of parameter we require bounds on the bias and variance bounds, provided in Appendix A.5. To obtain such bounds, we will make several assumptions on the absolutely continuous densities , , and support sets , , :
- (A.1)
- Each of the densities belong to with smoothness parameters and Lipschitz constant K.
- (A.2)
- The volumes of the support sets are finite, i.e., .
- (A.3)
- All densities are bounded i.e., there exist two sets of constants and such that , and .
The following theorem on the bias follows under assumptions (A.1) and (A.3):
Theorem 4.
For given , , , and the bias of the satisfies
where is the largest possible degree of any vertex of MST on . The explicit form of (13) is provided in Appendix A.5.
Please note that according to Theorem 13 in [44], the constant is lower bounded by , and is the binary entropy i.e.,
A proof of Theorem 4 is given in Appendix A.5. The next theorem gives an upper bound on the variance of the FR estimator . The proof of the variance result requires a different approach than the bias bound (the Efron–Stein inequality [45]). It is similar to arguments in ([46], Appendix A.3), and is omitted. In Theorem 5 we assume that the densities , , and are absolutely continuous and bounded (A.3).
Theorem 5.
Given , the variance of the estimator is bounded by
where is a constant depending only on the dimension d.
2.4. Minimax Parameter
Recall assumptions (A.1), (A.2), and (A.3) in Section 2.3. The constant can be chosen to minimize the maximum the MSE converges rate where the maximum is taken over the space of Hölder smooth joint densities .
Throughout this subsection we use the following notations:
- ,
- and ,
- ,
- and , where is the smoothness parameter,
- .
Now define by
Consider the following optimization problem:
where
and
Please note that in (18), are constants, and only depends on the dimension d. Also, in (19), and are constants. Let be the optimal i.e., be the solution of the optimization problem (16). Set
such that is (17) when . For , the optimal choice of in terms of maximizing the MSE is and the saddle point for the parameter , denoted by , is given as follows:
- , if .
- , if .
- , if .
Further details are given in Appendix A.6.
3. Simulation Study
In this section, numerical simulations are presented that illustrate the theory in Section 2. We perform multiple experiments to demonstrate the utility of the proposed GMI estimator of the HP-divergence in terms of the dimension d and the sample size n. Our proposed MST-based estimator of the GMI is compared to density plug-in estimators of the GMI, in particular the standard KDE density plug-in estimator of [22], where the convergence rates of Theorems 4 and 5 are validated. We use multivariate normal simulated data in the experiments. In this section, we also discuss the choice of the proportionality parameter and compare runtime of the proposed GMI estimator approach with KDE method.
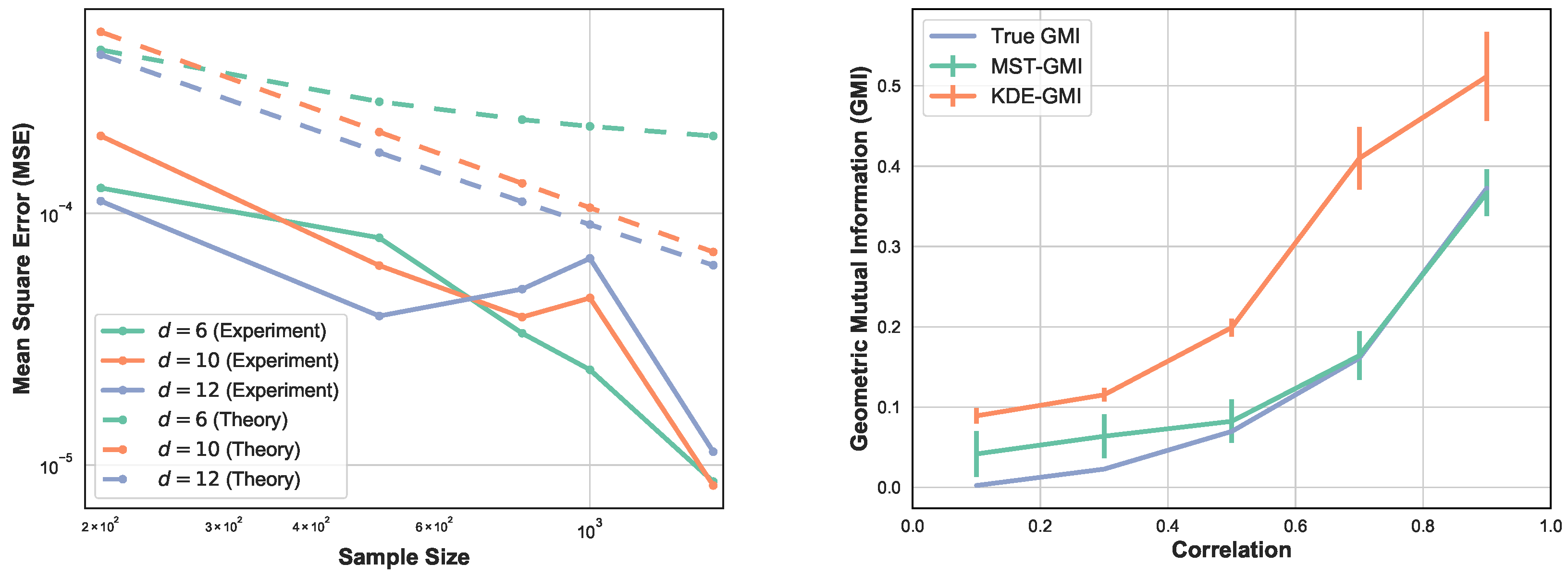
Here we perform four sets of experiments to illustrate the proposed GMI estimator. For the first set of experiments the MSE of the GMI estimator in Algorithm 1 is shown in Figure 2-left. The samples were drawn from d-dimensional normal distribution, with various sample sizes and dimensions . We selected the proportionality parameter and computed the MSE in terms of the sample size n. We show the log–log plot of MSE when n varies in . Please note that the empirically optimal proportion depends on n, so to avoid the computational complexity we fixed for this experiment. The experimental result shown in Figure 2-left validates the theoretical MSE growth rates derived from (13) and (14), i.e., decreasing sub-linearly in n and increasing exponentially in d.
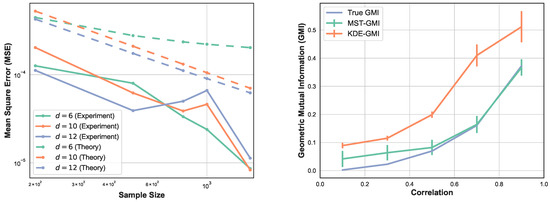
Figure 2.
(left) Log–log plot of theoretical and experimental MSE of the proposed MST-based GMI estimator as a function of sample size n for and fixed smoothness parameter . (right) The GMI estimator was implemented using two approaches, Algorithm 1 and KDE method where the KDE-GMI used KDE density estimators in the formula (2). In this experiment, samples are generated from the two-dimensional normal distribution with zero mean and covariance matrix (21) for various value of .
In Figure 2-right, we compare the proposed MST-based GMI estimator with the KDE-GMI estimator [22]. For the KDE approach, we estimated the joint and marginal densities and then plugged them into the proposed expression (2). The bandwidth h used for the KDE plug-in estimator was set as . The choice of h minimizes the bound on the MSE of the plug-in estimator. We generated data from the two-dimensional normal distribution with zero mean and covariance matrix
The coefficient is varied in range . The true GMI was computed by the Monte Carlo approximation to the integral (2). Please note that as increases, the MST-GMI outperforms the KDE-GMI approach. In this set of experiments .
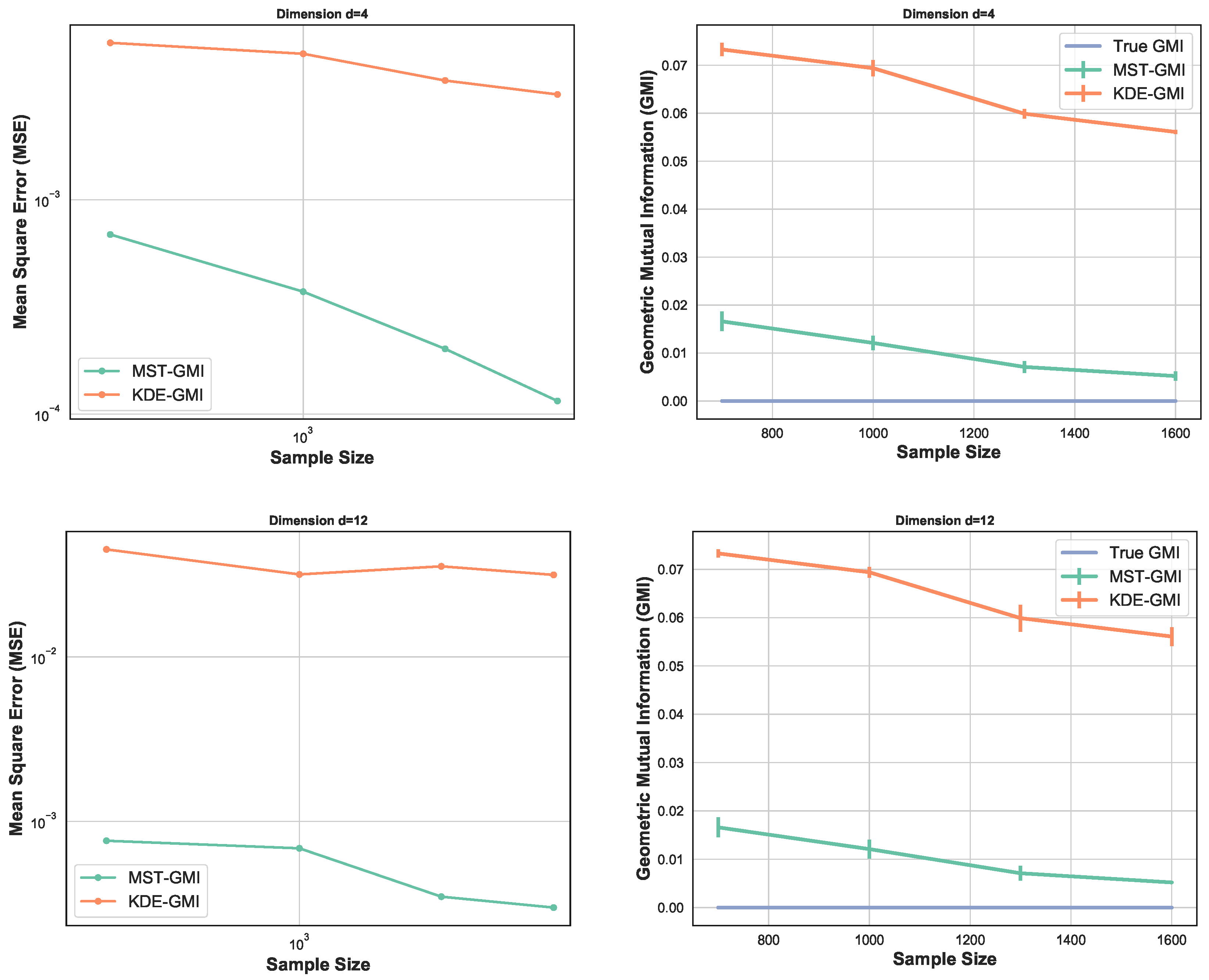
Figure 3 again compares the MST-GMI estimator with the KDE-GMI estimator. samples are drawn from the multivariate standard normal distribution with dimensions and . In both cases the proportionality parameter . The left plots in Figure 3 show the MSE (100 trials) of the GMI estimator implemented with an KDE estimator (with bandwidth as in Figure 2 i.e., ) for dimensions and various sample sizes. For all dimensions and sample sizes the MST-GMI estimator also outperforms the plug-in KDE-GMI estimator based on the estimated log–log MSE slope given in Figure 3 (left plots). The right plots in Figure 3 compares the MST-GMI with the KDE-GMI. In this experiment, the error bars denote standard deviations with 100 trials. We observe that for higher dimension and larger sample size n, the KDE-GMI approaches the true GMI at a slower rate than the MST-GMI estimator. This reflects the power of the proposed graph-based approach to estimating GMI.
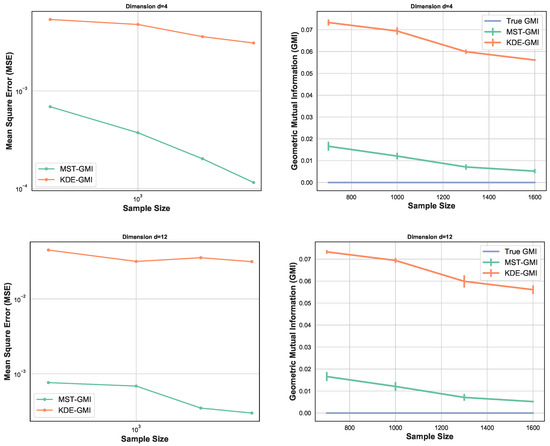
Figure 3.
MSE log–log plots as a function of sample size n (left) for the proposed MST-GMI estimator (“Estimated GMI”) and the standard KDE-GMI plug-in estimator of GMI. The right column of plots correspond to the GMI estimated for dimension (top) and (bottom). In both cases the proportionality parameter is . The MST-GMI estimator in both plots for sample size n in outperforms the KDE-GMI estimator, especially for larger dimensions.
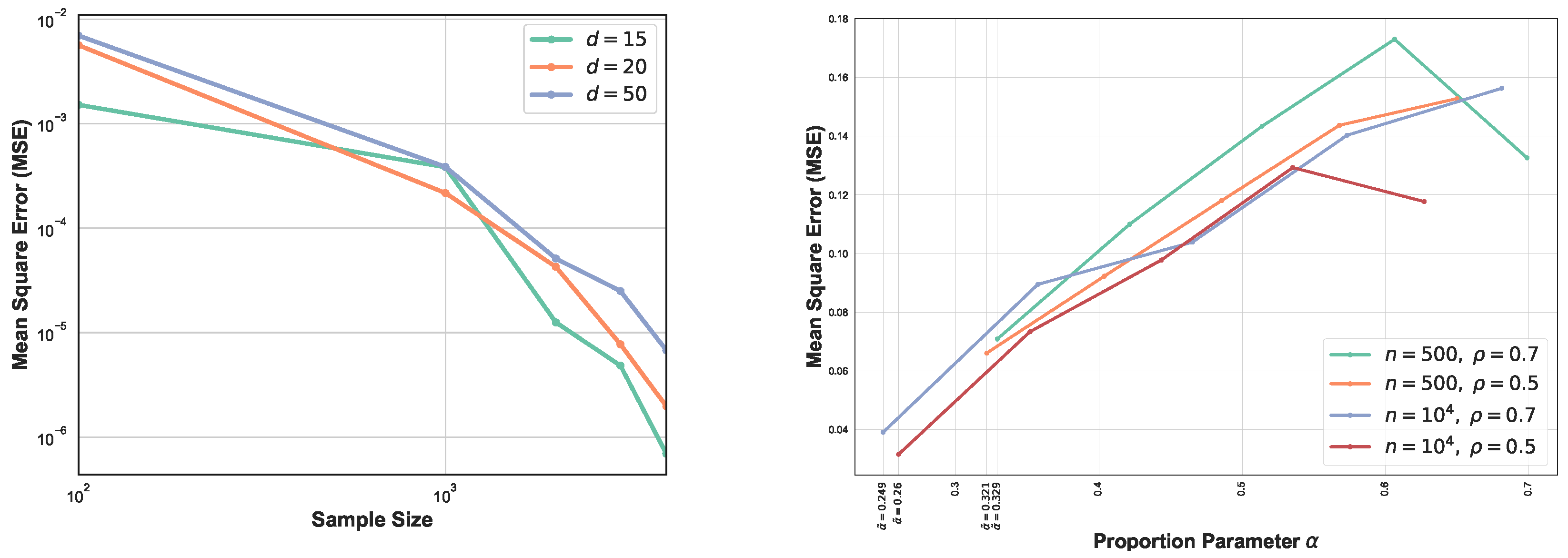
The comparison between MSEs for various dimension d is shown in Figure 4 (left). This experiment highlights the impact of higher dimension on the GMI estimators. As expected, for larger sample size n, MSE decreases while for higher dimension it increases. In this setting, we have generated samples from standard normal distribution with size and . From Figure 4 (left) we observe that for larger sample size, MSE curves are ordered based on their corresponding dimensions. Results in Section 2.4 strongly depend on the lower bounds and upper bounds and provide optimal parameter in the range , therefore in the experiment section we only analyze one case where the lower bounds and upper bounds are known and the optimal becomes . Figure 4 (right) illustrates the MSE vs proportion parameter when samples are generated from truncated normal distribution with . First, following Section 2.4, we compute the bound and then derive the optimal in this range. Therefore, each experiment with different sample size and provides different range . We observe that the MSE does not appeared a monotonic function in and its behavior strongly depends on sample size n, d, and density functions’ bounds. Additional study of the dependency is described in Appendix A.6. In this set of experiments , therefore following the results in Section 2.4, we have . In this experiment the optimal value of is always the lower bound and indicated in the Figure 4 (right).
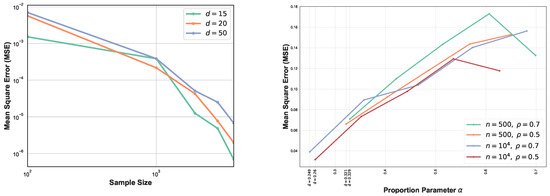
Figure 4.
MSE log–log plots as a function of sample size n for the proposed FR estimator. We compare the MSE of our proposed FR estimator for various dimensions (left). As d increases, the blue curve takes larger values than green and orange curves i.e., MSE increases as d grows. However, this is more evidential for large sample size n. The second experiment (right) focuses on optimal proportion for and . is the optimal for .
The parameter is studied further for three scenarios where the lower bounds and upper bounds are assumed unknown, therefore results in Section 2.4 are not applicable. In this set of experiments, we varied in the range to divide our original sample. We generated sample from an isotropic multivariate standard normal distribution in all three scenarios (all features are independent). Therefore, the true GMI is zero and in all scenarios the GMI column, corresponding to the MST-GMI, is compared with zero. In each scenario we fixed dimension d and sample size n and varied . The dimension and sample size in Scenarios 1,2, and 3 are and , respectively. In Table 1 the last column () stars the parameter with the minimum MSE and GMI in each scenario. Table 1 shows that in these sets of experiments when , the GMI estimator has less MSE (i.e., is more accurate) than when or . This experimentally demonstrates that if we split our training data, the proposed Algorithm 1 performs better with .

Table 1.
Comparison between different scenarios of various dimensions and sample sizes in terms of parameter . We applied the MST-GMI estimator to estimate the GMI () with . We varied dimension and sample size in each scenario. We observe that for , the MST-GMI estimator provides lowest MSE when indicated by star (*).
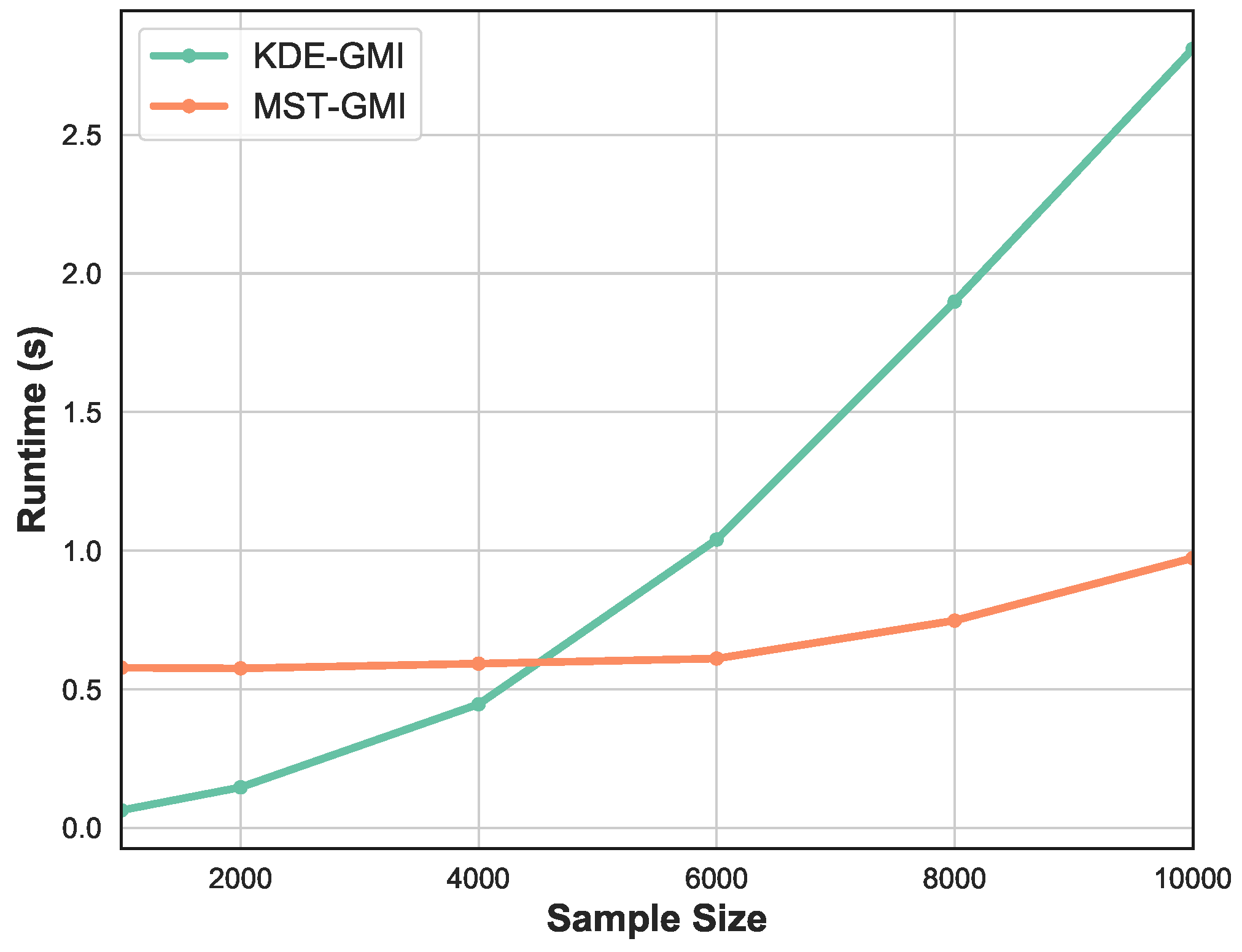
Finally, Figure 5 shows the runtime as a function of sample size n. We vary sample size in the range . Observe that for smaller number of samples the KDE-GMI method is slightly faster but as n becomes large we see significant relative speedup of the proposed MST-GMI method.
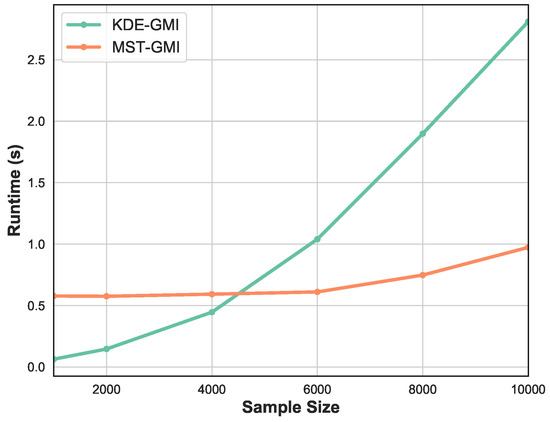
Figure 5.
Runtime of KDE approach and proposed MST-based estimator of GMI vs sample size. The proposed GMI estimator achieves significant speedup, while for small sample size, the KDE method becomes overly fast. Please note that in this experiment the sample is generated from the Gaussian distribution in dimension .
4. Conclusions
In this paper, we have proposed a new measure of mutual information, called Geometric MI (GMI), which is related to the Henze–Penrose divergence. The GMI can be viewed as dependency measure that is the limit of the Friedman–Rafsky test statistic, which depends on the MST over all data points. We established some properties of the GMI in terms of convexity/concavity, chain rule, and a type of data-processing inequality. A direct estimator of the GMI, called the MST-GMI, was introduced that uses random permutations of observed relationships between variables in the multivariate samples. An explicit form for the MSE convergence rate bound was derived that depends on a free parameter called the proportionality parameter. An asymptotically optimal form for this free parameter was given that minimizes the MSE convergence rate. Simulation studies were performed that illustrate and verify the theory.
Author Contributions
S.Y.S. wrote this article primarily under the supervision of A.O.H. as principle investigator (PI), and A.O.H. edited the paper. S.Y.S. provided the primary contributions for the proofs of all theorems and performed all experiments.
Funding
The work presented in this paper was partially supported by ARO grant W911NF-15-1-0479 and DOE grant DE-NA0002534.
Acknowledgments
The authors would like to thank Brandon Oselio for the helpful comments.
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Appendix A
We organize the appendices as follows: Theorem 1 which establishes convexity/concavity is proved in Appendix A.1. Appendix A.2 and Appendix A.3 establish the inequality (9) and (10) for given , respectively. In Appendix A.4, we first prove that the set which is randomly generated from original dependent data, contains asymptotically independent samples. Later by using the generated independent sample we show that for given the FR estimator of the GMI given in Algorithm 1 tends to . Appendix A.5 dedicates proof of Theorem 4. The proportionality parameter () optimization strategy is presented in Appendix A.6.
Appendix A.1. Theorem 1
Proof.
The proof is similar to the result for standard (Shannon) mutual information. However, we require the following lemma, proven in analogous manner to the log-sum inequality:
Lemma A1.
For non-negative real numbers and , given , ,
Notice this follows by using the convex function for any , , and the Jensen inequality.
Define the shorthand , , and for , and , respectively. To prove part (i) of Theorem 1, we represent the LHS of (5) as:
Furthermore, the RHS of (5) can be rewritten as
Thus, to prove , we use the inequality below:
In Lemma A1, let
and for ,
Then the claimed assertion (i) is obtained. Part (ii) follows by convexity of and the following expression:
Therefore, the claim in (6) is proved. □
Appendix A.2. Theorem 2
Proof.
We start with (9). Given and , we can easily check that for positive , , such that :
This implies
By substituting
we get
Consequently
Appendix A.3. Proposition 1
Proof.
Recall the Theorem 2, part (i). First from we have and then by applying the Jensen inequality,
where
Furthermore, we can easily show that if , we have and therefore . This together with (A3) proves that under both conditions and , the equality holds true. □
Appendix A.4. Theorem 3
Proof.
We first derive two required Lemmas A2 and A3 below:
Lemma A2.
Consider random vector with joint probability density function (pdf) . Let be a set of samples with pdf . Let and be two distinct subsets of such that and sample proportion is and . Next, let be a set of pairs such that , are selected at random from . Denote as the random vector corresponding to samples in . Then as such that also grows in a linked manner that then the distribution of convergences to i.e., random vectors and become mutually independent.
Proof. Consider two subsets , then we have
Here stands for the indicator function. Please note that
and and , are independent, therefore
this implies that
On the other hand, we know that , so we get
An immediate result of Lemma A2 is the following:
Lemma A3.
For given random vector from joint density function and with marginal density functions and let be realization of random vector as in Lemma A2 with parameter . Then for given points of at , we have
Now, we want to provide a proof of assertion (11). Consider two subsets and as described in Section 2.2. Assume that the components of sample follow density function . Therefore by owing to Lemmas A2 and A3, when then . Let and be Poisson variables with mean and independent of one another and and . Assume two Poisson processes and , and denote the FR statistic on these processes. Following the arguments in [13,46] we shall prove the following:
This follows due to , where is a constant defined in Lemma 1, [13] and . Thus, as . Let be independent variables with common density
for . Let be an independent Poisson variable with mean . Let a non-homogeneous Poisson process of rate . Assign mark 1 to a point in with probability
and mark 2 otherwise. By the marking theorem [13,47], the FR test statistic has the same distribution as . Given points of at and , the probability that they have different marks is given by (A7).
define
then
For fixed , consider the collection:
By the fact that , we have
Introduce
Then uniformly as and . Thus, using Proposition 1 in [13], we get
□
Appendix A.5. Theorem 4
Proof.
We begin by providing a family of bias rate bounds for the FR test statistic in terms of a parameter l. Assume , , and are in . Then by plugging the optimal l, we prove the bias rate bound given in (13).
Theorem A1.
Let be the FR test statistic. Then a bound on the bias rate of the estimator for , is given by
where is the Hölder smoothness parameter and is the largest possible degree of any vertex of MST.
Set
and
where
A more explicit form for the bound on the RHS is given below:
Proof. Consider two Poisson variables and with mean and respectively and independent of one another and and . Let and be the Poisson processes and . Likewise Appendix A.4, set . Applying Lemma 1, and (12) in [13], we can write
Here denotes the largest possible degree of any vertex of the MST in . Following the arguments in [46], we have and . Hence
Next let and be independent binomial random variables with marginal densities and such that are non-negative constants and . Therefore, using the subadditivity property in Lemma 2.2, [46], we can write
where , and is the Hölder smoothness parameter. Furthermore, for given , , let be independent variables with common densities for :
Denote be an independent Poisson variable with mean and a non-homogeneous Poisson of rate . Let be the non-Poisson point process . Assign a mark from the set to each point of . Let be the sets of points marked 1 with each probability and let be the set points with mark 2. Please note that owing to the marking theorem [47], and are independent Poisson processes with the same distribution as and , respectively. Considering as FR test statistic on nodes in , we have
By the fact that , we have
Here , , and is given in below:
Next set
By owing to the Lemma B.6 in [46] and applying analogous arguments, we can rewrite the expression in (A20):
where
Going back to Lemma A3, we know that
Therefore, the first term on the RHS of (A20) is less and equal to
and the second term is less and equal to
where
Recall the definition of the dual MST and FR statistic denoted by following [46]:
Definition A1.
(Dual MST, and dual FR statistic ) Let be the set of corner points associated with a particular subsection , of . Define the dual as the boundary MST graph in partition [48], which contains and points falling inside partition cell and those corner points in which minimize total MST length. Please note that it is allowed to connect the MSTs in and through points strictly contained in and and therefore corner points. Thus, the dual MST can connect the points in by direct edges to pair to another point in or by passing through the corner the corner points which are all connected in order to minimize the total weights. To clarify, assume that there are two points in , then the dual MST consists of the two edges connecting these points to the corner if they are closer to a corner point otherwise the dual MST connects them to each other.
Furthermore, is defined as the number of edges in the graph connecting nodes from different samples and number of edges connecting to the corner points. Please note that the edges connected to the corner nodes (regardless of the type of points) are always counted in the dual FR test statistic .
Similarly, consider the Poisson processes samples and the FR test statistic over these samples, denoted by . By superadditivity of the dual [46], we have
The first term of RHS in (A21) is greater or equal to
Furthermore,
where is the largest possible degree of any vertex of the MST in , as before. Consequently, we have
where is defined in (A16) and has been introduced in (A14). The last line in (A22) follows from the fact that
Here is given as (A14) by substituting in such that . Hence, the proof of Theorem A1 is completed. □
Going back to the proof of (13), without loss of generality assume that , for and . We select l as a function of n and to be the sequence increasing in n which minimizes the maximum of these rates:
Appendix A.6.
Our main goal in Section 2.4 was to find proportion such that the parametric MSE rate depending on the joint density and marginal densities is minimized. Recalling the explicit bias bound in (A16), it can be seen that this function will be a complicated function of , and . By rearrangement of terms in (A13), we first find an upper bound for in (A16), denoted by , as follows:
where . From Appendix A.5 we know that optimal l is given by (A23). One can check that for , the optimal provides a tighter bound. In (A24), the constants D and are
And the function is given as the following:
where is given in (A15). Next After all still the expression (A27) is complicated to optimize therefore we use the fact that to bound the function . Define the set
where
Here K is the smoothness constant in the Hölder class. Notice that set is a convex set. We bound by
Set ,
This simplifies to
Under the condition
is an increasing function in . Furthermore, for and
the function is strictly concave. Next, to find an optimal we consider the following optimization problem:
here , and , such that . We know that under conditions (A31) and (A32), the function is strictly concave and increasing in . We first solve the optimization problem:
where
The Lagrangian for this problem is
In this case, the optimum is bounded, and the Lagrangian multiplier is such that
Set In view of the concavity of and Lemma 1, page 227 in [49], maximizing over is equivalent to
for all , and
Denote the inverse function of . Since is strictly decreasing in (this is because is strictly concave, so that is continuous and strictly decreasing in ). From (A36) and (A37), we see immediately that on any interval , we have . We can write then
and . Next, we find the solution of
The function is increasing in and , and therefore it takes its minimum at . This implies that . Returning to our primary minimization over :
where and . We know that and , therefore the condition below
implies the constraint . Since the objective function (A38) is a complicated function in , it is not feasible to determine whether it is a convex function in . For this reason, let us solve the optimization problem in (A38) in a special case when . This implies . Under assumption the objective function in (A38) is convex in . Also, the case is equivalent to . Therefore, in the optimization problem we have constraint
We know that is convex over . Therefore, the problem becomes ordinary convex optimization problem. Let , and be any points that satisfy the KKT conditions for this problem:
Recall from (20):
where is given in (A28). So, the last condition in (A39) becomes . We then have
where is inverse function of . Since , at least one of or should be zero:
- , . Then and implies . Since , so this leads to .
- , . Then and implies . We know that , hence .
- , . Then and so .
Consequently, by following the behavior of with respect to and , we can often find the optimal , and . For instance, if is positive for all then we conclude that .
References
- Lewi, J.; Butera, R.; Paninski, L. Real-time adaptive information theoretic optimization of neurophysiology experiments. In Advances in Neural Information Processing Systems; The MIT Press: Cambridge, MA, USA, 2006; pp. 857–864. [Google Scholar]
- Peng, H.C.; Herskovits, E.H.; Davatzikos, C. Bayesian Clustering Methods for Morphological Analysis of MR Images. In Proceedings of the IEEE International Symposium on Biomedical Imaging, Washington, DC, USA, 7–10 July 2002; pp. 485–488. [Google Scholar]
- Moon, K.R.; Noshad, M.; Yasaei Sekeh, S.; Hero, A.O. Information theoretic structure learning with confidence. In Proceedings of the 2017 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), New Orleans, LA, USA, 5–9 March 2017. [Google Scholar]
- Brillinger, D.R. Some data analyses using mutual information. Braz. J. Probab. Stat. 2004, 18, 163–183. [Google Scholar]
- Torkkola, K. Feature extraction by non parametric mutual information maximization. J. Mach. Learn. Res. 2003, 3, 1415–1438. [Google Scholar]
- Vergara, J.R.; Estévez, P.A. A review of feature selection methods based on mutual information. Neural Comput. Appl. 2014, 24, 175–186. [Google Scholar] [CrossRef]
- Peng, H.; Long, F.; Ding, C. Feature selection based on mutual information criteria of max-dependency, max-relevance. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Long, F.; Ding, C. Evaluation, Application, and Small Sample Performance. IEEE Trans. Pattern Anal. Mach. Intell. 1997, 19, 153–158. [Google Scholar]
- Sorjamaa, A.; Hao, J.; Lendasse, A. Mutual information and knearest neighbors approximator for time series prediction. Lecture Notes Comput. Sci. 2005, 3697, 553–558. [Google Scholar]
- Mohamed, S.; Rezende, D.J. Variational information maximization for intrinsically motivated reinforcement learning. In Advances in Neural Information Processing Systems; The MIT Press: Cambridge, MA, USA, 2015; pp. 2116–2124. [Google Scholar]
- Neemuchwala, H.; Hero, A.O. Entropic graphs for registration. In Multi-Sensor Image Fusion and its Applications; CRC Press Book: Boca Raton, FL, USA, 2005; pp. 185–235. [Google Scholar]
- Neemuchwala, H.; Hero, A.O.; Zabuawala, S.; Carson, P. Image registration methods in high-dimensional space. Int. J. Imaging Syst. Technol. 2006, 16, 130–145. [Google Scholar] [CrossRef]
- Henze, N.; Penrose, M.D. On the multivarite runs test. Ann. Stat. 1999, 27, 290–298. [Google Scholar]
- Berisha, V.; Hero, A.O. Empirical non-parametric estimation of the Fisher information. IEEE Signal Process. Lett. 2015, 22, 988–992. [Google Scholar] [CrossRef]
- Berisha, V.; Wisler, A.; Hero, A.O.; Spanias, A. Empirically estimable classification bounds based on a nonparametric divergence measure. IEEE Trans. Signal Process. 2016, 64, 580–591. [Google Scholar] [CrossRef]
- Kailath, T. The divergence and Bhattacharyya distance measures in signal selection. IEEE Trans. Commun. Technol. 1967, 15, 52–60. [Google Scholar] [CrossRef]
- Yasaei Sekeh, S.; Oselio, B.; Hero, A.O. Multi-class Bayes error estimation with a global minimal spanning tree. In Proceedings of the 2018 56th Annual Allerton Conference on Communication, Control, and Computing (Allerton), Monticello, IL, USA, 2–5 October 2018. [Google Scholar]
- Noshad, M.; Moon, K.R.; Yasaei Sekeh, S.; Hero, A.O. Direct Estimation of Information Divergence Using Nearest Neighbor Ratios. In Proceedings of the IEEE International Symposium on Information Theory (ISIT), Aachen, Germany, 25–30 June 2017. [Google Scholar]
- March, W.; Ram, P.; Gray, A. Fast Euclidean minimum spanning tree: Algorithm, analysis, and applications. In Proceedings of the 16th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Boston, MA, USA, 20–23 August 2010; pp. 603–612. [Google Scholar]
- Borůvka, O. O jistém problému minimálním. Práce Moravské Pridovedecké Spolecnosti 1926, 3, 37–58. [Google Scholar]
- Kraskov, A.; Stögbauer, H.; Grassberger, P. Estimating mutual information. Phys. Rev. E 2004, 69, 066–138. [Google Scholar] [CrossRef] [PubMed]
- Moon, K.R.; Sricharan, K.; Hero, A.O. Ensemble Estimation of Mutual Information. In Proceedings of the IEEE International Symposium on Information Theory (ISIT), Aachen, Germany, 25–30 June 2017; pp. 3030–3034. [Google Scholar]
- Moon, K.R.; Hero, A.O. Multivariate f-divergence estimation with confidence. In Proceedings of the Advances in Neural Information Processing Systems 27 (NIPS 2014), Montreal, QC, Canada, 8–13 December 2014; pp. 2420–2428. [Google Scholar]
- Moon, K.R.; Sricharan, K.; Greenewald, K.; Hero, A.O. Improving convergence of divergence functional ensemble estimators. In Proceedings of the IEEE International Symposium on Information Theory (ISIT), Barcelona, Spain, 10–15 July 2016. [Google Scholar]
- Leonenko, N.; Pronzato, L.; Savani, V. A class of Rényi information estimators for multidimensional densities. Ann. Stat. 2008, 36, 2153–2182. [Google Scholar] [CrossRef]
- Gao, S.; Ver Steeg, G.; Galstyan, A. Efficient estimation of mutual information for strongly dependent variables. In Proceedings of the Eighteenth International Conference on Artificial Intelligence and Statistics, San Diego, CA, USA, 9–12 May 2015; pp. 277–286. [Google Scholar]
- Pál, D.; Póczos, B.; Szapesvári, C. Estimation of Rényi entropy and mutual information based on generalized nearest-neighbor graphs. In Proceedings of the 24th Annual Conference on Neural Information Processing Systems 2010, Vancouver, BC, Canada, 6–9 December 2010. [Google Scholar]
- Krishnamurthy, A.; Kandasamy, K.; Póczos, B.; Wasserman, L. Nonparametric estimation of Rényi divergence and friends. In Proceedings of the 31st International Conference on Machine Learning, Beijing, China, 21–26 June 2014; pp. 919–927. [Google Scholar]
- Kandasamy, K.; Krishnamurthy, A.; Póczos, B.; Wasserman, L.; Robins, J. Nonparametric von mises estimators for entropies, divergences and mutual informations. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 7–12 December 2015; pp. 397–405. [Google Scholar]
- Singh, S.; Póczos, B. Analysis of k nearest neighbor distances with application to entropy estimation. In Proceedings of the Advances in Neural Information Processing Systems, Barcelona, Spain, 5–10 December 2016; pp. 1217–1225. [Google Scholar]
- Sugiyama, M. Machine learning with squared-loss mutual information. Entropy 2012, 15, 80–112. [Google Scholar] [CrossRef]
- Costa, A.; Hero, A.O. Geodesic entropic graphs for dimension and entropy estimation in manifold learning. IEEE Trans. Signal Process. 2004, 52, 2210–2221. [Google Scholar] [CrossRef]
- Friedman, J.H.; Rafsky, L.C. Multivariate generalizations of the Wald-Wolfowitz and Smirnov two-sample tests. Ann. Stat. 1979, 7, 697–717. [Google Scholar] [CrossRef]
- Smirnov, N.V. On the estimation of the discrepancy between empirical curves of distribution for two independent samples. Bull. Moscow Univ. 1939, 2, 3–6. [Google Scholar]
- Wald, A.; Wolfowitz, J. On a test whether two samples are from the same population. Ann. Math. Stat. 1940, 11, 147–162. [Google Scholar] [CrossRef]
- Noshad, M.; Hero, A.O. Scalable mutual information estimation using dependence graphs. In Proceedings of the International Conference on Artificial Intelligence and Statistics (AISTAT), Brighton, UK, 12–17 May 2019. [Google Scholar]
- Yasaei Sekeh, S.; Oselio, B.; Hero, A.O. A Dimension-Independent discriminant between distributions. In Proceedings of the 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Calgary, AB, Canada, 15–20 April 2018. [Google Scholar]
- Csiszár, I. Information-type measures of difference of probability distributions and indirect observations. Studia Sci. Math. Hungar. 1967, 2, 299–318. [Google Scholar]
- Ali, S.; Silvey, S.D. A general class of coefficients of divergence of one distribution from another. J. R. Stat. Soc. Ser. B (Methodol.) 1966, 28, 131–142. [Google Scholar] [CrossRef]
- Cover, T.; Thomas, J. Elements of Information Theory, 1st ed.; John Wiley & Sons: Chichester, UK, 1991. [Google Scholar]
- Härdle, W. Applied Nonparametric Regression; Cambridge University Press: Cambridge, UK, 1991. [Google Scholar]
- Lorentz, G.G. Approximation of Functions; Holt, Rinehart and Winston: New York, NY, USA; Chicago, IL, USA; Toronto, ON, Canada, 1966. [Google Scholar]
- Andersson, P. Characterization of pointwise Hölder regularity. Appl. Comput. Harmon. Anal. 1997, 4, 429–443. [Google Scholar] [CrossRef][Green Version]
- Robins, G.; Salowe, J.S. On the maximum degree of minimum spanning trees. In Proceedings of the SCG 94 Tenth Annual Symposium on Computational Geometry, Stony Brook, NY, USA, 6–8 June 1994; pp. 250–258. [Google Scholar]
- Efron, B.; Stein, C. The jackknife estimate of variance. Ann. Stat. 1981, 9, 586–596. [Google Scholar] [CrossRef]
- Yasaei Sekeh, S.; Noshad, M.; Moon, K.R.; Hero, A.O. Convergence Rates for Empirical Estimation of Binary Classification Bounds. arXiv 2018, arXiv:1810.01015. [Google Scholar]
- Kingman, J.F.C. Poisson Processes; Oxford Univ. Press: Oxford, UK, 1993. [Google Scholar]
- Yukich, J.E. Probability Theory of Classical Euclidean Optimization; Vol. 1675 of Lecture Notes in Mathematics; Springer: Berlin, Germany, 1998. [Google Scholar]
- Luenberger, D.G. Optimization by Vector Space Methods; Wiley Professional Paperback Series; Wiley-Interscience: Hoboken, NJ, USA, 1969. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).