An Improved Method of Handling Missing Values in the Analysis of Sample Entropy for Continuous Monitoring of Physiological Signals
Abstract
:1. Introduction
2. Methods
2.1. Sample Entropy
2.2. KeepSampEn, SkipSampEn, LinearSampEn and BootSampEn
2.3. Experimental Datasets
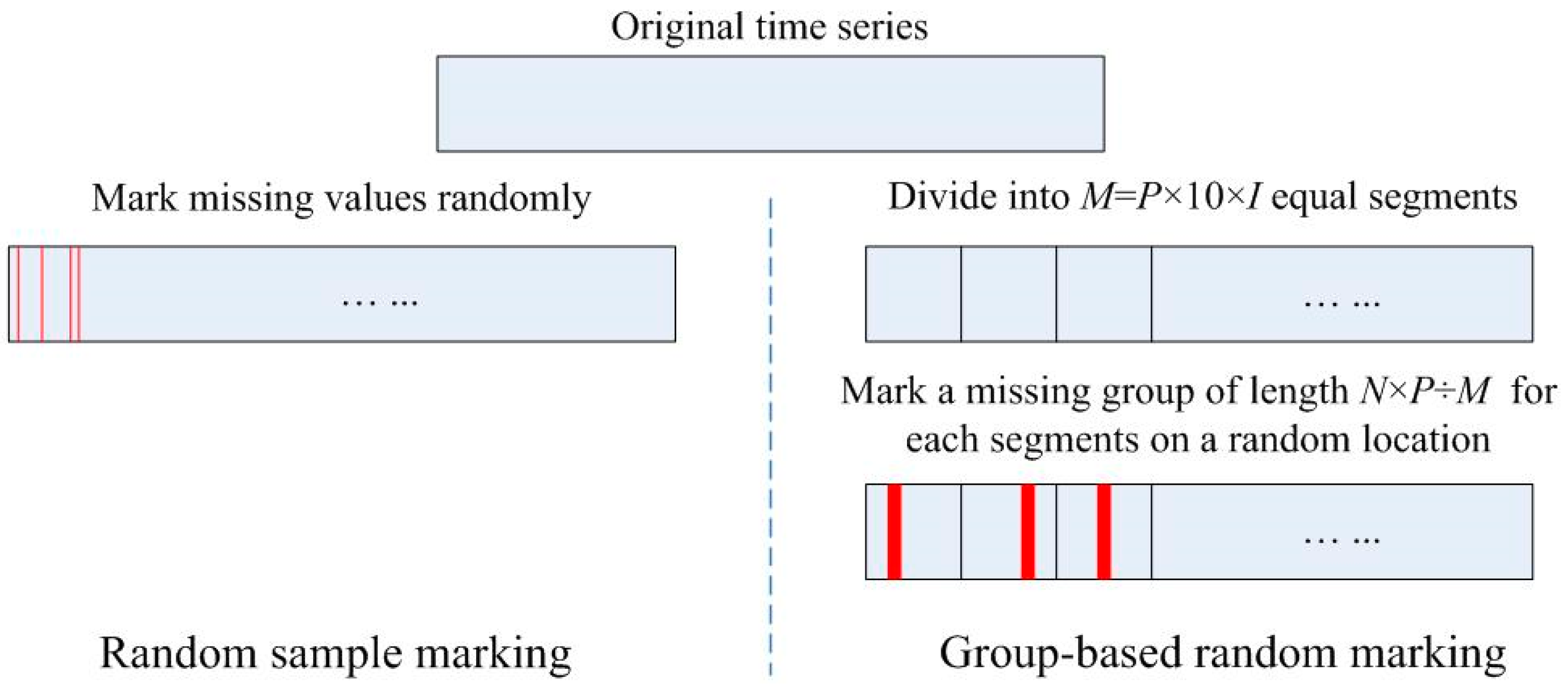
2.4. Scheme for Generating Missing Values
3. Results
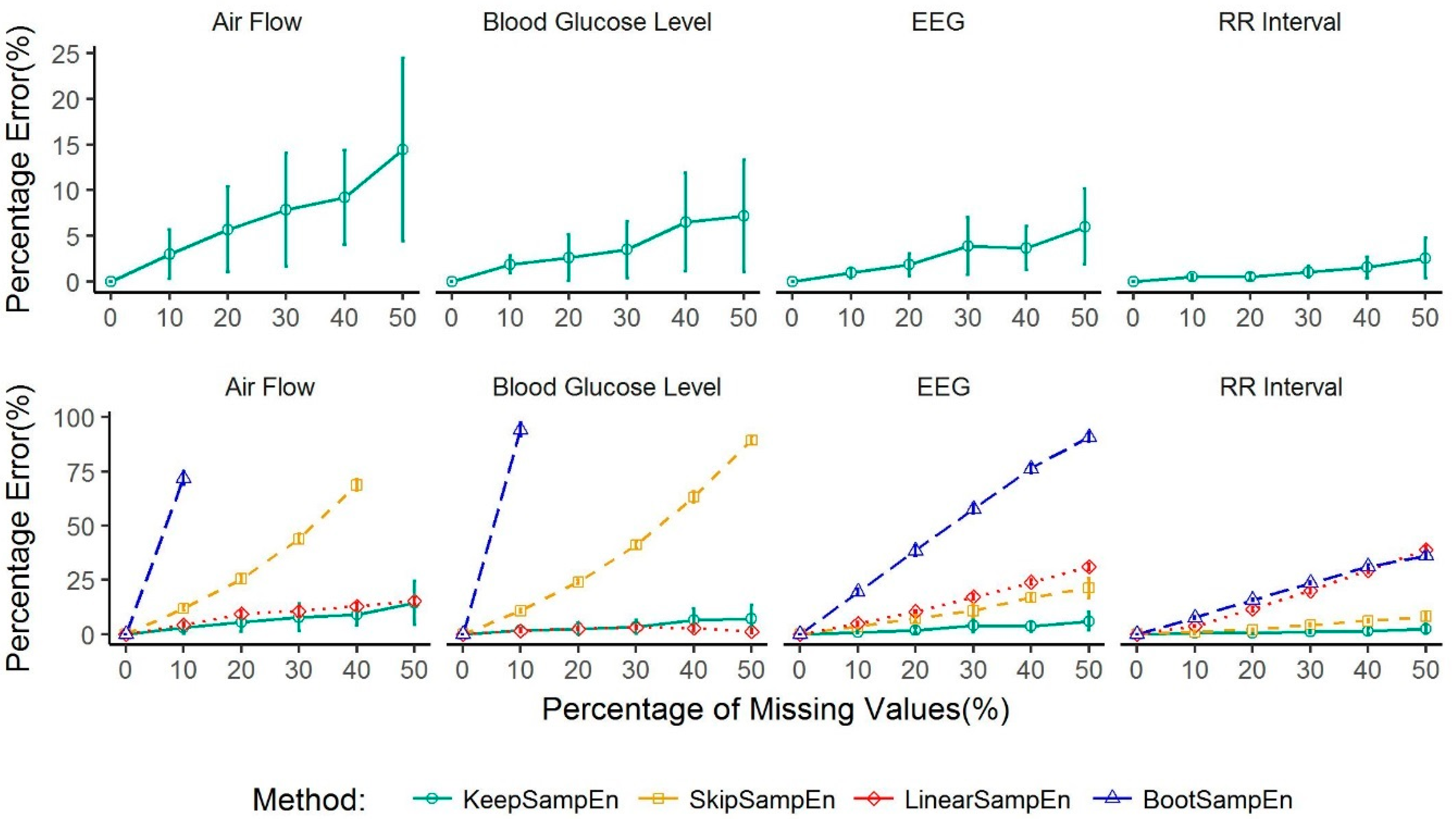
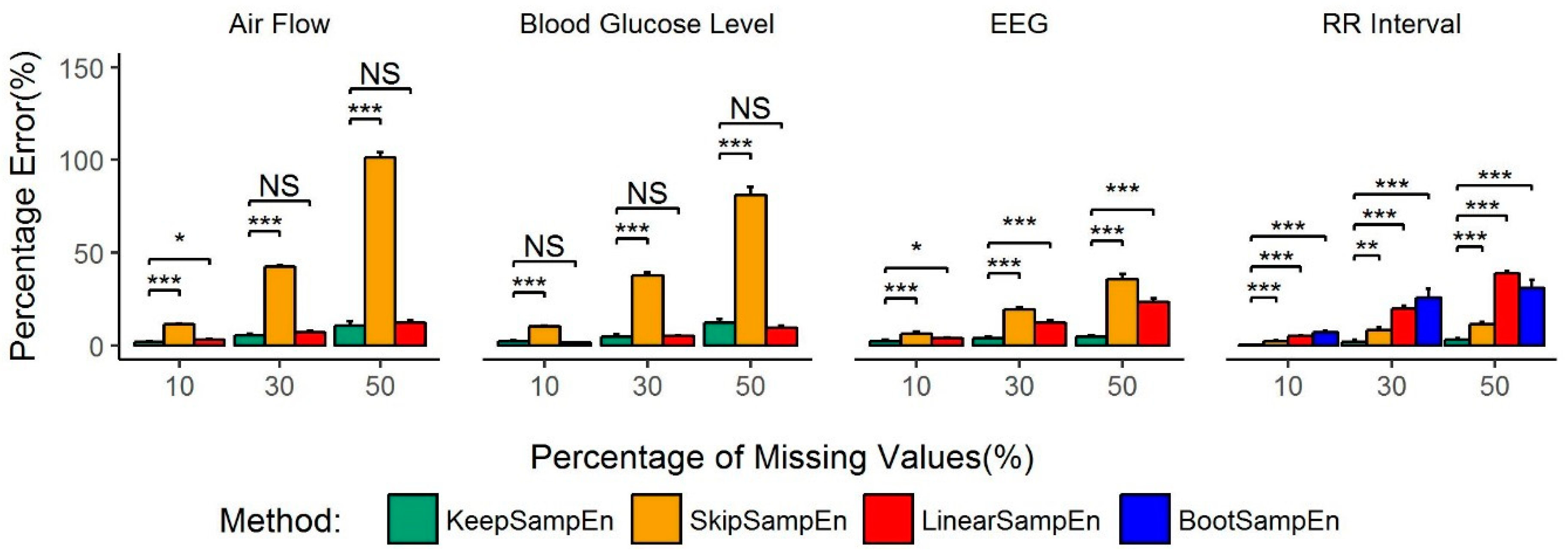
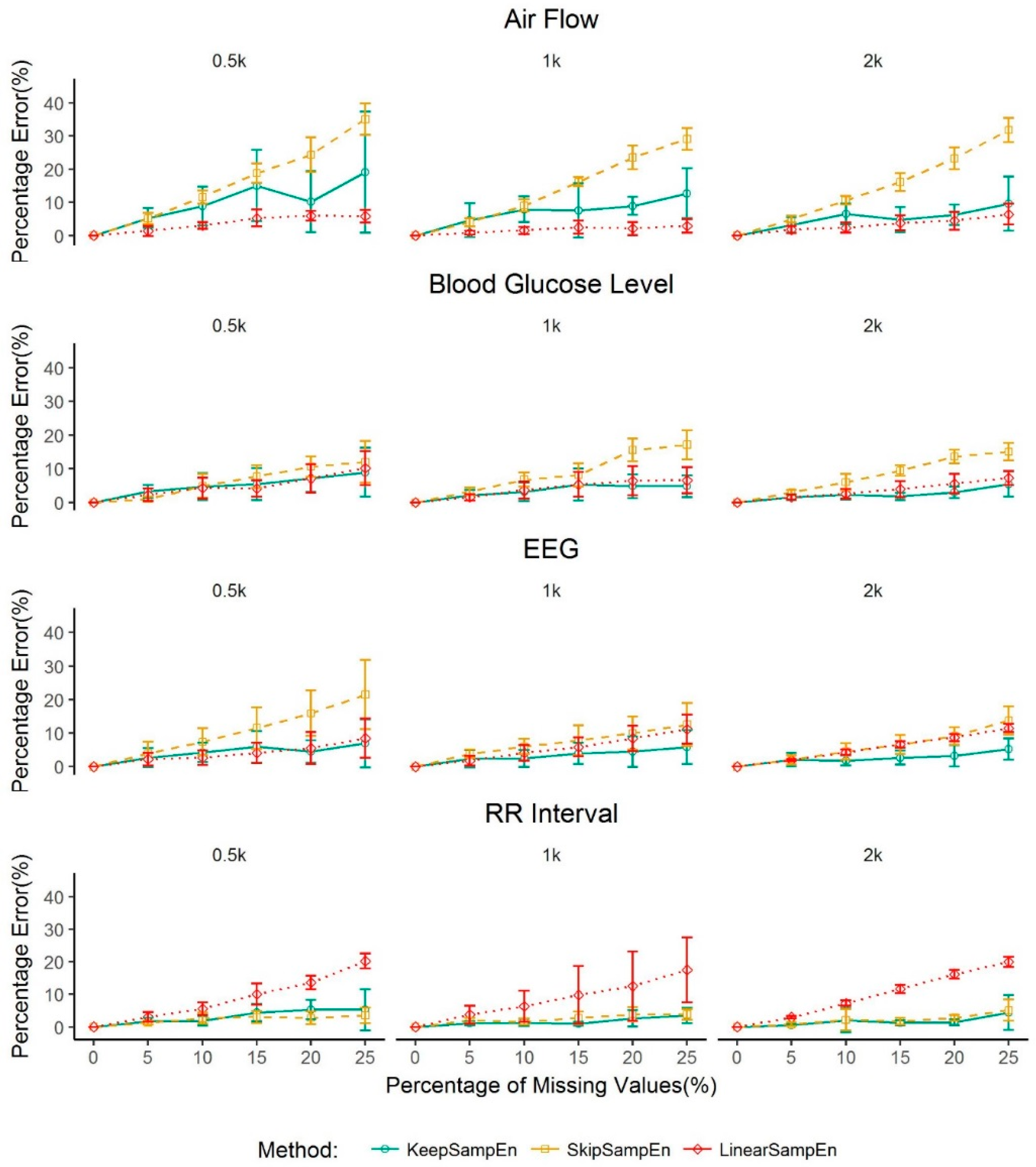
3.1. Robustness to the Impact of Physiological Types
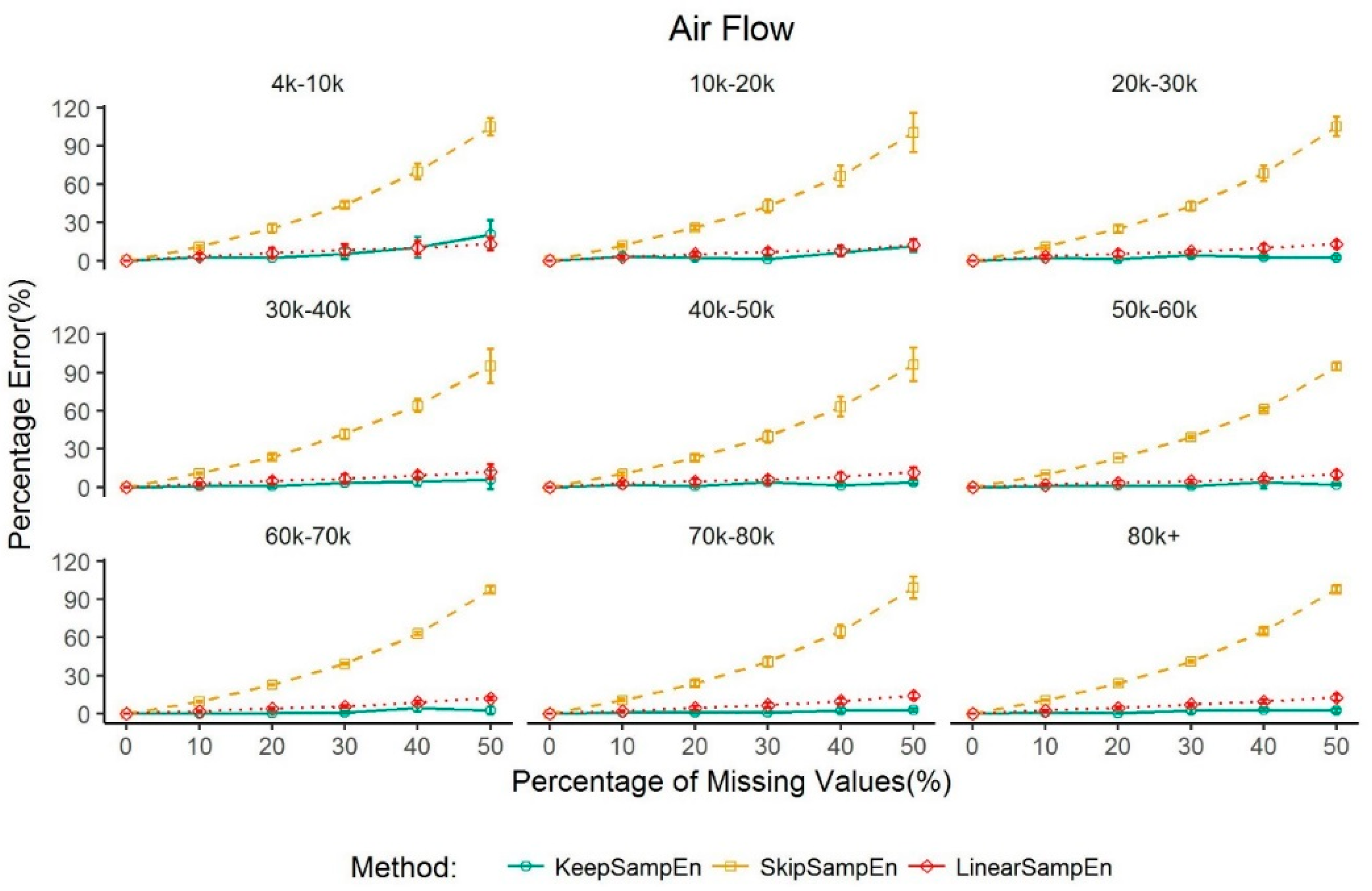
3.2. Robustness to the Impact of Data Size
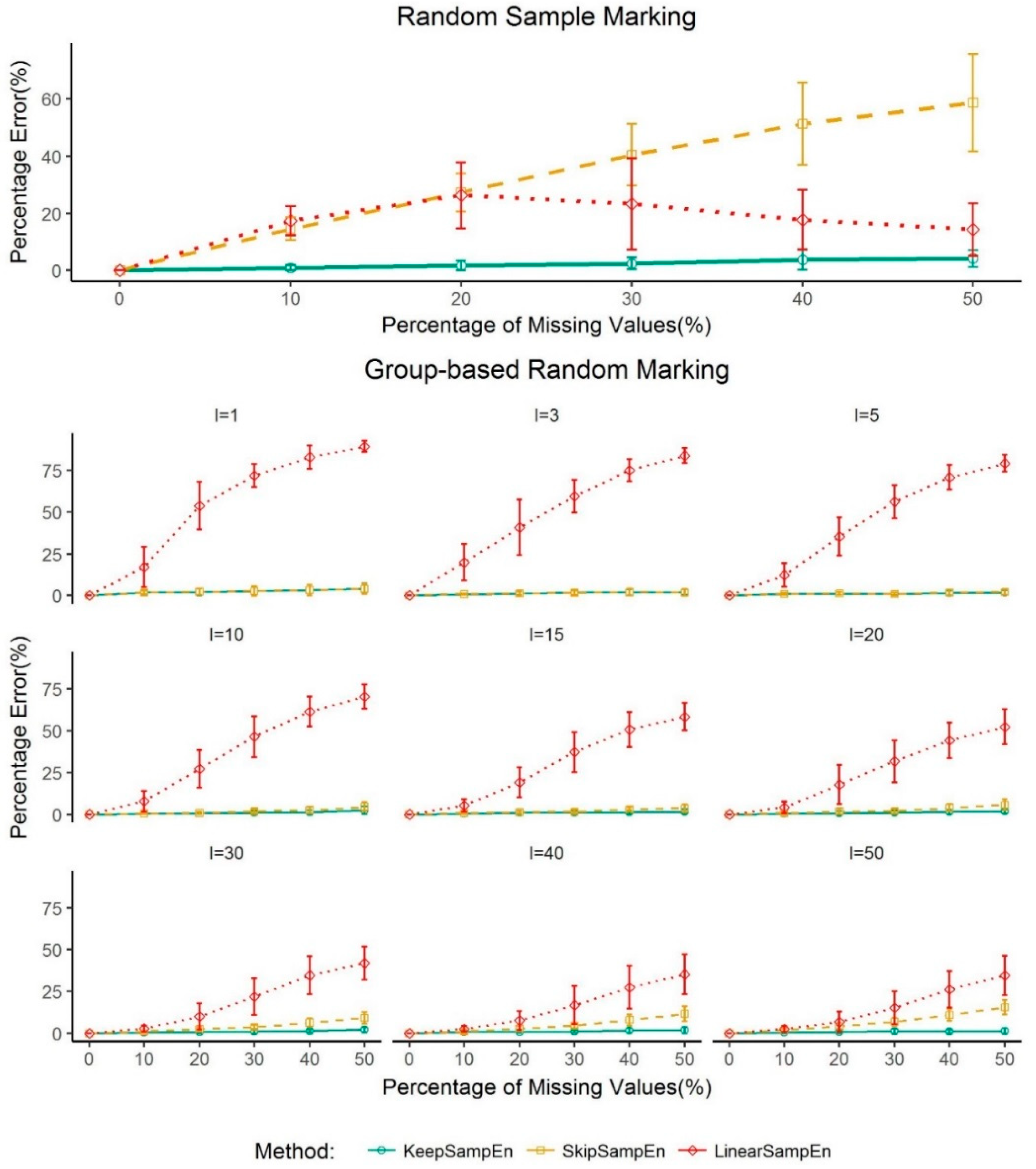
3.3. Robustness to the Impact of Schemes for Generating Missing Values
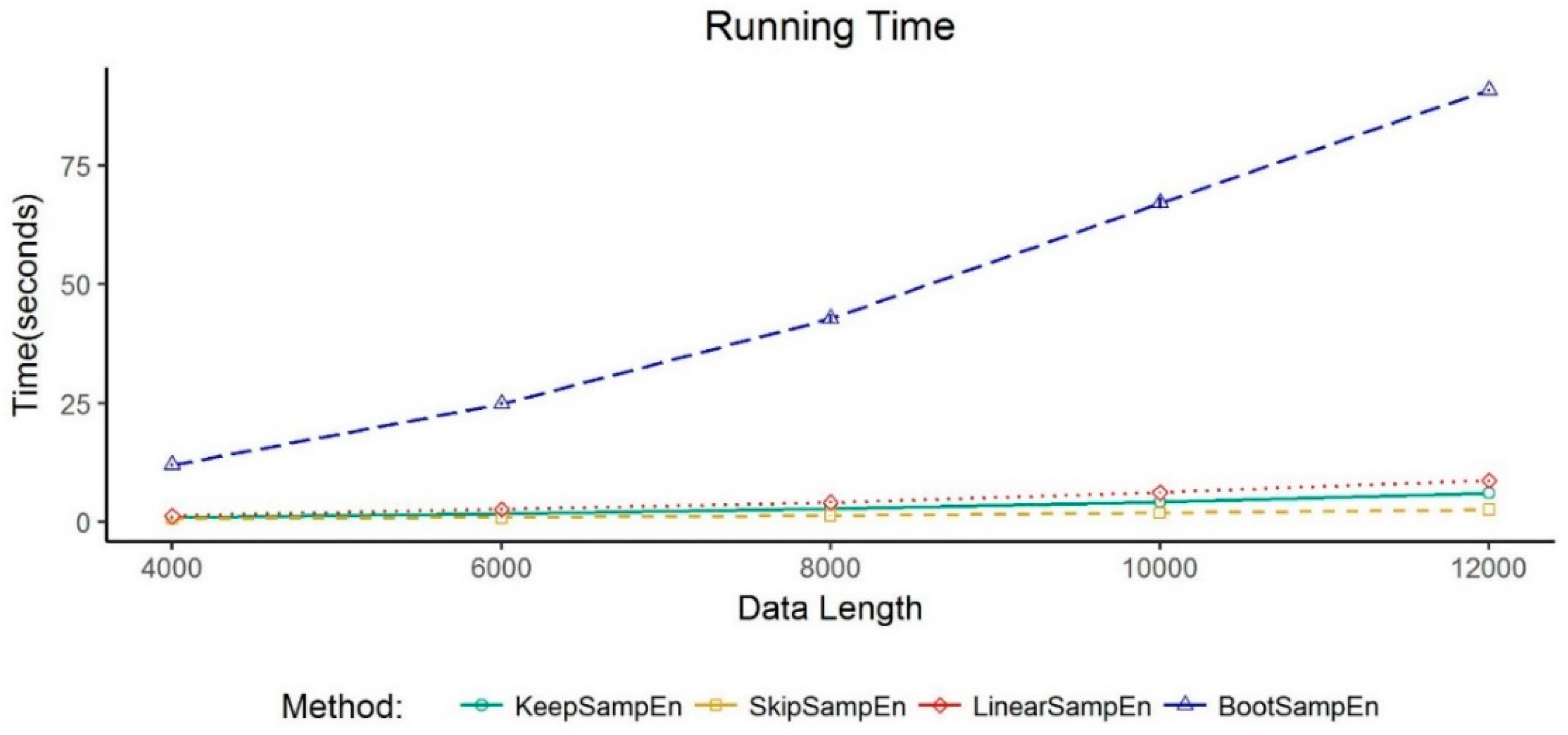
3.4. Exploration of the Computational Complexity of KeepSampEn
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Appendix A

References
- Shannon, C.E.; Weaver, W. The Mathematical Theory of Information. Math. Gazette 1949, 97, 170–180. [Google Scholar]
- Ivanov, P.C.; Amaral, L.A.; Goldberger, A.L.; Havlin, S.; Rosenblum, M.G.; Struzik, Z.R.; Stanley, H.E. Multifractality in Human Heartbeat Dynamics. Nature 1999, 399, 461. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Jin, Y.; Lo, I.L.; Zhao, H.; Sun, B.; Zhao, Q.; Zheng, J.; Zhang, X.D. Complexity Change in Cardiovascular Disease. Int. J. Biol. Sci. 2017, 13, 1320–1328. [Google Scholar] [CrossRef] [PubMed]
- Henriques, T.S.; Costa, M.D.; Mathur, P.; Mathur, P.; Davis, R.B.; Mittleman, M.A.; Khabbaz, K.R.; Goldberger, A.L.; Subramaniam, B. Complexity of Preoperative Blood Pressure Dynamics: Possible Utility in Cardiac Surgical Risk Assessment. J. Clin. Monit. Comput. 2018, 33, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Kohnert, K.D.; Heinke, P.; Vogt, L.; Augstein, P.; Thomas, A.; Salzsieder, E. Associations of Blood Glucose Dynamics with Antihyperglycemic Treatment and Glycemic Variability in Type 1 and Type 2 Diabetes. J. Endocrinol. Investig. 2017, 40, 1201–1207. [Google Scholar] [CrossRef]
- Investigators, F.S.T. Glucose Variability in a 26-Week Randomized Comparison of Mealtime Treatment with Rapid-Acting Insulin Versus GLP-1 Agonist in Participants with Type 2 Diabetes at High Cardiovascular Risk. Diabetes Care 2016, 39, 973–981. [Google Scholar]
- Kumar, R.B.; Goren, N.D.; Stark, D.E.; Wall, D.P.; Longhurst, C.A. Automated Integration of Continuous Glucose Monitor Data in the Electronic Health Record Using Consumer Technology. J. Am. Med. Inform. Assoc. 2016, 23, 532–537. [Google Scholar] [CrossRef]
- Wu, H.T.; Hsu, P.C.; Lin, C.F.; Wang, H.J.; Sun, C.K.; Liu, A.B.; Lo, M.T.; Tang, C.J. Multiscale Entropy Analysis of Pulse Wave Velocity for Assessing Atherosclerosis in the Aged and Diabetic. IEEE Trans. Biomed. Eng. 2011, 58, 2978–2981. [Google Scholar]
- Watanabe, E.; Kiyono, K.; Hayano, J.; Yamamoto, Y.; Inamasu, J.; Yamamoto, M.; Ichikawa, T.; Sobue, Y.; Harada, M.; Ozaki, Y. Multiscale Entropy of the Heart Rate Variability for the Prediction of an Ischemic Stroke in Patients with Permanent Atrial Fibrillation. PLoS ONE 2015, 10, e0137144. [Google Scholar] [CrossRef]
- Jin, Y.; Chen, C.; Cao, Z.; Sun, B.; Lo, I.L.; Liu, T.M.; Zheng, J.; Sun, S.; Shi, Y.; Zhang, X.D. Entropy Change of Biological Dynamics in COPD. Int. J. Chronic Obstr. Pulm. Disease 2017, 12, 2997. [Google Scholar] [CrossRef]
- Xue, M.; Wang, D.; Zhang, Z.; Cao, Z.; Luo, Z.; Zheng, Y.; Lu, J.; Zhao, Q.; Zhang, X.D. Demonstrating the Potential of Using Transcutaneous Oxygen and Carbon Dioxide Tensions to Assess the Risk of Pressure Injuries. Int. J. Biol. Sci. 2018, 14, 1466–1471. [Google Scholar] [CrossRef]
- Sun, S.; Jin, Y.; Chen, C.; Sun, B.; Cao, Z.; Lo, I.L.; Zhao, Q.; Zheng, J.; Shi, Y.; Zhang, X.D. Entropy Change of Biological Dynamics in Asthmatic Patients and Its Diagnostic Value in Individualized Treatment: A Systematic Review. Entropy 2018, 20, 402. [Google Scholar] [CrossRef]
- Wang, D.; Zheng, P.; Zhang, T.; Li, C.; Chen, L.; Zhai, Y.; Leng, D.; Jin, Y.; Sun, B.; Zhang, X.D. Analysis of the Complexity Patterns in Respiratory Data. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine, Madrid, Spain, 3–6 December 2018; pp. 2556–2560. [Google Scholar]
- Sahoo, S.S.; Jayapandian, C.; Garg, G.; Kaffashi, F.; Chung, S.; Bozorgi, A.; Chen, C.H.; Loparo, K.; Lhatoo, S.D.; Zhang, G.Q. Heart Beats in the Cloud: Distributed Analysis of Electrophysiological ‘Big Datac Using Cloud Computing for Epilepsy Clinical Research. J. Am. Med. Inform. Assoc. Jamia 2014, 21, 263–271. [Google Scholar] [CrossRef]
- Zhang, X.D.; Zhang, Z.; Wang, D. CGManalyzer: an R package for Analyzing Continuous Glucose Monitoring Studies. Bioinformatics 2018, 34, 1609–1611. [Google Scholar] [CrossRef]
- Cirugedaroldan, E.; Cuestafrau, D.; Miromartinez, P.; Oltracrespo, S. Comparative Study of Entropy Sensitivity to Missing Biosignal Data. Entropy 2014, 16, 5901–5918. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Li, J.; Xu, W.; Zhang, P.; Zhang, X. Detecting Complexity Changes for Heart Rate Variability in the Case of Missing Data. Sci. Sin. Inf. 2015, 45, 1015. [Google Scholar] [CrossRef]
- Zhang, X.D.; Pechter, D.; Yang, L.; Ping, X.; Yao, Z.; Zhang, R.; Shen, X.; Li, N.X.; Connick, J.; Nawrocki, A.R. Decreased Complexity of Glucose Dynamics Preceding the Onset of Diabetes in Mice and Rats. PLoS ONE 2017, 12, e0182810. [Google Scholar] [CrossRef]
- Cho, K.; Miyano, T. Entropy Test for Complexity in Chaotic Time Series. Nonlinear Theory Its Appl. 2016, 7, 21–29. [Google Scholar] [CrossRef]
- Costa, M.; Goldberger, A.L.; Peng, C.K. Multiscale entropy analysis of biological signals. Phys. Rev. E 2005, 71, 021906. [Google Scholar] [CrossRef]
- Richman, J.S.; Moorman, J.R. Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol. Heart Circ. Physiol. 2000, 278, H2039–H2049. [Google Scholar] [CrossRef]
- Pincus, S.M. Approximate Entropy as a Measure of System Complexity. Proc. Natl. Acad. Sci. USA 1991, 88, 2297–2301. [Google Scholar] [CrossRef]
- Lake, D.E.; Richman, J.S.; Griffin, M.P.; Moorman, J.R. Sample Entropy Analysis of Neonatal Heart Rate Variability. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002, 283, R789–R797. [Google Scholar] [CrossRef]
- Shara, N.M.; Umans, J.G.; Wang, W.; Howard, B.V.; Resnick, H.E. Assessing the Impact of Different Imputation Methods on Serial Measures of Renal Function: The Strong Heart Study. Kidney Int. 2007, 71, 701–705. [Google Scholar] [CrossRef]
- Kim, K.K.; Baek, H.J.; Yong, G.L.; Park, K.S. Effect of Missing RR-interval Data on Nonlinear Heart Rate Variability Analysis. Comput. Methods Programs Biomed. 2012, 106, 210–218. [Google Scholar] [CrossRef]
- Dai, X.; Zhang, D. Research of Interpolation Methods of Missing RR-interval Data. Chin. J. Med. Phys. 2013, 30, 3903–3905. [Google Scholar]
- Costa, M.D.; Peng, C.K.; Goldberger, A.L. Multiscale Analysis of Heart Rate Dynamics: Entropy and Time Irreversibility Measures. Cardiovasc. Eng. 2008, 8, 88–93. [Google Scholar] [CrossRef]
- Cao, Z.; Luo, Z.; Hou, A.; Nie, Q.; Xie, B.; An, X.; Wan, Z.; Ye, X.; Xu, Y.; Chen, X. Volume-Targeted Versus Pressure-Limited Noninvasive Ventilation in Subjects with Acute Hypercapnic Respiratory Failure: A Multicenter Randomized Controlled Trial. Respir. Care 2016, 61, 1440–1450. [Google Scholar] [CrossRef]
- Yentes, J.M.; Hunt, N.; Schmid, K.K.; Kaipust, J.P.; McGrath, D.; Stergiou, N. The Appropriate Use of Approximate Entropy and Sample Entropy with Short Data Sets. Ann. Biomed. Eng. 2013, 41, 349–365. [Google Scholar] [CrossRef]
- Voss, A.; Schulz, S.; Schroeder, R.; Baumert, M.; Caminal, P. Methods Derived from Nonlinear Dynamics for Analysing Heart Rate Variability. Philos. Trans. A Math. Phys. Eng. Sci. 2009, 367, 277–296. [Google Scholar] [CrossRef]
- Bao, Y.; Li, H.; Sun, X.; Yu, Y.; Ou, J. Compressive Sampling-based Data Loss Recovery for Wireless Sensor Networks Used in Civil Structural Health Monitoring. Struct. Health Monit. 2013, 12, 78–95. [Google Scholar] [CrossRef]
- Xu, Y.; Lee, W.C.; Xu, J. Analysis of a Loss-Resilient Proactive Data Transmission Protocol in Wireless Sensor Networks. In Proceedings of the IEEE International Conference on Computer Communications, Barcelona, Spain, 6–12 May 2007; pp. 1712–1720. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, X.; Chen, C.; Geng, Q.; Cao, Z.; Chen, X.; Lin, J.; Jin, Y.; Zhang, Z.; Shi, Y.; Zhang, X.D. An Improved Method of Handling Missing Values in the Analysis of Sample Entropy for Continuous Monitoring of Physiological Signals. Entropy 2019, 21, 274. https://doi.org/10.3390/e21030274
Dong X, Chen C, Geng Q, Cao Z, Chen X, Lin J, Jin Y, Zhang Z, Shi Y, Zhang XD. An Improved Method of Handling Missing Values in the Analysis of Sample Entropy for Continuous Monitoring of Physiological Signals. Entropy. 2019; 21(3):274. https://doi.org/10.3390/e21030274
Chicago/Turabian StyleDong, Xinzheng, Chang Chen, Qingshan Geng, Zhixin Cao, Xiaoyan Chen, Jinxiang Lin, Yu Jin, Zhaozhi Zhang, Yan Shi, and Xiaohua Douglas Zhang. 2019. "An Improved Method of Handling Missing Values in the Analysis of Sample Entropy for Continuous Monitoring of Physiological Signals" Entropy 21, no. 3: 274. https://doi.org/10.3390/e21030274
APA StyleDong, X., Chen, C., Geng, Q., Cao, Z., Chen, X., Lin, J., Jin, Y., Zhang, Z., Shi, Y., & Zhang, X. D. (2019). An Improved Method of Handling Missing Values in the Analysis of Sample Entropy for Continuous Monitoring of Physiological Signals. Entropy, 21(3), 274. https://doi.org/10.3390/e21030274





