Performance Evaluation of Fixed Sample Entropy in Myographic Signals for Inspiratory Muscle Activity Estimation
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Protocol
2.3. Data Analysis
2.3.1. Pre-processing and Segmentation of Myographic Signals
2.3.2. Individual and Global SD Calculation
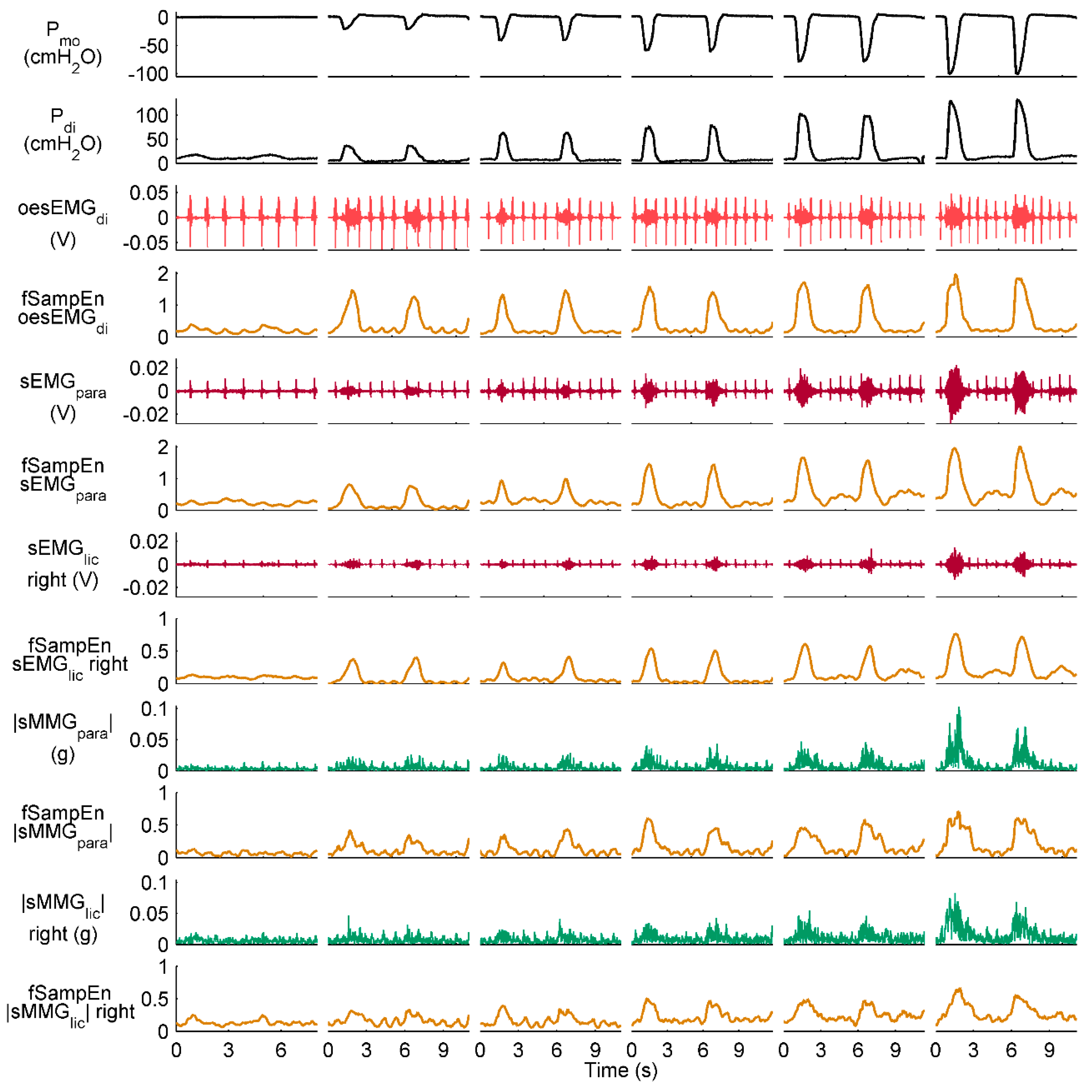
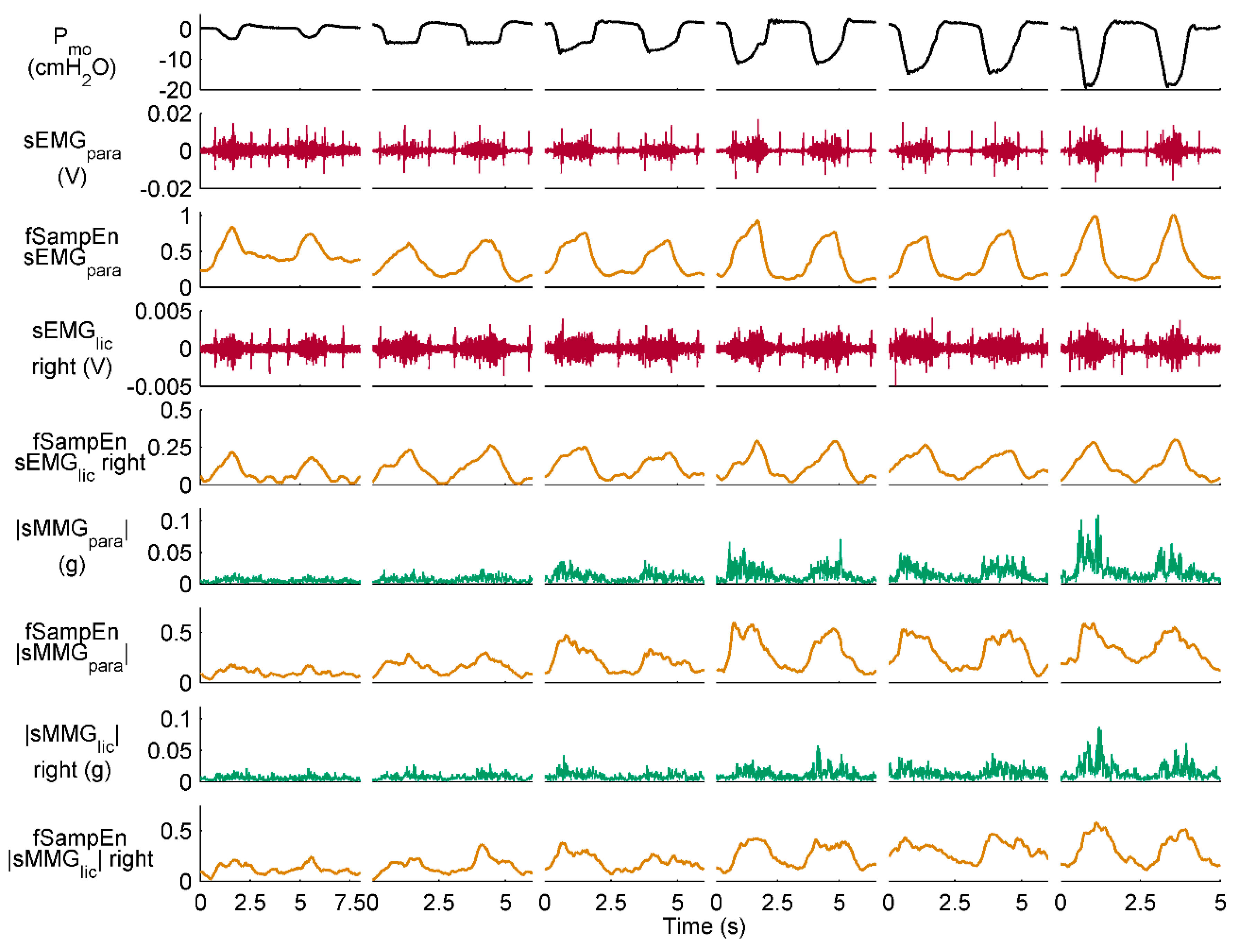
2.3.3. fSampEn Time-Series Calculation and Evaluation
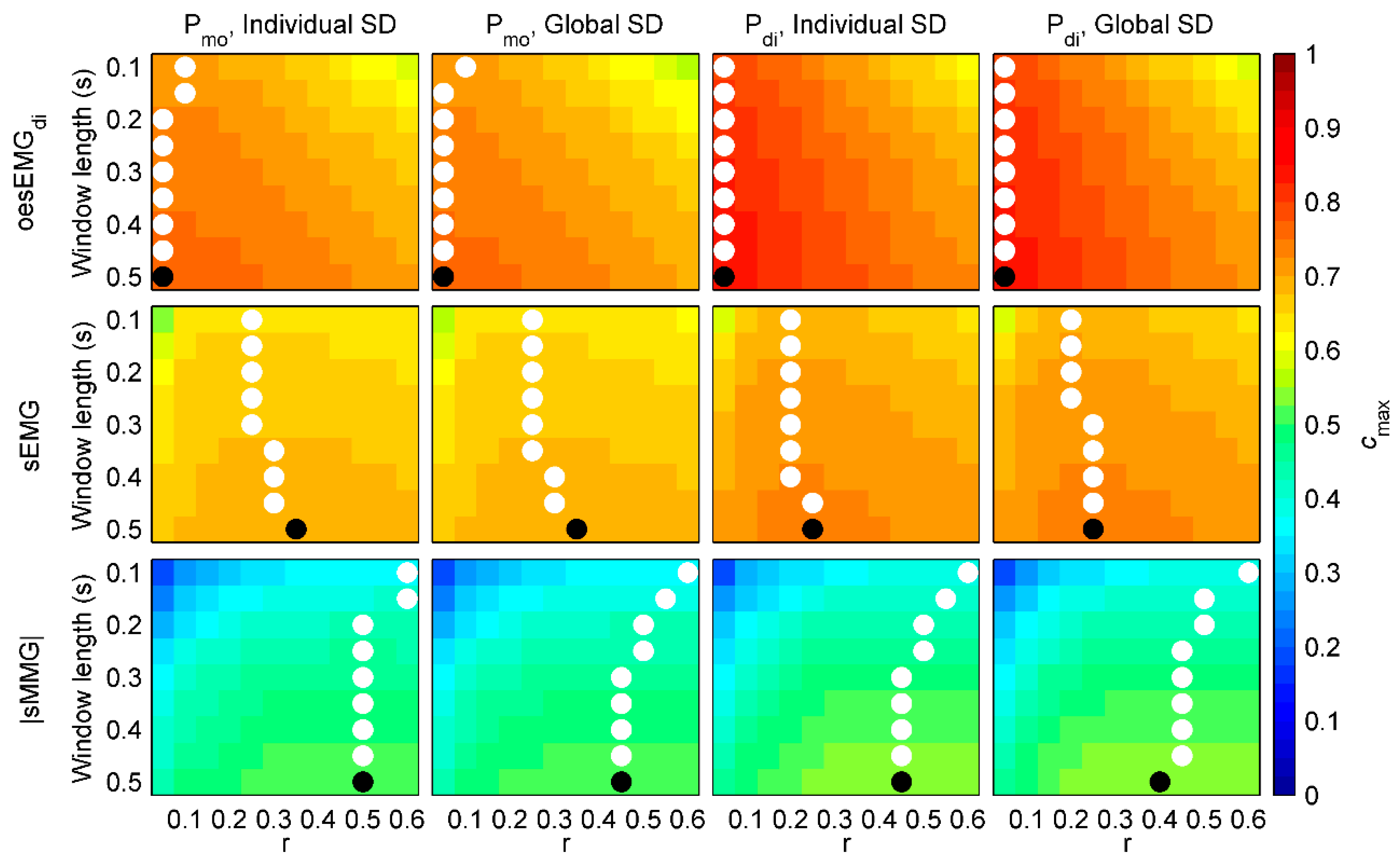
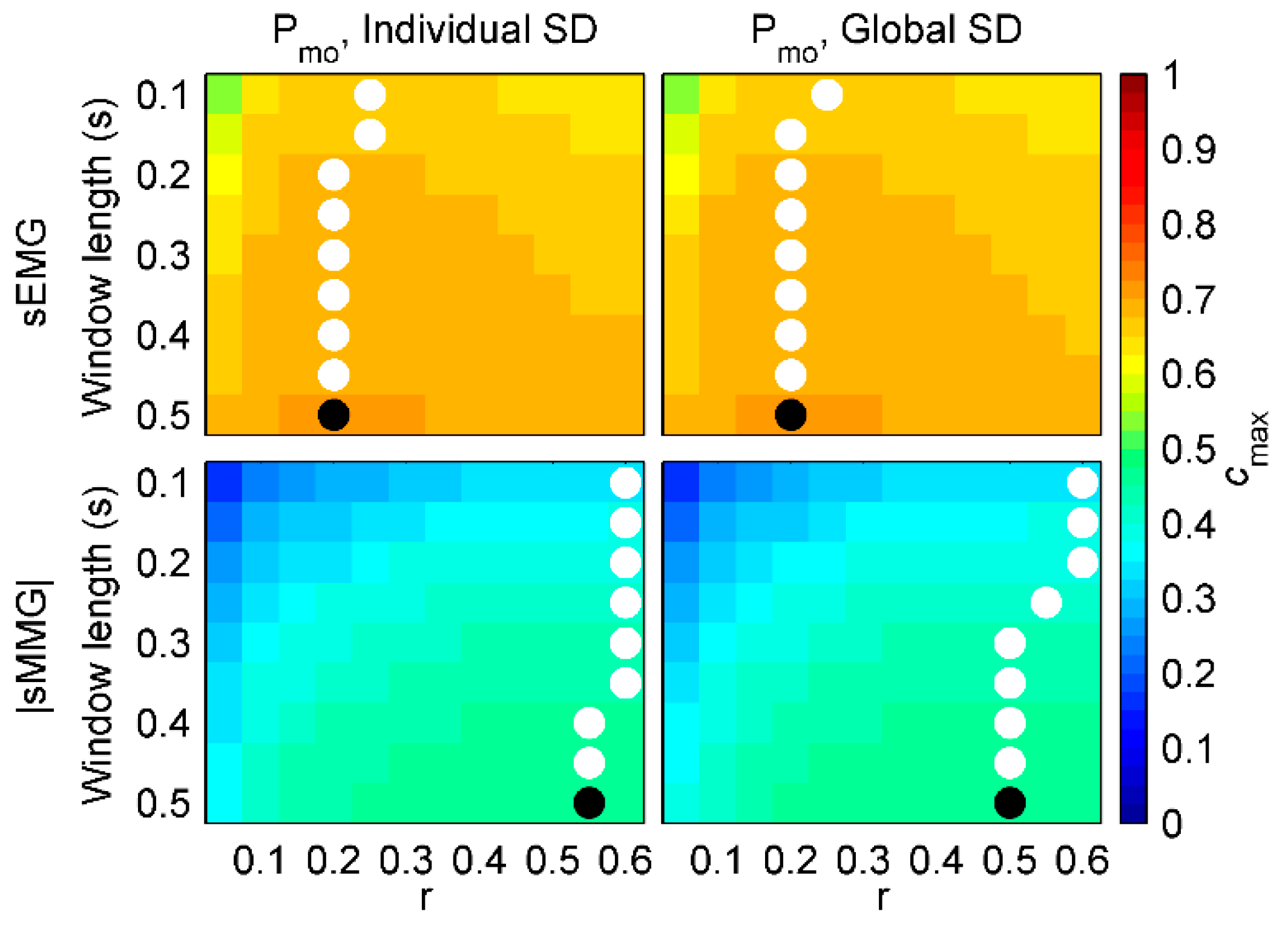
3. Results
3.1. Individual and Global SDs
3.2. Performance of fSampEn in Healthy Subjects
3.3. Performance of fSampEn in COPD Patients
3.4. General fSampEn Parameters
4. Discussion and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Jolley, C.J.; Luo, Y.M.; Steier, J.; Reilly, C.; Seymour, J.; Lunt, A.; Ward, K.; Rafferty, G.F.; Polkey, M.I.; Moxham, J. Neural respiratory drive in healthy subjects and in COPD. Eur. Respir. J. 2009, 33, 289–297. [Google Scholar] [CrossRef] [PubMed]
- American Thoracic Society/European Respiratory Society. ATS/ERS statement on respiratory muscle testing. Am. J. Respir. Crit. Care Med. 2002, 166, 518–624. [Google Scholar] [CrossRef] [PubMed]
- Mead, J.; Loring, S.H. Analysis of volume displacement and length changes of the diaphragm during breathing. J. Appl. Physiol. 1982, 53, 750–755. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.M.; Moxham, J.; Polkey, M.I. Diaphragm electromyography using an oesophageal catheter: Current concepts. Clin. Sci. (Lond.) 2008, 115, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Sarlabous, L.; Torres, A.; Fiz, J.A.; Martínez-Llorens, J.M.; Gea, J.; Jané, R. Inspiratory muscle activation increases with COPD severity as confirmed by non-invasive mechanomyographic analysis. PLoS ONE 2017, 12, e0177730. [Google Scholar] [CrossRef] [PubMed]
- Lozano-García, M.; Sarlabous, L.; Moxham, J.; Rafferty, G.F.; Torres, A.; Jané, R.; Jolley, C.J. Surface mechanomyography and electromyography provide non-invasive indices of inspiratory muscle force and activation in healthy subjects. Sci. Rep. 2018, 8, 16921. [Google Scholar] [CrossRef] [PubMed]
- Reilly, C.C.; Ward, K.; Jolley, C.J.; Lunt, A.C.; Steier, J.; Elston, C.; Polkey, M.I.; Rafferty, G.F.; Moxham, J. Neural respiratory drive, pulmonary mechanics and breathlessness in patients with cystic fibrosis. Thorax 2011, 66, 240–246. [Google Scholar] [CrossRef] [PubMed]
- MacBean, V.; Hughes, C.; Nicol, G.; Reilly, C.C.; Rafferty, G.F. Measurement of neural respiratory drive via parasternal intercostal electromyography in healthy adult subjects. Physiol. Meas. 2016, 37, 2050–2063. [Google Scholar] [CrossRef]
- Estrada, L.; Torres, A.; Sarlabous, L.; Jané, R. Improvement in neural respiratory drive estimation from diaphragm electromyographic signals using fixed sample entropy. IEEE J. Biomed. Health Inform. 2016, 20, 476–485. [Google Scholar] [CrossRef]
- Lin, L.; Guan, L.; Wu, W.; Chen, R. Correlation of surface respiratory electromyography with esophageal diaphragm electromyography. Respir. Physiol. Neurobiol. 2018. [Google Scholar] [CrossRef]
- González-Izal, M.; Malanda, A.; Gorostiaga, E.; Izquierdo, M. Electromyographic models to assess muscle fatigue. J. Electromyogr. Kinesiol. 2012, 22, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Ibitoye, M.O.; Hamzaid, N.A.; Zuniga, J.M.; Hasnan, N.; Wahab, A.K.A. Mechanomyographic parameter extraction methods: An appraisal for clinical applications. Sensors 2014, 14, 22940–22970. [Google Scholar] [CrossRef] [PubMed]
- Sinderby, C.; Lindström, L.; Grassino, A.E. Automatic assessment of electromyogram quality. J. Appl. Physiol. 1995, 79, 1803–1815. [Google Scholar] [CrossRef] [PubMed]
- Sinderby, C.A.; Beck, J.C.; Lindström, L.H.; Grassino, A.E. Enhancement of signal quality in esophageal recordings of diaphragm EMG. J. Appl. Physiol. 1997, 82, 1370–1377. [Google Scholar] [CrossRef] [PubMed]
- Sarlabous, L.; Torres, A.; Fiz, J.A.; Jané, R. Evidence towards improved estimation of respiratory muscle effort from diaphragm mechanomyographic signals with cardiac vibration interference using sample entropy with fixed tolerance values. PLoS ONE 2014, 9, e88902. [Google Scholar] [CrossRef] [PubMed]
- Richman, J.S.; Moorman, J.R. Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol. Heart Circ. Physiol. 2000, 278, H2039–H2049. [Google Scholar] [CrossRef] [PubMed]
- Ràfols-de-Urquía, M.; Estrada, L.; Estévez-Piorno, J.; Sarlabous, L.; Jané, R.; Torres, A. Evaluation of a wearable device to determine cardiorespiratory parameters from surface diaphragm electromyography. IEEE J. Biomed. Health Inform. 2018. [Google Scholar] [CrossRef] [PubMed]
- Estrada, L.; Torres, A.; Sarlabous, L.; Jané, R. Onset and Offset Estimation of the Neural Inspiratory Time in Surface Diaphragm Electromyography: A Pilot Study in Healthy Subjects. IEEE J. Biomed. Health Inform. 2018, 22, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Sarlabous, L.; Torres, A.; Fiz, J.A.; Gea, J.; Martinez-Llorens, J.M.; Jané, R. Efficiency of mechanical activation of inspiratory muscles in COPD using sample entropy. Eur. Respir. J. 2015, 46, 1808–1811. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhou, P. Sample entropy analysis of surface EMG for improved muscle activity onset detection against spurious background spikes. J. Electromyogr. Kinesiol. 2012, 22, 901–907. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Zhang, X. A novel technique for muscle onset detection using surface EMG signals without removal of ECG artifacts. Physiol. Meas. 2014, 35, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, Q. Use of the integrated profile for voluntary muscle activity detection using EMG signals with spurious background spikes: A study with incomplete spinal cord injury. Biomed. Signal Process. Control 2016, 24, 19–24. [Google Scholar] [CrossRef]
- Kamavuako, E.N.; Farina, D.; Yoshida, K.; Jensen, W. Estimation of grasping force from features of intramuscular EMG signals with mirrored bilateral training. Ann. Biomed. Eng. 2012, 40, 648–656. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ren, X.; Gao, X.; Chen, X.; Zhou, P. Complexity analysis of surface EMG for overcoming ECG interference toward proportional myoelectric control. Entropy 2016, 18, 106. [Google Scholar] [CrossRef]
- Lozano-García, M.; Sarlabous, L.; Moxham, J.; Rafferty, G.F.; Torres, A.; Jolley, C.J.; Jané, R. Assessment of Inspiratory Muscle Activation using Surface Diaphragm Mechanomyography and Crural Diaphragm Electromyography. In Proceedings of the 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Honolulu, HI, USA, 18–21 July 2018; pp. 3342–3345. [Google Scholar]
- Estrada, L.; Torres, A.; Sarlabous, L.; Jané, R. Influence of parameter selection in fixed sample entropy of surface diaphragm electromyography for estimating respiratory activity. Entropy 2017, 19, 460. [Google Scholar] [CrossRef]
- Petitjean, M.; Bellemare, F. Phonomyogram of the diaphragm during unilateral and bilateral phrenic nerve stimulation and changes with fatigue. Muscle Nerve 1994, 17, 1201–1209. [Google Scholar] [CrossRef] [PubMed]
- Reilly, C.C.; Jolley, C.J.; Ward, K.; MacBean, V.; Moxham, J.; Rafferty, G.F. Neural respiratory drive measured during inspiratory threshold loading and acute hypercapnia in healthy individuals. Exp. Physiol. 2013, 98, 1190–1198. [Google Scholar] [CrossRef]
- Baydur, A.; Behrakis, P.K.; Zin, W.A.; Jaeger, M.; Milic-Emili, J. A simple method for assessing the validity of the esophageal balloon technique. Am. Rev. Respir. Dis. 1982, 126, 788–791. [Google Scholar] [CrossRef]
- Watson, A.C.; Hughes, P.D.; Louise Harris, M.; Hart, N.; Ware, R.J.; Wendon, J.; Green, M.; Moxham, J. Measurement of twitch transdiaphragmatic, esophageal, and endotracheal tube pressure with bilateral anterolateral magnetic phrenic nerve stimulation in patients in the intensive care unit. Crit. Care Med. 2001, 29, 1325–1331. [Google Scholar] [CrossRef]
- Luo, Y.M.; Moxham, J. Measurement of neural respiratory drive in patients with COPD. Respir. Physiol. Neurobiol. 2005, 146, 165–174. [Google Scholar] [CrossRef]
- Lake, D.E.; Richman, J.S.; Griffin, M.P.; Moorman, J.R. Sample entropy analysis of neonatal heart rate variability. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002, 283, R789–R797. [Google Scholar] [CrossRef] [PubMed]
- Gonem, S.; Umar, I.; Burke, D.; Desai, D.; Corkill, S.; Owers-Bradley, J.; Brightling, C.E.; Siddiqui, S. Airway impedance entropy and exacerbations in severe asthma. Eur. Respir. J. 2012, 40, 1156–1163. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Liu, Q.; Fan, S.Z.; Lu, C.W.; Lin, T.Y.; Abbod, M.F.; Shieh, J.S. Analysis of EEG via multivariate empirical mode decomposition for depth of anesthesia based on sample entropy. Entropy 2013, 15, 3458–3470. [Google Scholar] [CrossRef]
- Duiverman, M.L.; van Eykern, L.A.; Vennik, P.W.; Koëter, G.H.; Maarsingh, E.J.W.; Wijkstra, P.J. Reproducibility and responsiveness of a noninvasive EMG technique of the respiratory muscles in COPD patients and in healthy subjects. J. Appl. Physiol. 2004, 96, 1723–1729. [Google Scholar] [CrossRef] [PubMed]
- Yentes, J.M.; Hunt, N.; Schmid, K.K.; Kaipust, J.P.; McGrath, D.; Stergiou, N. The appropriate use of approximate entropy and sample entropy with short data sets. Ann. Biomed. Eng. 2013, 41, 349–365. [Google Scholar] [CrossRef]
- Richman, J.S.; Lake, D.E.; Moorman, J.R. Sample Entropy. Methods Enzymol. 2004, 384, 172–184. [Google Scholar]



| Subject | oesEMGdi vs. Pmo | oesEMGdi vs. Pdi | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ind. SD | Glob. SD | Δcmax (%) | Ind. SD | Glob. SD | Δcmax (%) | |||||
| Ind. r | cmax | Glob. r | cmax | Ind. r | cmax | Glob. r | cmax | |||
| H1 | 0.20 | 0.767 | 0.05 | 0.758 | 1.15 | 0.05 | 0.869 | 0.05 | 0.870 | 0.06 |
| H2 | 0.05 | 0.690 | 0.05 | 0.691 | 0.22 | 0.05 | 0.750 | 0.05 | 0.751 | 0.08 |
| H3 | 0.10 | 0.776 | 0.05 | 0.775 | 0.09 | 0.05 | 0.782 | 0.05 | 0.779 | 0.32 |
| H4 | 0.20 | 0.751 | 0.05 | 0.742 | 1.18 | 0.10 | 0.879 | 0.05 | 0.871 | 0.84 |
| H5 | 0.40 | 0.723 | 0.05 | 0.691 | 4.53 | 0.05 | 0.883 | 0.05 | 0.883 | 0.05 |
| H6 | 0.10 | 0.823 | 0.05 | 0.818 | 0.60 | 0.05 | 0.784 | 0.05 | 0.784 | 0.03 |
| H7 | 0.25 | 0.743 | 0.05 | 0.725 | 2.36 | 0.05 | 0.941 | 0.05 | 0.942 | 0.06 |
| H8 | 0.05 | 0.814 | 0.05 | 0.814 | 0.05 | 0.05 | 0.907 | 0.05 | 0.909 | 0.18 |
| H9 | 0.05 | 0.799 | 0.05 | 0.798 | 0.10 | 0.05 | 0.848 | 0.05 | 0.845 | 0.43 |
| H10 | 0.15 | 0.822 | 0.05 | 0.811 | 1.27 | 0.05 | 0.860 | 0.05 | 0.860 | 0.00 |
| H11 | 0.30 | 0.726 | 0.05 | 0.716 | 1.32 | 0.05 | 0.868 | 0.05 | 0.869 | 0.10 |
| H12 | 0.05 | 0.774 | 0.05 | 0.772 | 0.28 | 0.05 | 0.728 | 0.05 | 0.725 | 0.42 |
| Median (IQR) | 0.13 (0.05–0.21) | 0.770 (0.739–0.803) | 0.05 (0.05–0.05) | 0.765 (0.723–0.801) | 0.88 (0.19–1.28) | 0.05 (0.05–0.05) | 0.864 (0.784–0.880) | 0.05 (0.05–0.05) | 0.865 (0.783–0.874) | 0.09 (0.06–0.35) |
| Subject | sEMG vs. Pmo | sEMG vs. Pdi | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ind. SD | Glob. SD | Δcmax (%) | Ind. SD | Glob. SD | Δcmax (%) | |||||
| Ind. r | cmax | Glob. r | cmax | Ind. r | cmax | Glob. r | cmax | |||
| H1 | 0.60 | 0.765 | 0.35 | 0.747 | 2.32 | 0.40 | 0.850 | 0.25 | 0.829 | 2.54 |
| H2 | 0.60 | 0.575 | 0.35 | 0.572 | 0.47 | 0.60 | 0.639 | 0.25 | 0.619 | 3.18 |
| H3 | 0.60 | 0.663 | 0.35 | 0.664 | 0.12 | 0.60 | 0.658 | 0.25 | 0.653 | 0.80 |
| H4 | 0.30 | 0.666 | 0.35 | 0.666 | 0.07 | 0.30 | 0.647 | 0.25 | 0.643 | 0.72 |
| H5 | 0.60 | 0.707 | 0.35 | 0.703 | 0.57 | 0.35 | 0.801 | 0.25 | 0.800 | 0.04 |
| H6 | 0.35 | 0.736 | 0.35 | 0.732 | 0.63 | 0.30 | 0.697 | 0.25 | 0.696 | 0.18 |
| H7 | 0.50 | 0.714 | 0.35 | 0.706 | 1.12 | 0.20 | 0.842 | 0.25 | 0.842 | 0.00 |
| H8 | 0.30 | 0.822 | 0.35 | 0.822 | 0.01 | 0.30 | 0.864 | 0.25 | 0.862 | 0.18 |
| H9 | 0.10 | 0.709 | 0.35 | 0.689 | 2.69 | 0.15 | 0.720 | 0.25 | 0.718 | 0.35 |
| H10 | 0.20 | 0.779 | 0.35 | 0.779 | 0.10 | 0.10 | 0.840 | 0.25 | 0.834 | 0.65 |
| H11 | 0.45 | 0.585 | 0.35 | 0.568 | 2.87 | 0.25 | 0.701 | 0.25 | 0.699 | 0.27 |
| H12 | 0.10 | 0.680 | 0.35 | 0.606 | 11.01 | 0.10 | 0.643 | 0.25 | 0.604 | 5.97 |
| Median (IQR) | 0.40 (0.28–0.60) | 0.708 (0.665–0.744) | 0.35 (0.35–0.35) | 0.696 (0.649–0.736) | 0.60 (0.11–2.41) | 0.30 (0.19–0.36) | 0.711 (0.655–0.840) | 0.25 (0.25–0.25) | 0.708 (0.650–0.830) | 0.50 (0.18–1.23) |
| Subject | |sMMG| vs. Pmo | |sMMG| vs. Pdi | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ind. SD | Glob. SD | Δcmax (%) | Ind. SD | Glob. SD | Δcmax (%) | |||||
| Ind. r | cmax | Glob. r | cmax | Ind. r | cmax | Glob. r | cmax | |||
| H1 | 0.55 | 0.642 | 0.45 | 0.641 | 0.10 | 0.50 | 0.649 | 0.40 | 0.648 | 0.13 |
| H2 | 0.50 | 0.388 | 0.45 | 0.385 | 0.62 | 0.50 | 0.431 | 0.40 | 0.427 | 0.91 |
| H3 | 0.50 | 0.443 | 0.45 | 0.439 | 0.94 | 0.60 | 0.431 | 0.40 | 0.432 | 0.21 |
| H4 | 0.55 | 0.394 | 0.45 | 0.392 | 0.52 | 0.25 | 0.471 | 0.40 | 0.469 | 0.51 |
| H5 | 0.55 | 0.529 | 0.45 | 0.529 | 0.02 | 0.50 | 0.562 | 0.40 | 0.562 | 0.03 |
| H6 | 0.40 | 0.532 | 0.45 | 0.530 | 0.20 | 0.50 | 0.480 | 0.40 | 0.479 | 0.16 |
| H7 | 0.15 | 0.530 | 0.45 | 0.524 | 1.29 | 0.20 | 0.653 | 0.40 | 0.650 | 0.33 |
| H8 | 0.60 | 0.666 | 0.45 | 0.652 | 2.02 | 0.60 | 0.717 | 0.40 | 0.700 | 2.39 |
| H9 | 0.45 | 0.470 | 0.45 | 0.469 | 0.24 | 0.50 | 0.478 | 0.40 | 0.478 | 0.06 |
| H10 | 0.40 | 0.654 | 0.45 | 0.654 | 0.01 | 0.35 | 0.690 | 0.40 | 0.690 | 0.05 |
| H11 | 0.55 | 0.536 | 0.45 | 0.532 | 0.62 | 0.50 | 0.642 | 0.40 | 0.639 | 0.47 |
| H12 | 0.50 | 0.471 | 0.45 | 0.468 | 0.62 | 0.10 | 0.408 | 0.40 | 0.400 | 2.04 |
| Median (IQR) | 0.50 (0.44–0.55) | 0.530 (0.463–0.562) | 0.45 (0.45–0.45) | 0.526 (0.461–0.560) | 0.57 (0.18–0.70) | 0.50 (0.33–0.50) | 0.521 (0.461–0.650) | 0.40 (0.40–0.40) | 0.520 (0.460–0.649) | 0.27 (0.11–0.61) |
| Subject | sEMG vs. Pmo | |sMMG| vs. Pmo | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ind. SD | Glob. SD | Δcmax (%) | Ind. SD | Glob. SD | Δcmax (%) | |||||
| Ind. r | cmax | Glob. r | cmax | Ind. r | cmax | Glob. r | cmax | |||
| P1 | 0.20 | 0.649 | 0.20 | 0.648 | 0.10 | 0.60 | 0.376 | 0.50 | 0.377 | 0.22 |
| P2 | 0.15 | 0.634 | 0.20 | 0.634 | 0.07 | 0.60 | 0.334 | 0.50 | 0.330 | 1.29 |
| P3 | 0.15 | 0.555 | 0.20 | 0.545 | 1.72 | 0.60 | 0.425 | 0.50 | 0.425 | 0.11 |
| P4 | 0.10 | 0.571 | 0.20 | 0.569 | 0.35 | 0.35 | 0.434 | 0.50 | 0.428 | 1.26 |
| P5 | 0.15 | 0.822 | 0.20 | 0.822 | 0.00 | 0.60 | 0.551 | 0.50 | 0.550 | 0.29 |
| P6 | 0.55 | 0.705 | 0.20 | 0.698 | 1.11 | 0.40 | 0.553 | 0.50 | 0.551 | 0.26 |
| P7 | 0.40 | 0.756 | 0.20 | 0.753 | 0.32 | 0.60 | 0.630 | 0.50 | 0.632 | 0.24 |
| P8 | 0.20 | 0.793 | 0.20 | 0.791 | 0.15 | 0.40 | 0.585 | 0.50 | 0.584 | 0.29 |
| P9 | 0.50 | 0.665 | 0.20 | 0.659 | 0.82 | 0.20 | 0.247 | 0.50 | 0.233 | 5.75 |
| P10 | 0.15 | 0.815 | 0.20 | 0.815 | 0.01 | 0.30 | 0.445 | 0.50 | 0.442 | 0.81 |
| P11 | 0.30 | 0.794 | 0.20 | 0.793 | 0.11 | 0.60 | 0.523 | 0.50 | 0.524 | 0.21 |
| P12 | 0.25 | 0.692 | 0.20 | 0.691 | 0.03 | 0.60 | 0.450 | 0.50 | 0.446 | 0.84 |
| P13 | 0.15 | 0.628 | 0.20 | 0.627 | 0.19 | 0.50 | 0.554 | 0.50 | 0.553 | 0.14 |
| P14 | 0.30 | 0.792 | 0.20 | 0.783 | 1.17 | 0.30 | 0.574 | 0.50 | 0.574 | 0.11 |
| Median (IQR) | 0.20 (0.15–0.30) | 0.698 (0.638–0.793) | 0.20 (0.20–0.20) | 0.694 (0.637–0.789) | 0.17 (0.08–0.7) | 0.55 (0.36–0.60) | 0.487 (0.427–0.553) | 0.50 (0.50–0.50) | 0.485 (0.426–0.552) | 0.27 (0.21–0.83) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lozano-García, M.; Estrada, L.; Jané, R. Performance Evaluation of Fixed Sample Entropy in Myographic Signals for Inspiratory Muscle Activity Estimation. Entropy 2019, 21, 183. https://doi.org/10.3390/e21020183
Lozano-García M, Estrada L, Jané R. Performance Evaluation of Fixed Sample Entropy in Myographic Signals for Inspiratory Muscle Activity Estimation. Entropy. 2019; 21(2):183. https://doi.org/10.3390/e21020183
Chicago/Turabian StyleLozano-García, Manuel, Luis Estrada, and Raimon Jané. 2019. "Performance Evaluation of Fixed Sample Entropy in Myographic Signals for Inspiratory Muscle Activity Estimation" Entropy 21, no. 2: 183. https://doi.org/10.3390/e21020183
APA StyleLozano-García, M., Estrada, L., & Jané, R. (2019). Performance Evaluation of Fixed Sample Entropy in Myographic Signals for Inspiratory Muscle Activity Estimation. Entropy, 21(2), 183. https://doi.org/10.3390/e21020183







