1. Introduction
The human brain shows complex spatiotemporal behaviors when executing physiological functions. Characterizing dynamics of the complex spatiotemporal behaviors is of great significance in understanding the human brain. Since blood oxygenation level-dependent (BOLD) signals of different brain regions can be measured by the functional magnetic resonance imaging (fMRI) technique at high spatial and temporal resolutions, BOLD signals have been widely used to characterize dynamics of the spatiotemporal behaviors of the human brain [
1,
2]. For instance, the temporal correlation in BOLD signals of two distinct brain regions is commonly employed to describe the functional connectivity (FC) between them [
3]. A positive and strong temporal correlation corresponds to a strong FC, and some brain regions with strong FCs among them constitute a brain functional network [
4,
5,
6]. Alterations of some FCs in a brain functional network are often associated with brain disorder, such as schizophrenia [
7], major depression [
8], autism [
9], Alzheimer’s Disease [
10], and attention deficit hyperactivity disorder [
11]. For example, Cheng et al. evaluated the FC between different brain regions in subjects with autism and found a key system in the middle temporal gyrus with reduced FC and a key system in the precuneus with reduced FC [
12].
In most previous research on FC, only one correlation coefficient is acquired using entire BOLD signals of two distinct brain regions. The one correlation coefficient is called the static FC between the two brain regions. Recently, to understand dynamics of the spatiotemporal behaviors of the human brain more deeply, some researchers acquired a sequence of correlation coefficients by applying the sliding-window approach to BOLD signals of two distinct brain regions [
13,
14,
15,
16,
17,
18,
19,
20,
21,
22,
23]. These correlation coefficients form a time series which is called the dynamic FC between the two brain regions. The dynamic FC exhibits complex characteristics which are effective in describing properties of the brain functional networks of patients with brain disorder. For instance, in one of our recent studies, complex characteristics of dynamic FC were described by sample entropy (SampEn), and the effects of schizophrenia on such complex characteristics were investigated. It was shown that the visual cortex of the patients with schizophrenia exhibited significantly higher SampEn than that of the healthy controls [
24]. As introduced above, both the static FC and the SampEn of dynamic FC are effective in describing properties of the brain functional networks of patients with brain disorder. However, the effectivenesses of the static FC and the dynamic FC have not been compared directly.
Studies on the static FC or the dynamic FC are often carried out by first extracting BOLD signals of different brain regions and then evaluating the static or the dynamic FC between different brain regions for further analysis. Different brain regions are often determined by a brain template, such as the Dosenbach’s template [
25]. The Dosenbach’s template includes 160 regions of interest (ROIs) determined by a sequence of meta-analyses of task-based fMRI studies which cover much of the human brain [
25]. Furthermore, the 160 ROIs can be separated into six functional networks including the default, the frontal-parietal, the cingulo-opercular, the sensorimotor, the occipital, and the cerebellum networks, which were identified by performing modularity optimization on the average FC matrix across a large cohort of healthy subjects [
25]. The six functional networks have been used in predicting brain maturity across development [
25,
26], parcellating cortical or subcortical regions [
27], examining the influence of temporal properties of BOLD signals on FC [
28] and so on. For instance, Zhong et al. parcellated the hippocampus based on the FC, and showed that both the left and right hippocampus were divided into three subregions exhibiting different FC profiles with the six functional networks [
27]. However, machine learning algorithms have not been used to identify the six functional networks.
The K-means clustering algorithm is one of the unsupervised learning algorithms [
29]. Since the K-means clustering algorithm can cluster different observations into different clusters in a simple and easy way, it has been widely used in fMRI studies [
30,
31,
32,
33,
34,
35,
36,
37,
38]. For instance, Fan et al. used the K-means clustering algorithm to parcellate the thalamus based on the static FC and found that the thalamus could be divided into seven symmetric thalamic clusters [
36]. Park et al. parcellated the primary and secondary visual cortices (V1 and V2) into several subregions by applying the K-means clustering algorithm to the static FC and found that V1 and V2 could be separated into anterior and posterior subregions [
38].
The present study intends to cluster the Dosenbach’s 160 ROIs into six clusters by applying the K-means clustering algorithm to the static FC and the SampEn of dynamic FC, to analyze the overlap and consistency between the six clusters and the six functional networks, and to compare the effectivenesses of the static FC and the dynamic FC. It is shown that applying the K-means clustering algorithm to FC is feasible to identify the six functional networks, and the SampEn of dynamic FC is more effective than the static FC as the six clusters obtained from the SampEn of dynamic FC show higher overlap and consistency ratios with the six functional networks.
This paper is organized as follows. The experiments and methods are presented in
Section 2. The cluster results for the static FC and the SampEn of dynamic FC and the comparisons between them are shown in
Section 3. The conclusion and discussion are described in
Section 4. Some supplementary tables are presented in the appendix.
2. Experiments and Methods
2.1. Participants
FMRI data for this study were acquired at Olin Neuropsychiatry Research Center and have been made publicly available
http://fcon_1000.projects.nitrc.org/indi/abide. The data were acquired from 31 healthy participants (18 males and 13 females) over the age range 18–30 years. This sample was retained after applying criteria for head motion, from a total of 35 healthy participants. Informed consent was obtained from all participants in accordance with Olin Neuropsychiatry Research Center Institutional Review Board oversight.
2.2. Data Acquisition and Preprocessing
BOLD signals are extracted from three-dimensional functional images collected on a Siemens 3T MRI scanner with the following parameters: repetition time (TR), 475 ms; echo time, 30 ms; field of view, ; slices, 48; slice thickness, 3 mm; flip angle, . During the data collection, all participants were instructed to rest but not fall asleep. For each participant, 947 three-dimensional functional images were collected.
The functional images are preprocessed using SPM8 and DPABI softwares [
39,
40]. Firstly, the first 4 images are discarded to reduce the negative effects of scanner’s stabilization on the analysis results. Secondly, the images are corrected for time delay in slice acquisition and rigid-body head motion. Thirdly, several confounding factors are regressed out from the images, including 6 head motion parameters and the cerebrospinal, the white matter, and the global brain signals. Fourthly, temporal band-pass filtering (0.01–0.08 Hz) of the images are performed to reduce the negative effects of low-frequency drift and high-frequency physiological noise on the analysis results. Fifthly, the images are spatially normalized to the Montreal Neurological Institute space and are resampled to voxels of size
. Sixthly, the images are smoothed with a Gaussian kernel of 8 mm full-width at half-maximum. Finally, the BOLD signal of each voxel is extracted from the functional images.
2.3. The Dosenbach’s Template and the 6 Functional Networks
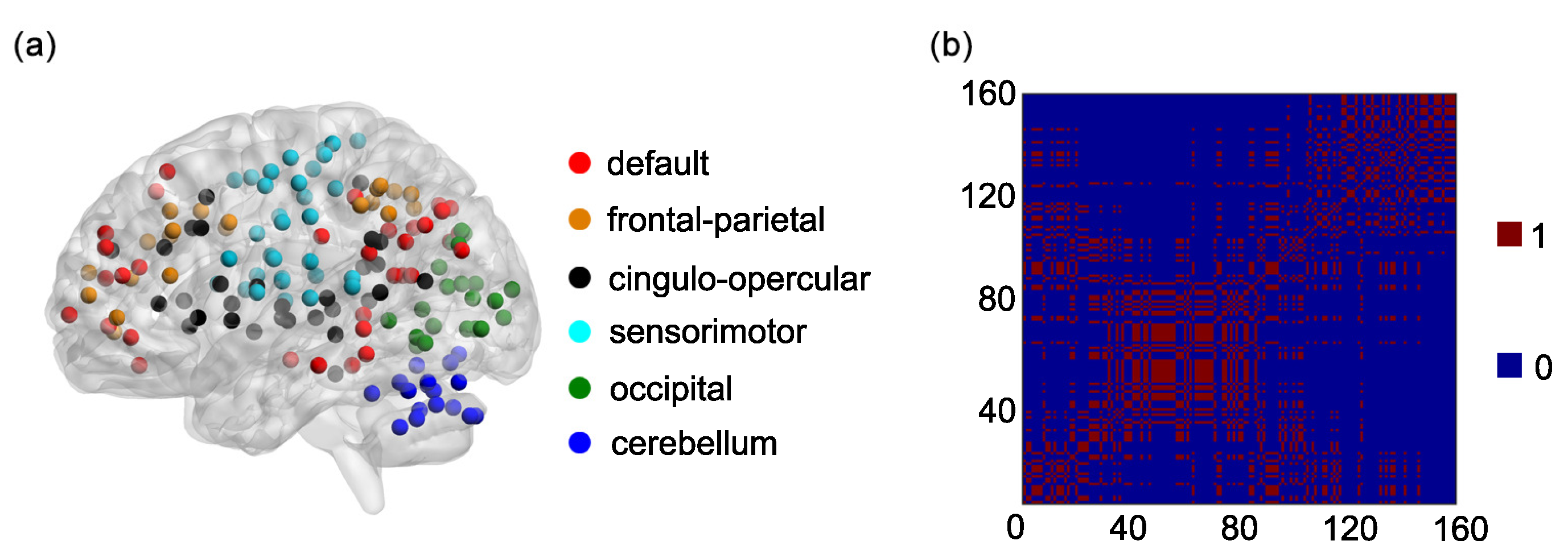
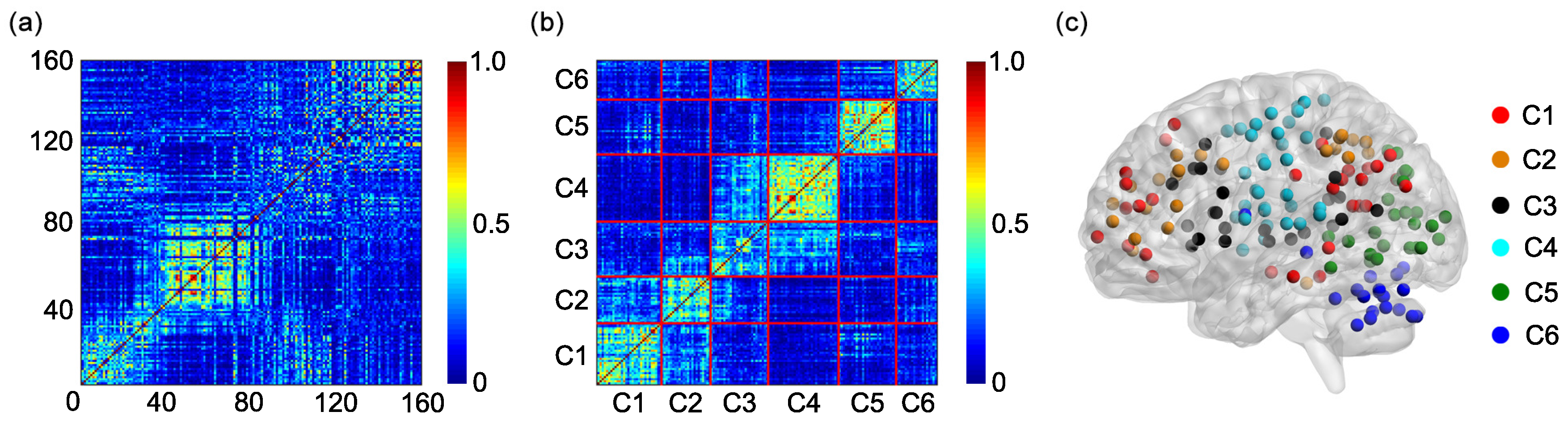
One hundred and sixty regions of interest (ROIs) are selected based on the Dosenbach’s template [
25]. The centroid of each ROI is derived from a sequence of meta-analyses of task-based fMRI studies (
Figure 1a). The radius of each ROI equals 5 mm (
Figure 1a). The name and the sequential number of each ROI can be found in
Table A1 in
Appendix A. The 160 ROIs can further be grouped into 6 functional networks, including the default, the frontal-parietal, the cingulo-opercular, the sensorimotor, the occipital, and the cerebellum networks (
Figure 1a). The name and the sequential number of each ROI in each functional network can be found in the first and second columns of
Table A2,
Table A3,
Table A4,
Table A5,
Table A6 and
Table A7 in
Appendix A.
Based on the 6 functional networks, an adjacent matrix can be generated [
36,
41,
42]. The adjacent matrix is labeled as
Each of the elements on the main diagonal of
A is 1. Other elements of
A are defined as follows:
if the
ith ROI and the
jth ROI are contained in the same functional network and
otherwise
(
Figure 1b).
2.4. The Static FC and the Dynamic FC
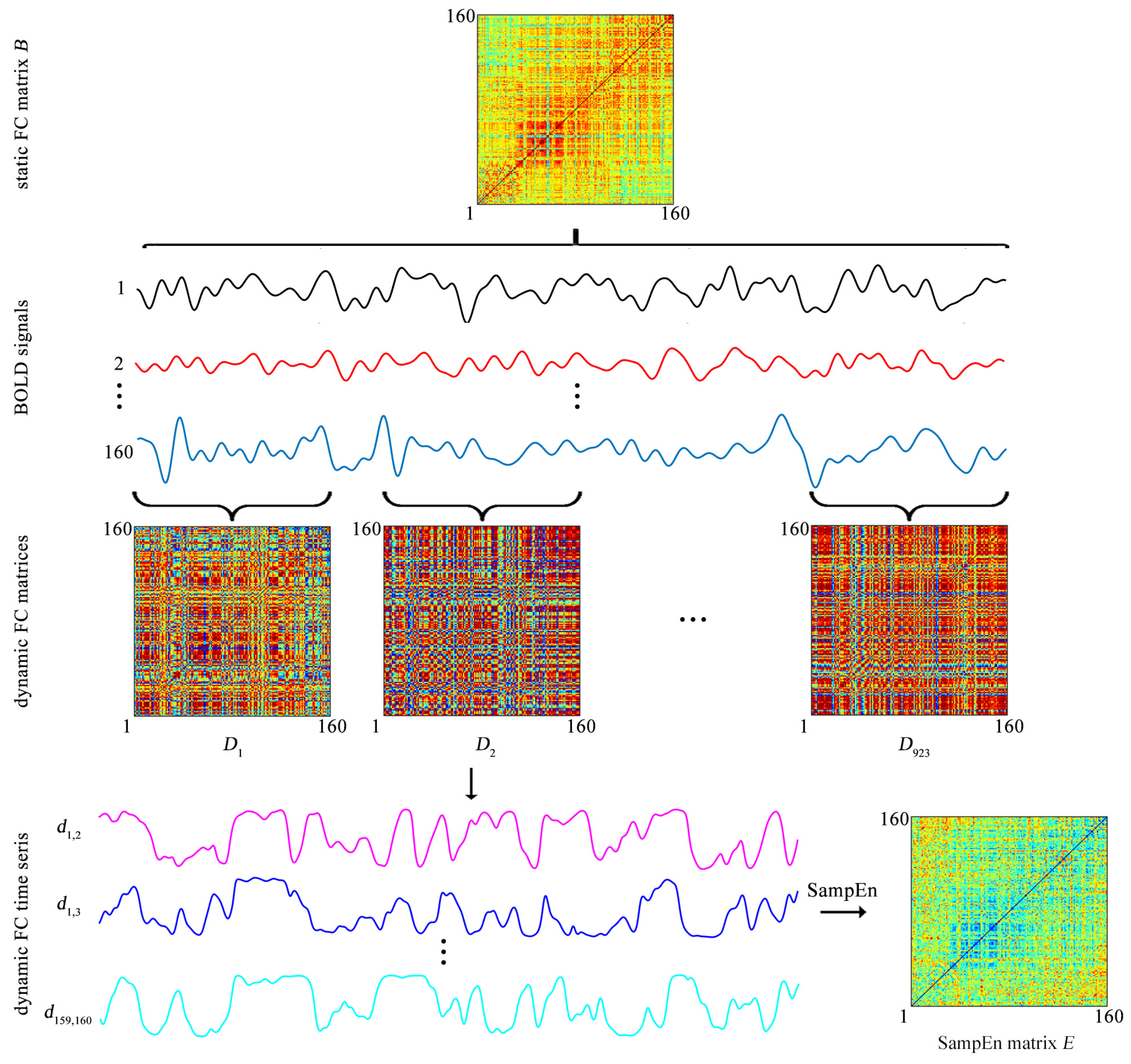
The BOLD signal of each ROI is extracted by averaging the BOLD signals over all voxels in this ROI. Then both the static FC and the dynamic FC are evaluated (
Figure 2).
The static FC between each pair of ROIs is assessed by a Pearson correlation coefficient. For each of the 31 participants, after the static FC between each pair of ROIs is evaluated, a static FC matrix of size
is obtained (
Figure 2), which is labeled as
The
ith row
represents the static FC between the
ith ROI and all the other ROIs
The matrix
B is used to cluster the 160 ROIs into 6 clusters.
Dynamic FC is assessed by the sliding-window approach. Specifically, a tapered window is created by convolving a rectangle window (size = 20 TRs = 9.5 s) with a Gaussian curve (standard deviation = 3 TRs) [
14,
15,
23]. The window is used to extract BOLD signals in a step of 1 TR, leading to 923 time windows per subject (
Figure 2). For the
kth time window
a Pearson correlation coefficient is used to evaluate the FC between each pair of ROIs and thus a FC matrix of size
which is labeled as
which is obtained for each subject (
Figure 2). As
k increases from 1 to 923,
forms a time series
which represents the temporal evolution of the FC between the
ith and
jth ROIs and is named as the dynamic FC (
Figure 2). Since previous studies showed that the window of size 20 TRs captures more transient patterns in dynamic FC [
23], the window size is fixed at 20 TRs throughout the study.
2.5. SampEn of a Dynamic FC Time Series
For each dynamic FC time series,
the SampEn is calculated. For convenience, time series
is denoted by
SampEn of
is computed as follows [
24,
43,
44,
45,
46].
Firstly, constructing embedding vectors in which m stands for the dimension of
Secondly, define
r stands for a tolerance value which is defined as
, where
is a small parameter and
is the standard deviation of
.
, the Heaviside function, which is defined as
represents the Chebyshev distance, i.e.,
Similarly, define
Thirdly, in view of Equations (
4) and (
7), we define
and
Finally, calculate SampEn of
as
The value of SampEn is not less than 0, and a larger value of SampEn means more complexity [
47]. Similar to our previous study [
24,
43],
m and
are fixed at 2 and 0.2, respectively.
In addition, because
the SampEn of
equals 0
Thus, for each participant, a SampEn matrix of size
is obtained (
Figure 2). The SampEn matrix is labeled as
The element
represents the SampEn of dynamic FC between the
ith ROI and
jth ROI
equals 0
The matrix
E is used to cluster the 160 ROIs into 6 clusters.
2.6. Clustering ROIs into 6 Clusters by Applying the K-Means Clustering Algorithm to the Static FC Matrix
For each of the 31 participants, there exists a static FC matrix B of size The ith row represents the static FC between the ith ROI and all the other ROIs.
The K-means clustering algorithm is commonly used to cluster different observations into different clusters based on the distance between these observations [
29]. In the present paper, the K-means clustering algorithm is applied to the matrix
B to cluster 160 ROIs into 6 clusters. Procedures of the algorithm are briefly described as follows.
First, select 6 rows from the matrix B and use these 6 rows as initial cluster centroids.
Secondly, calculate the squared Euclidean distance between each row and each initial cluster centroid, and then assign each row to the cluster with the closest centroid.
Thirdly, when all rows have been assigned, calculate the average of the rows in each cluster to obtain 6 new cluster centroids.
Finally, repeat the second and the third steps until the centroids no longer change.
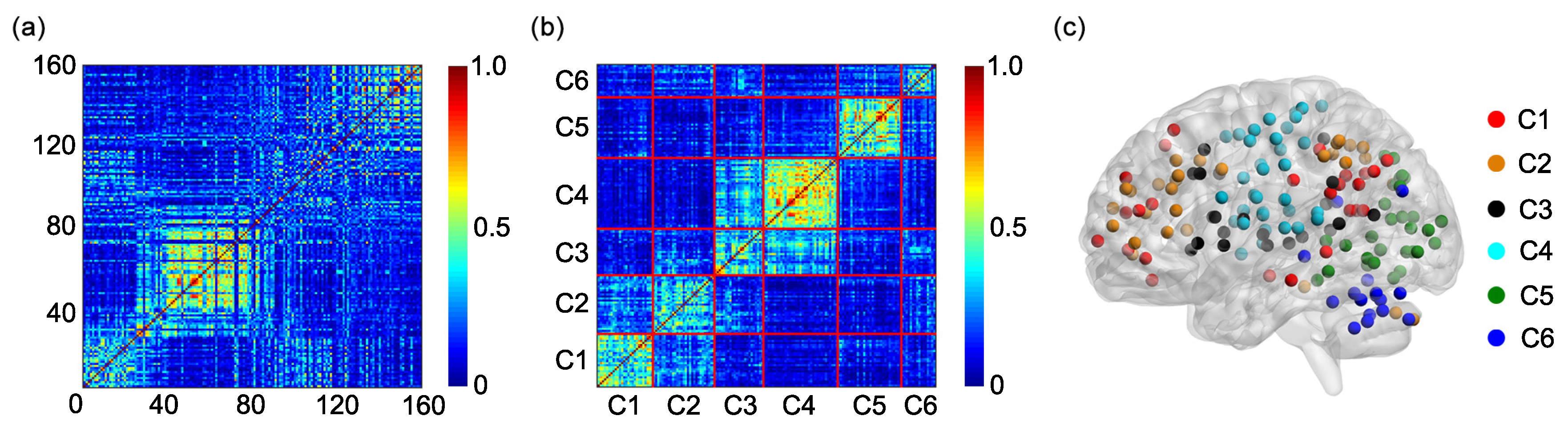
The algorithm generates 6 clusters, and each cluster is composed of different rows of the matrix
B (or of different ROIs). Based on the 6 clusters, an individual adjacent matrix of size
is generated [
36,
41,
42]. The individual adjacent matrix is labeled as
Each of the elements on the main diagonal of
F is 1, and other elements of
F are defined as follows:
if the
ith ROI and the
jth ROI are contained in the same cluster and
otherwise.
Since the study includes 31 participants, 31 individual adjacent matrices are obtained. A group adjacent matrix of size
is obtained by averaging 31 individual adjacent matrices. The group adjacent matrix is labeled as
The K-means clustering algorithm is further applied to the matrix
G to obtain the group cluster result [
36,
41,
42] and the 6 clusters of the group cluster result are compared with the 6 functional networks shown in
Figure 1a.
The detailed clustering procedure is performed by MATLAB software (MATLAB R2014b). Considering that the K-means clustering algorithm is sensitive to the initial cluster centroids, we repeat each clustering procedure 500 times, and the cluster result with the lowest within-cluster distance is adopted.
2.7. Clustering ROIs into 6 Clusters by Applying the K-Means Clustering Algorithm to the SampEn Matrix
The procedures described in
Section 2.6 are also applied to the SampEn matrix
E, and 6 clusters are obtained.
4. Conclusions and Discussion
Different brain regions in the human brain functionally interact with each other to construct multiple functional networks. Identifying the function of each functional network and the brain regions contained in each functional network is very important for understanding the human brain. The present study tests the feasibility of using the K-means clustering algorithm to identify the functional networks based on the FC, including the static FC and the dynamic FC. By applying the K-means clustering algorithm to the static FC or the SampEn of dynamic FC between different ROIs determined by the Dosenbach’s template, we show that the Dosenbach’s 160 ROIs can be divided into six clusters which show high overlap and consistency ratios with the six functional networks identified by applying modularity optimization on the average FC matrix across a large cohort of healthy subjects. The results indicate that the combination of the K-means clustering algorithm and the FC can identify the functional networks of the human brain. The K-means algorithm has been commonly used to parcellate cortical or subcortical regions based on the static FC [
30,
31,
32,
33,
34,
35,
36,
37,
38]. These previous studies along with the present study extend the application of machine learning methods in brain sciences.
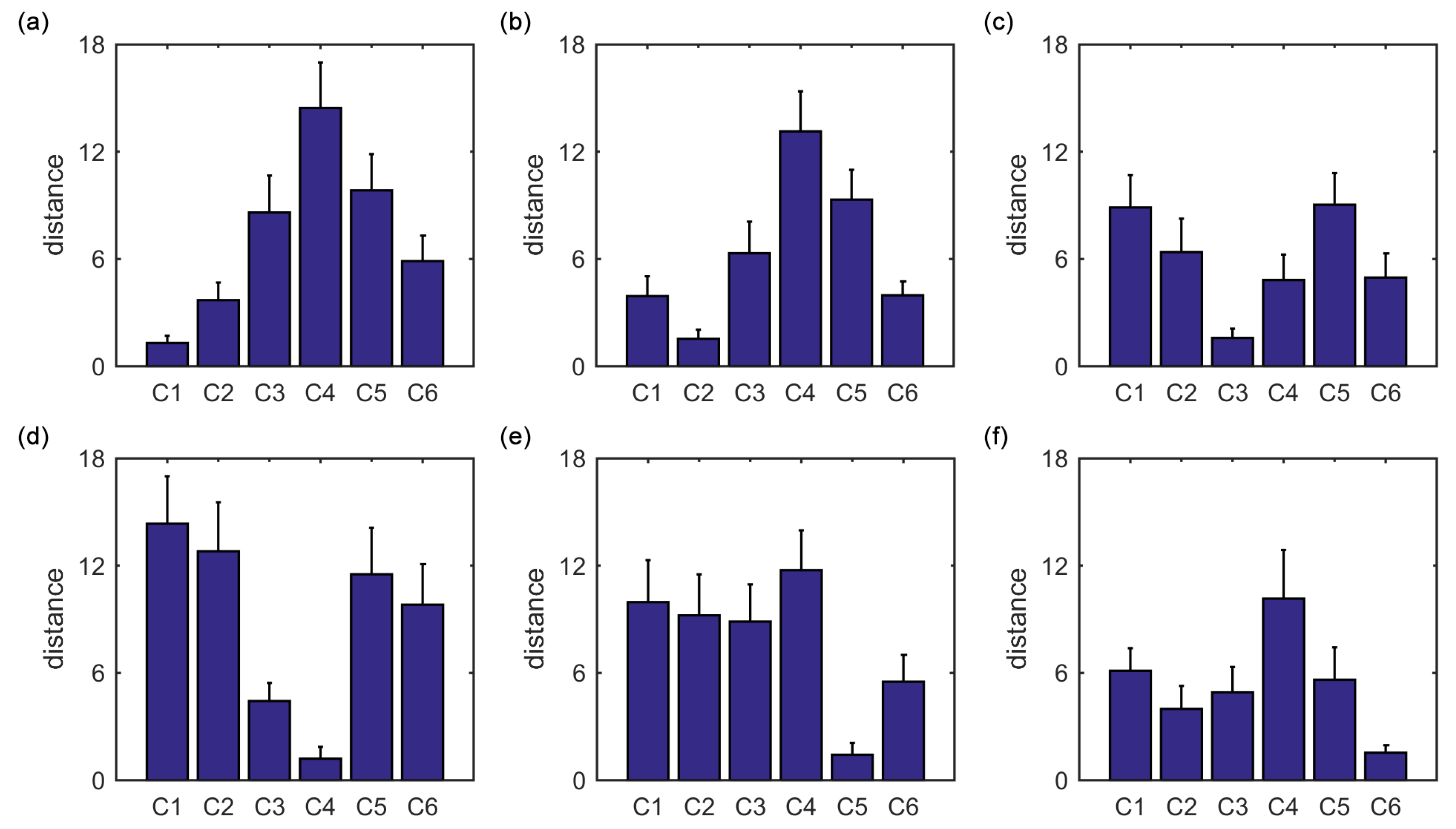
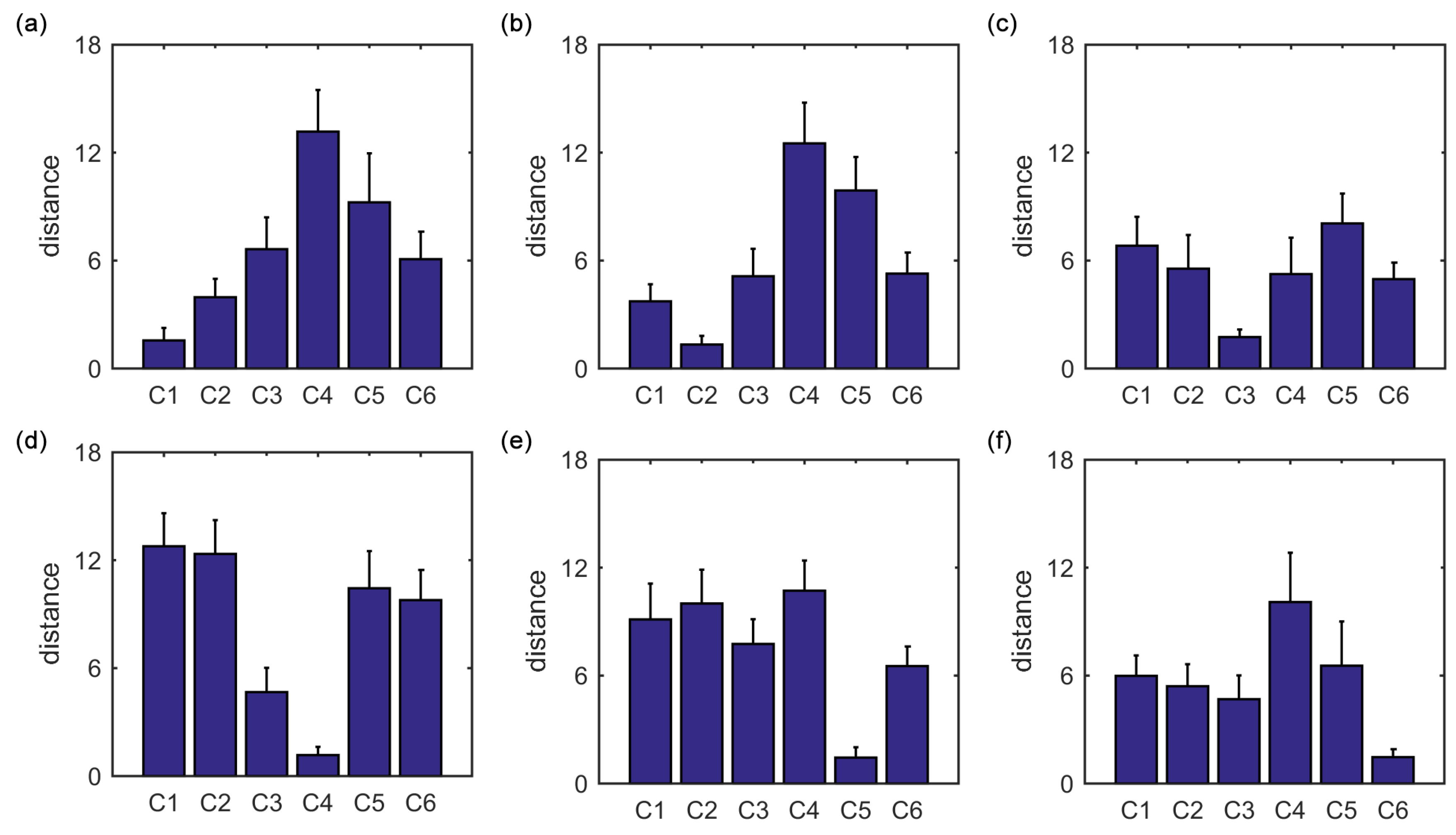
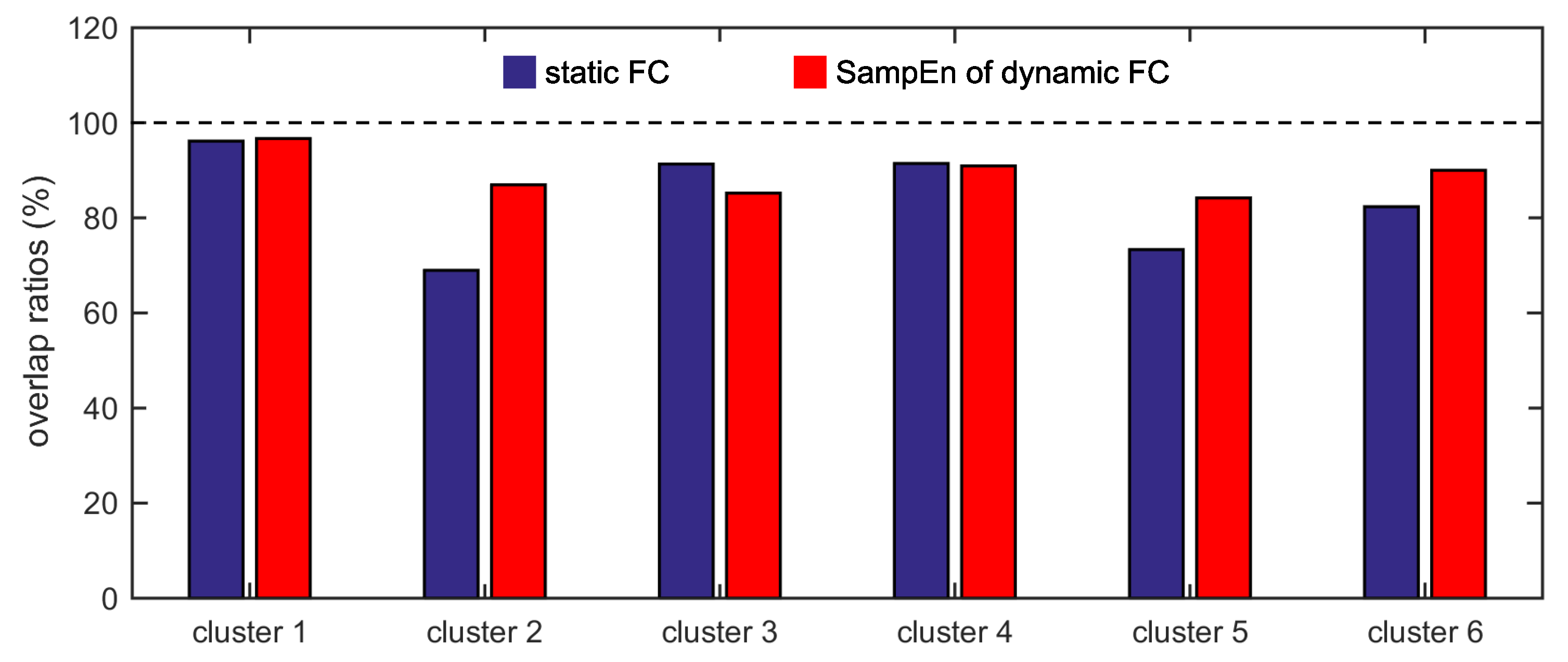
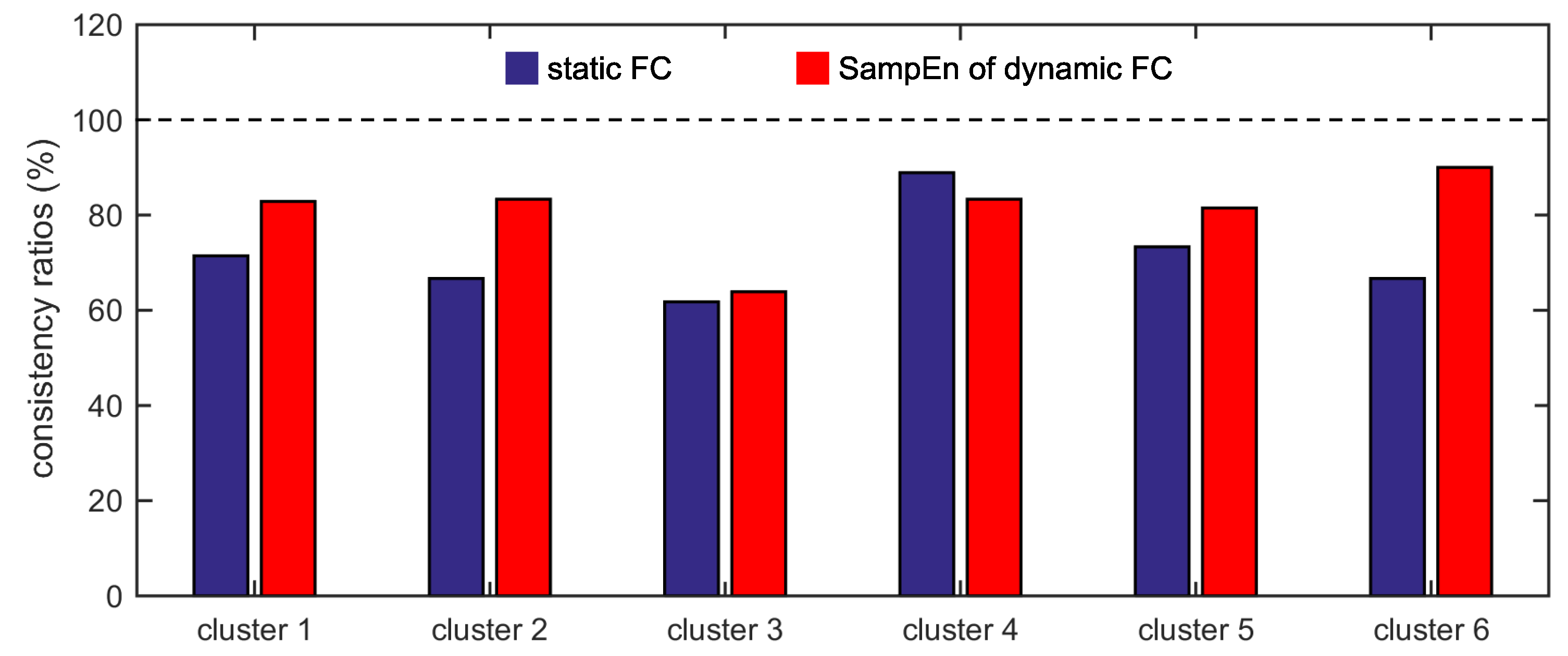
Furthermore, we show that, for four of six clusters, the overlap ratios corresponding to the SampEn of dynamic FC are larger than that corresponding to the static FC, and for five of six clusters, the consistency ratios corresponding to the SampEn of dynamic FC are larger than that corresponding to the static FC. This indicates that nonlinear dynamic characteristics of the FC is more effective than the static characteristics of the FC in identifying brain functional networks. In our previous studies, by characterizing the nonlinear characteristics of dynamic FC in healthy subjects and patients with schizophrenia, we have shown that SampEn of the amygdala-cortical FC in healthy subjects decreased with age increasing, and the visual cortex of the patients with schizophrenia exhibited significantly higher SampEn than that of the healthy subjects [
24,
43]. In the future, nonlinear characteristics of dynamic FC should be deeply used to characterize properties of brain functional networks and the complexity of the human brain.