Complexity and Vulnerability Analysis of the C. Elegans Gap Junction Connectome
Abstract
:1. Introduction
2. Results
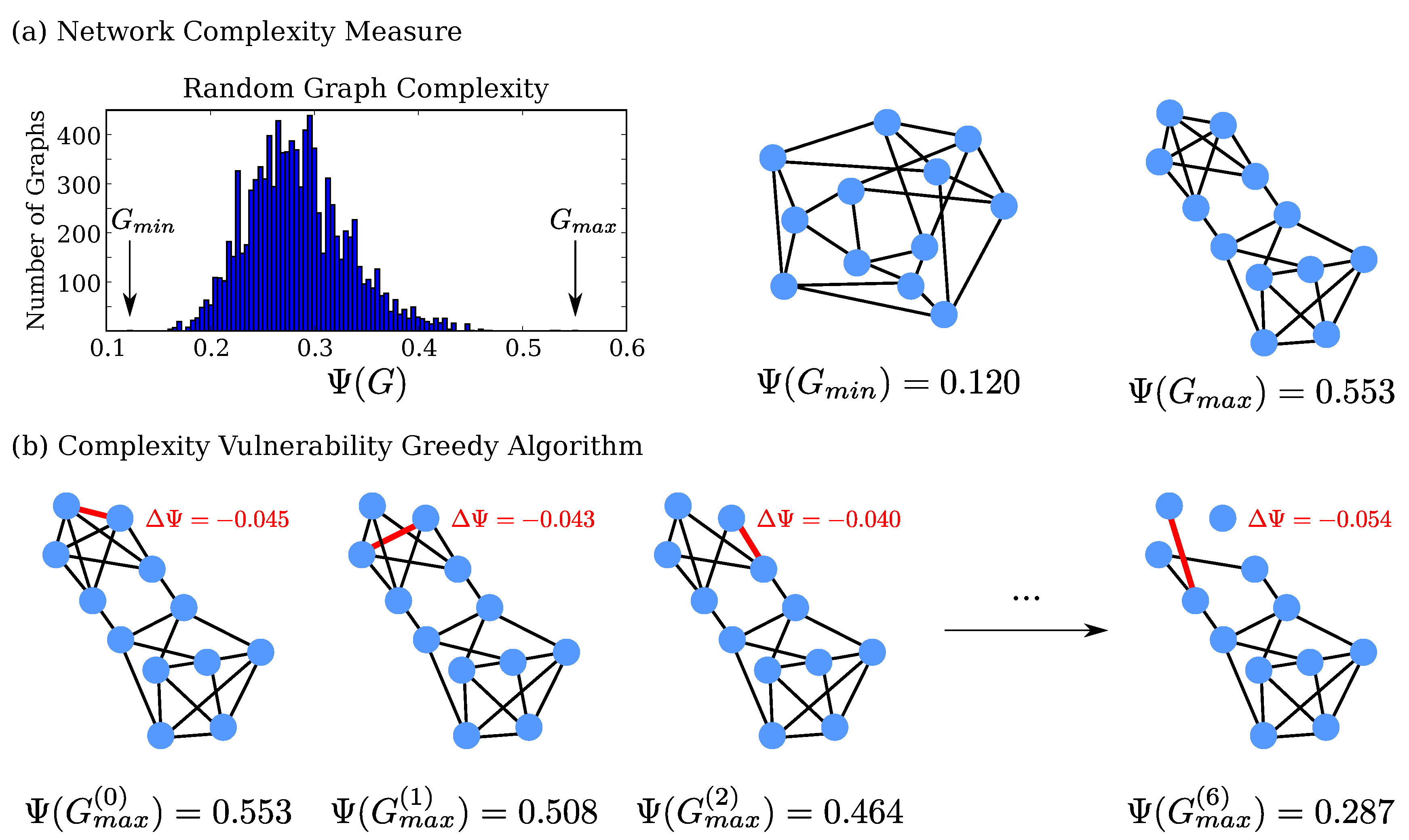
2.1. Investigation of Small, Random Networks
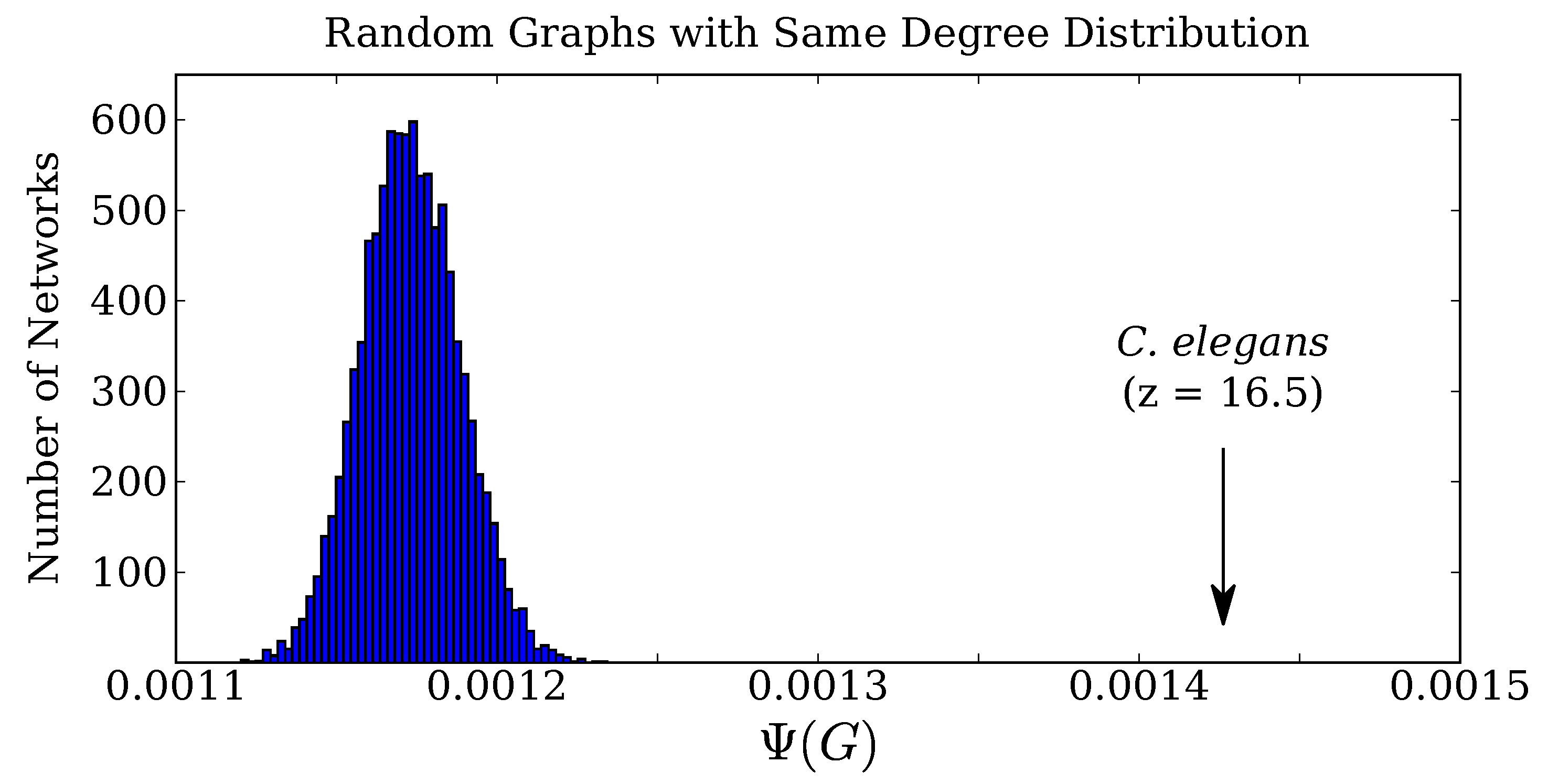
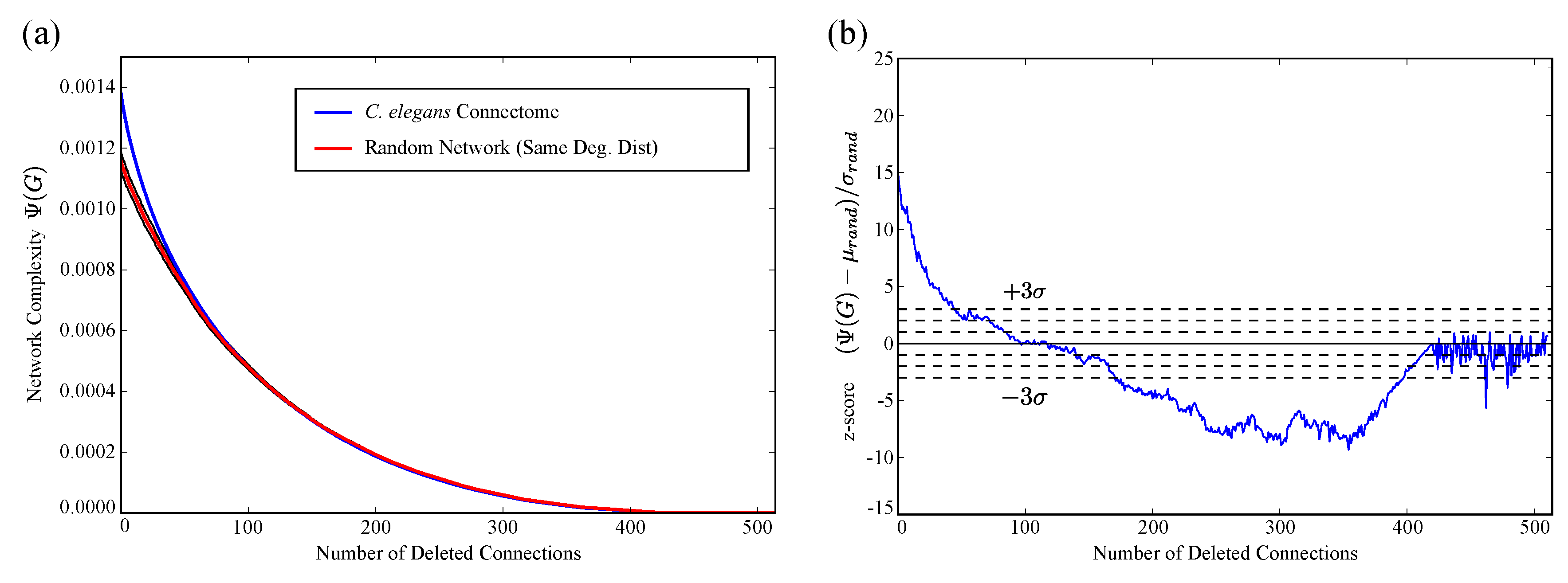
2.2. Application to C. Elegans Data
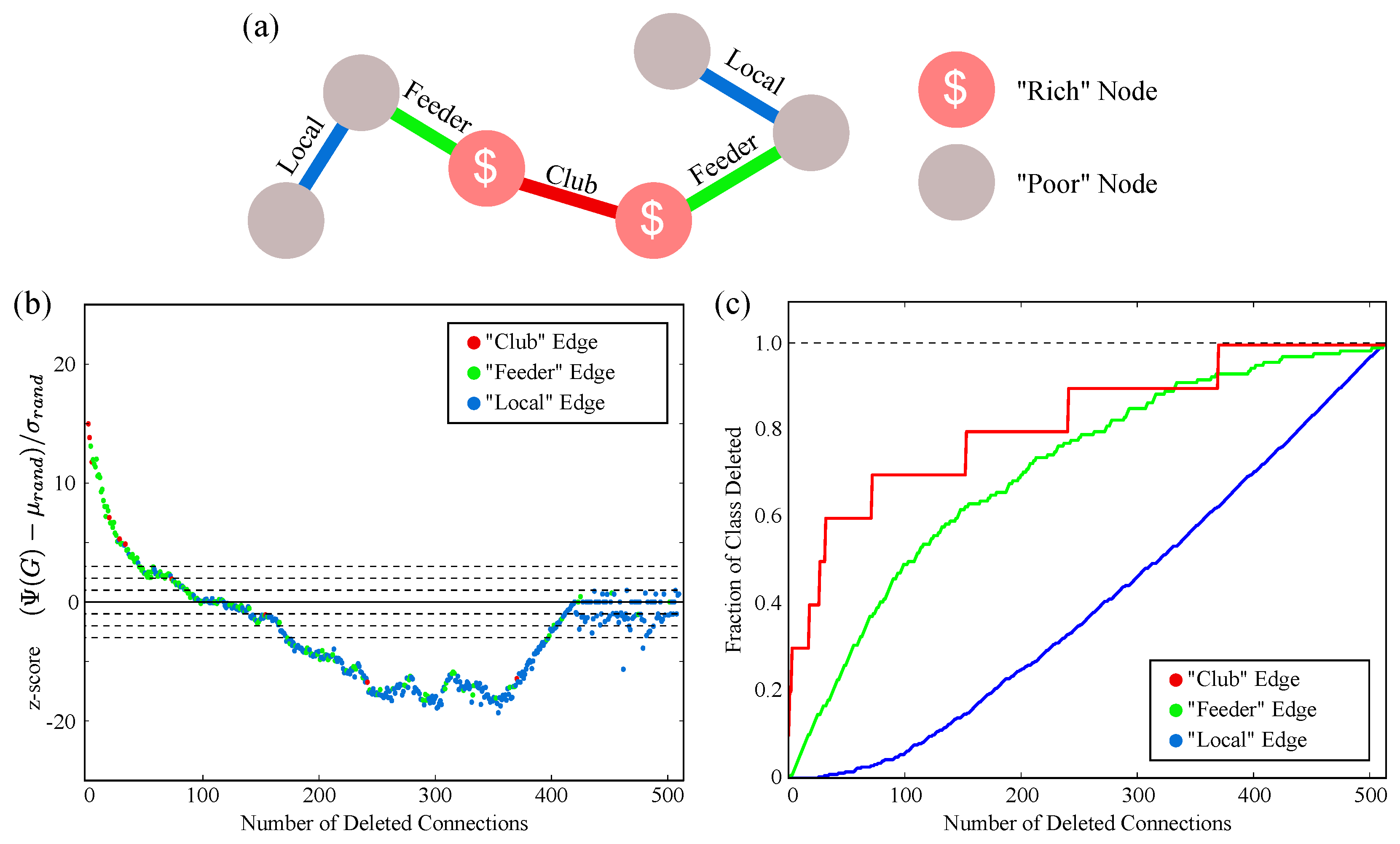
2.3. Complex Structure and the Rich Club
2.4. Complex Structure beyond Degree
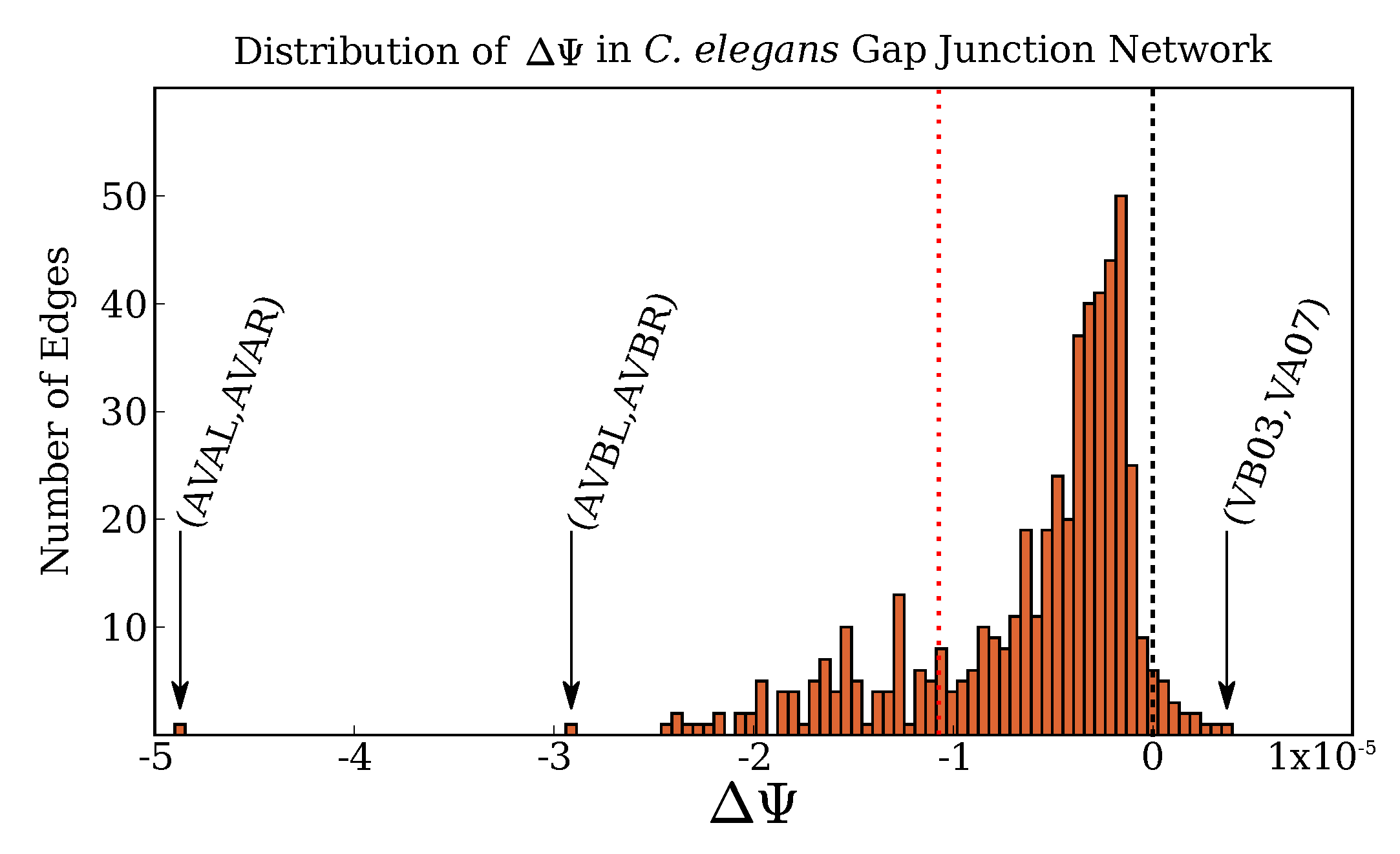
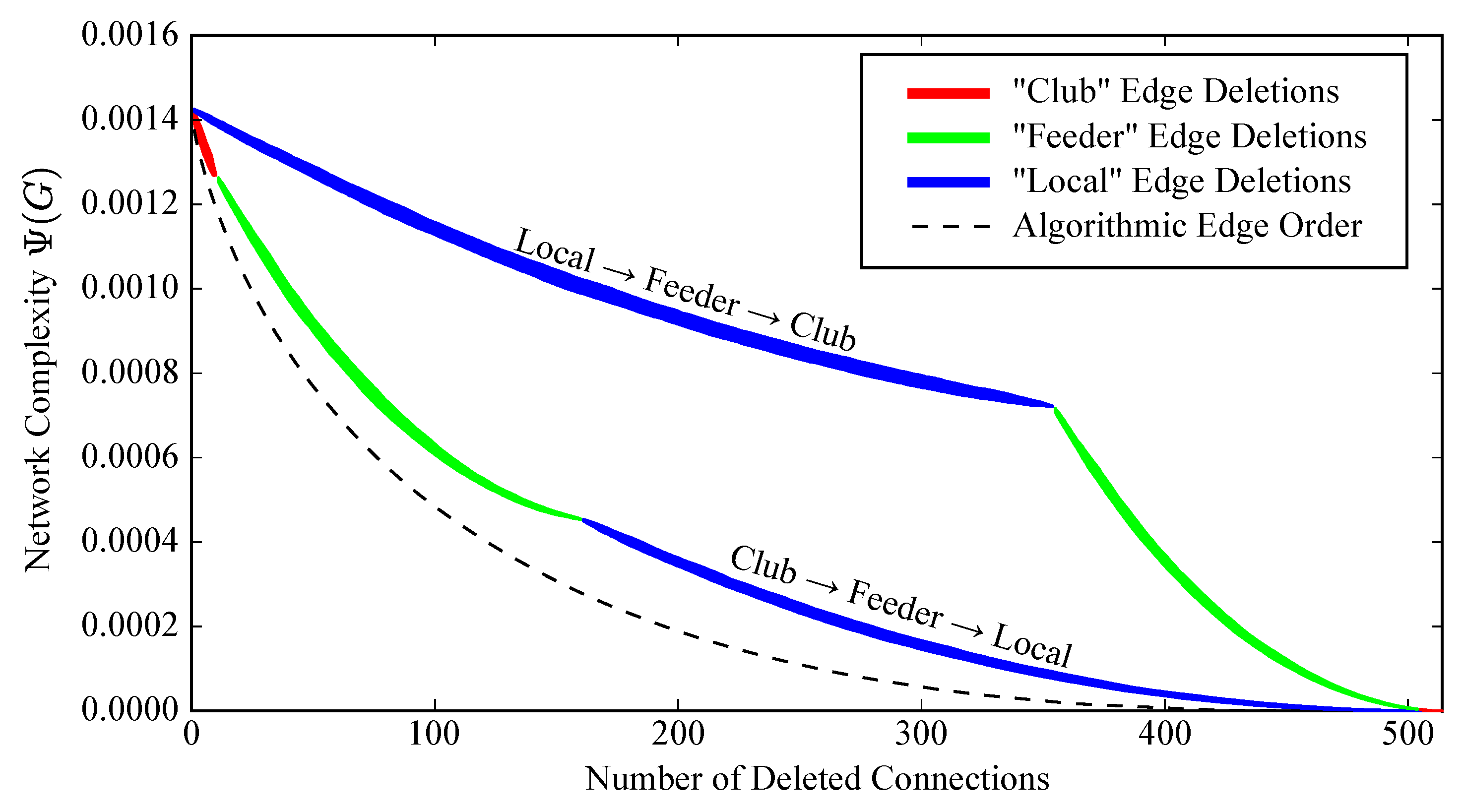
2.5. Robustness to Specific Elimination Order
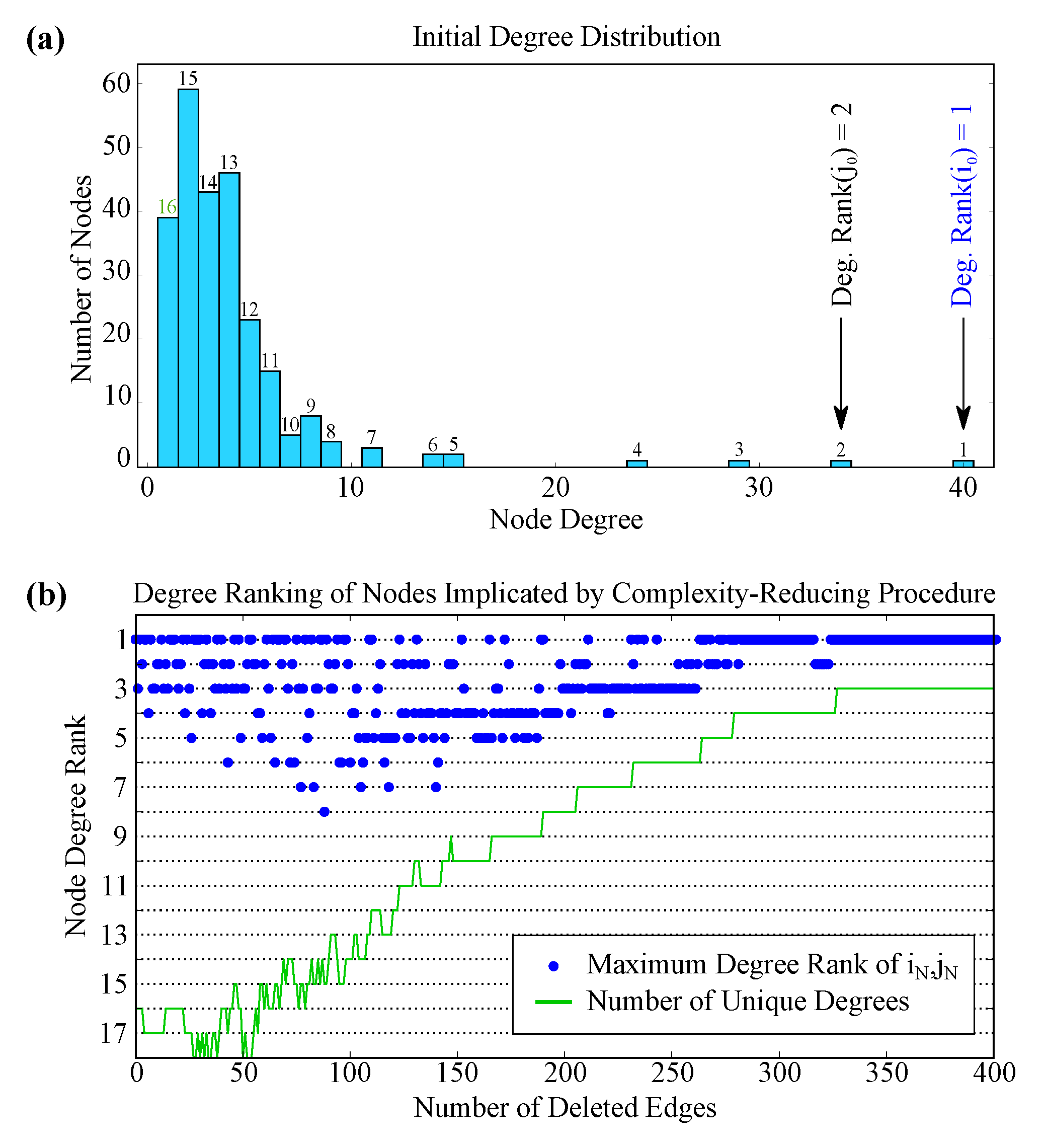
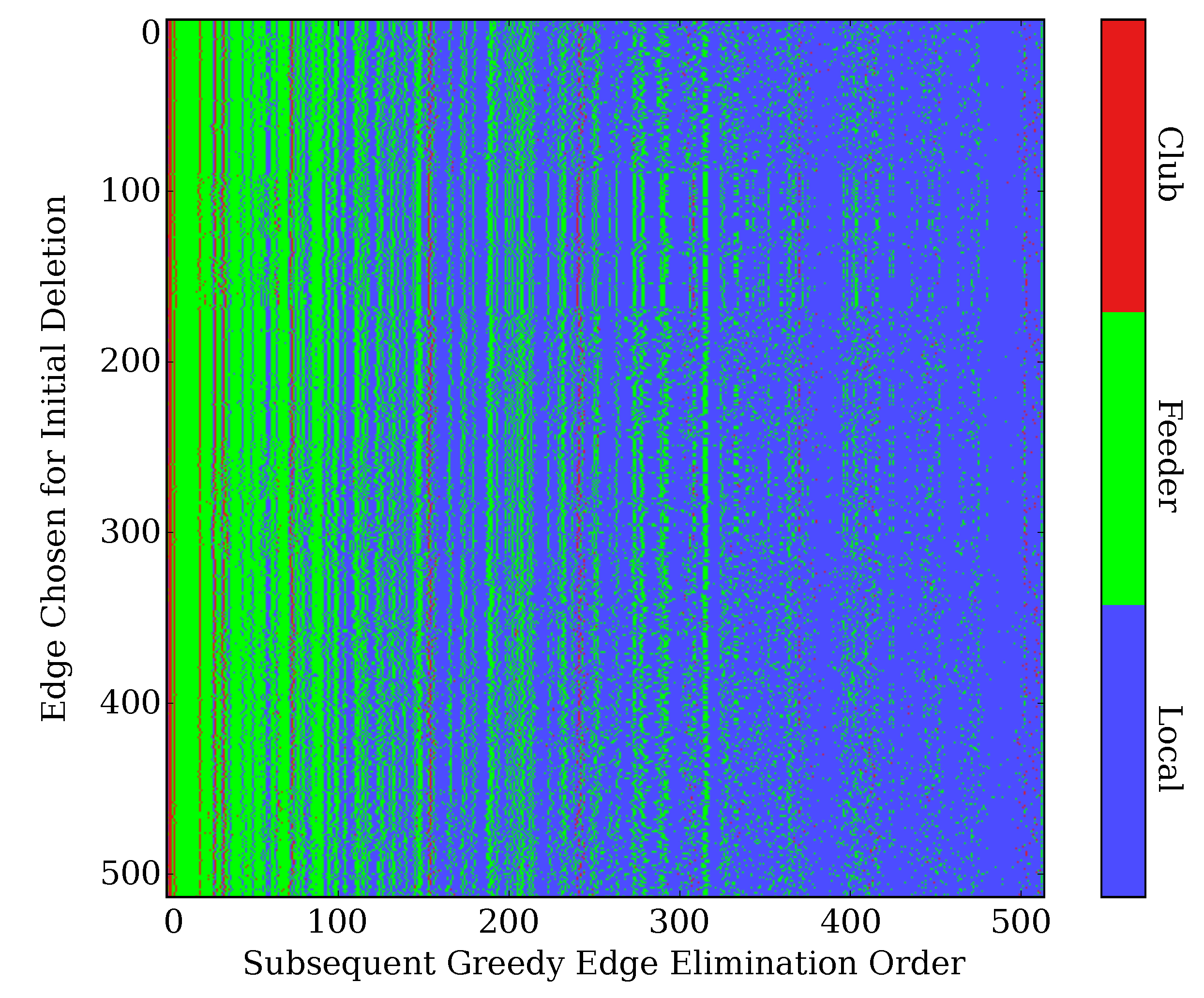
2.6. Robustness to Initial Edge Choice
3. Discussion
4. Materials and Methods
4.1. Network Complexity
4.2. and Iterative Edge Removal
4.3. Illustrative Example: 12 Nodes of Four Degrees
4.4. The C. Elegans Connectome
4.5. Comparison of C. elegans to Random Connectivity
4.6. C. elegans Rich Club
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sporns, O. The Non-Random Brain: Efficiency, Economy, and Complex Dynamics. Front. Comput. Neurosci. 2011, 5. [Google Scholar] [CrossRef] [PubMed]
- Gollo, L.L.; Zalesky, A.; Hutchison, R.M.; van den Heuvel, M.; Breakspear, M. Dwelling quietly in the rich club: Brain network determinants of slow cortical fluctuations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370. [Google Scholar] [CrossRef] [PubMed]
- Dehmer, M.; Barbarini, N.; Varmuza, K.; Graber, A. A Large Scale Analysis of Information-Theoretic Network Complexity Measures Using Chemical Structures. PLoS ONE 2009, 4, e8057. [Google Scholar] [CrossRef] [PubMed]
- Emmert-Streib, F.; Dehmer, M. Exploring Statistical and Population Aspects of Network Complexity. PLoS ONE 2012, 7, e34523. [Google Scholar] [CrossRef] [PubMed]
- Sakhanenko, N.A.; Galas, D.J. Complexity of Networks I: The SetComplexity of Binary Graphs. Complexity 2011, 17, 51–64. [Google Scholar] [CrossRef]
- Ignac, T.M.; Sakhanenko, N.A.; Galas, D.J. Complexity of Networks II: The Set Complexity of Edge-colored Graphs. Complexity 2012, 17, 23–36. [Google Scholar] [CrossRef]
- Varshney, L.R.; Chen, B.L.; Paniagua, E.; Hall, D.H.; Chklovskii, D.B. Structural Properties of the Caenorhabditis elegans Neuronal Network. PLoS Comput. Biol. 2011, 7, e1001066. [Google Scholar] [CrossRef] [PubMed]
- Chalfie, M.; Sulston, J.E.; White, J.G.; Southgate, E.; Thomson, J.N.; Brenner, S. The neural circuit for touch sensitivity in Caenorhabditis elegans. J. Neurosci. 1985, 5, 956–964. [Google Scholar] [PubMed]
- Sawin, E. Genetic and Cellular Analysis of Modulated Behaviors in Caenorhabditis elegans. Ph.D. Thesis, Massachusetts Institute of Technology, Cambridge, MA, USA, 1996. [Google Scholar]
- Sawin, E.R.; Ranganathan, R.; Horvitz, H.R. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron 2000, 26, 619–631. [Google Scholar] [CrossRef]
- Gray, J.M.; Hill, J.J.; Bargmann, C.I. A circuit for navigation in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2005, 102, 3184–3191. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.S.; Sternberg, P.W. Sensory regulation of male mating behavior in Caenorhabditis elegans. Neuron 1995, 14, 79–89. [Google Scholar] [CrossRef]
- Macosko, E.Z.; Pokala, N.; Feinberg, E.H.; Chalasani, S.H.; Butcher, R.A.; Clardy, J.; Bargmann, C.I. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature 2009, 458, 1171–1175. [Google Scholar] [CrossRef] [PubMed]
- Bany, I.A.; Dong, M.Q.; Koelle, M.R. Genetic and cellular basis for acetylcholine inhibition of Caenorhabditis elegans egg-laying behavior. J. Neurosci. 2003, 23, 8060–8069. [Google Scholar] [PubMed]
- Hardaker, L.A.; Singer, E.; Kerr, R.; Zhou, G.; Schafer, W.R. Serotonin modulates locomotory behavior and coordinates egg-laying and movement in Caenorhabditis elegans. J. Neurobiol. 2001, 49, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Bassett, D.S.; Greenfield, D.L.; Meyer-Lindenberg, A.; Weinberger, D.R.; Moore, S.W.; Bullmore, E.T. Efficient physical embedding of topologically complex information processing networks in brains and computer circuits. PLoS Comput. Biol. 2010, 6, e1000748. [Google Scholar] [CrossRef] [PubMed]
- Van den Heuvel, M.P.; Sporns, O. Rich-Club Organization of the Human Connectome. J. Neurosci. 2011, 31, 15775–15786. [Google Scholar] [CrossRef] [PubMed]
- Bullmore, E.; Sporns, O. The economy of brain network organization. Nat. Rev. Neurosci. 2012, 13, 336–349. [Google Scholar] [CrossRef] [PubMed]
- Harriger, L.; van den Heuvel, M.P.; Sporns, O. Rich club organization of macaque cerebral cortex and its role in network communication. PLoS ONE 2012, 7, e46497. [Google Scholar] [CrossRef] [PubMed]
- Ball, G.; Aljabar, P.; Zebari, S.; Tusor, N.; Arichi, T.; Merchant, N.; Robinson, E.C.; Ogundipe, E.; Rueckert, D.; Edwards, A.D.; et al. Rich-club organization of the newborn human brain. Proc. Natl. Acad. Sci. USA 2014, 111, 7456–7461. [Google Scholar] [CrossRef] [PubMed]
- Schroeter, M.S.; Charlesworth, P.; Kitzbichler, M.G.; Paulsen, O.; Bullmore, E.T. Emergence of rich-club topology and coordinated dynamics in development of hippocampal functional networks in vitro. J. Neurosci. 2015, 35, 5459–5470. [Google Scholar] [CrossRef] [PubMed]
- Nigam, S.; Shimono, M.; Ito, S.; Yeh, F.C.; Timme, N.; Myroshnychenko, M.; Lapish, C.C.; Tosi, Z.; Hottowy, P.; Smith, W.C.; et al. Rich-club organization in effective connectivity among cortical neurons. J. Neurosci. 2016, 36, 670–684. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, M.; Omidvarnia, A. Further Insight into the Brain’s Rich-Club Architecture. J. Neurosci. 2016, 36, 5675–5676. [Google Scholar] [CrossRef] [PubMed]
- Towlson, E.K.; Vértes, P.E.; Ahnert, S.E.; Schafer, W.R.; Bullmore, E.T. The Rich Club of the C. elegans Neuronal Connectome. J. Neurosci. 2013, 33, 6380–6387. [Google Scholar] [CrossRef] [PubMed]
- Kunert, J.; Shlizerman, E.; Kutz, J.N. Low-dimensional functionality of complex network dynamics: Neurosensory integration in the Caenorhabditis elegans connectome. Phys. Rev. E 2014, 89, 052805. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, H.; Kralik, J.D.; Jeong, J. Vulnerability-Based Critical Neurons, Synapses, and Pathways in the Caenorhabditis elegans Connectome. PLoS Comput. Biol. 2016, 12, e1005084. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Brunton, S.L.; Cain, N.; Mihalas, S.; Kutz, J.N.; Shea-Brown, E. Feedback through graph motifs relates structure and function in complex networks. arXiv, 2016; arXiv:1605.09073. [Google Scholar]
- Colizza, F.; Flammini, A.; Serrano, M.; Vespignani, A. Detecting rich-club ordering in complex networks. Nat. Phys. 2006, 2, 110–115. [Google Scholar] [CrossRef]
- Milo, R.; Shen-Orr, S.; Itzkovitz, S.; Kashtan, N.; Chklovskii, D.; Alon, U. Network Motifs: Simple Building Blocks of Complex Networks. Science 2002, 298, 824–827. [Google Scholar] [CrossRef] [PubMed]
- Sporns, O.; Kötter, R. Motifs in Brain Networks. PLoS Biol. 2004, 2, e369. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Hintze, A.; Adami, C. Colored Motifs Reveal Computational Building Blocks in the C. elegans Brain. PLoS ONE 2011, 6, e17013. [Google Scholar] [CrossRef] [PubMed]
- Galas, D.J.; Sakhanenko, N.A.; Skupin, A.; Ignac, T. Describing the Complexity of Systems: Multivariable “Set Complexity” and the Information Basis of Systems Biology. J. Comput. Biol. 2014, 21, 118–140. [Google Scholar] [CrossRef] [PubMed]
- Sakhanenko, N.; Galas, D. Biological Data Analysis as an Information Theory Problem: Multivariable Dependence Measures and the Shadows Algorithm. J. Comput. Biol. 2015, 22, 1005–1024. [Google Scholar] [CrossRef] [PubMed]
- Bohland, J.W.; Wu, C.; Barbas, H.; Bokil, H.; Bota, M.; Breiter, H.C.; Cline, H.T.; Doyle, J.C.; Freed, P.J.; Greenspan, R.J.; et al. A Proposal for a Coordinated Effort for the Determination of Brainwide Neuroanatomical Connectivity in Model Organisms at a Mesoscopic Scale. PLoS Comput. Biol. 2009, 5, e1000334. [Google Scholar] [CrossRef] [PubMed]
- Chiang, A.S.; Lin, C.Y.; Chuang, C.C.; Chang, H.M.; Hsieh, C.H.; Yeh, C.W.; Shih, C.T.; Wu, J.J.; Wang, G.T.; Chen, Y.C.; et al. Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution. Curr. Biol. 2011, 21, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Harris, J.; Ng, L.; Winslow, B.; Cain, N.; Mihalas, S.; Wang, Q.; Lau, C.; Kuan, L.; Henry, A.; et al. A mesoscale connectome of the mouse brain. Nature 2014, 508, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, T.P. The graph-tool python library. Figshare 2014. [Google Scholar] [CrossRef]
- White, J.G.; Southgate, E.; Thomson, J.N.; Brenner, S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1986, 314. [Google Scholar] [CrossRef]
- Hall, D.H.; Russell, R.L. The posterior nervous system of the nematode Caenorhabditis elegans: Serial reconstruction of identified neurons and complete pattern of synaptic interactions. J. Neurosci. 1991, 11, 1–22. [Google Scholar] [PubMed]
- Durbin, R. Studies on the Development and Organisation of the Nervous System of Caenorhabditis elegans. Ph.D. Thesis, University of Cambridge, Cambridge, UK, 1987. [Google Scholar]
- Zhou, S.; Mondragon, R. The rich-club phenomenon in the internet topology. IEEE Commun. Lett. 2004, 8, 180–182. [Google Scholar] [CrossRef]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kunert-Graf, J.M.; Sakhanenko, N.A.; Galas, D.J. Complexity and Vulnerability Analysis of the C. Elegans Gap Junction Connectome. Entropy 2017, 19, 104. https://doi.org/10.3390/e19030104
Kunert-Graf JM, Sakhanenko NA, Galas DJ. Complexity and Vulnerability Analysis of the C. Elegans Gap Junction Connectome. Entropy. 2017; 19(3):104. https://doi.org/10.3390/e19030104
Chicago/Turabian StyleKunert-Graf, James M., Nikita A. Sakhanenko, and David J. Galas. 2017. "Complexity and Vulnerability Analysis of the C. Elegans Gap Junction Connectome" Entropy 19, no. 3: 104. https://doi.org/10.3390/e19030104
APA StyleKunert-Graf, J. M., Sakhanenko, N. A., & Galas, D. J. (2017). Complexity and Vulnerability Analysis of the C. Elegans Gap Junction Connectome. Entropy, 19(3), 104. https://doi.org/10.3390/e19030104






