Comparative Genomic Analysis of Arctic Permafrost Bacterium Nesterenkonia sp. PF2B19 to Gain Insights into Its Cold Adaptation Tactic and Diverse Biotechnological Potential
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Site, Bacterial Strain and Growth Conditions
2.2. Genomic DNA Preparation and Genome Sequencing
2.3. Comparative Genomics
2.4. Functional Annotation
2.5. Accession Nnumber
3. Results and Discussion
3.1. Characterization and Phylogeny of PF2B19
3.2. General Genome Features of Permafrost Bacterium Nesterenkonia sp. PF2B19
3.3. General Genome Comparisons of PF2B19 with Its Closest Phylogenetic Affiliates
3.4. Comparative Genomics Identifies Unique Genes/Proteins in Nesterenkonia sp. PF2B19
3.5. Identification of Virulence Determinants
3.6. Genes Involved in Resistance to Antibiotics
3.6.1. Cold Stress Response
3.6.2. Oxidative Stress Response
3.6.3. Osmo-Protection
3.6.4. General Stress Response
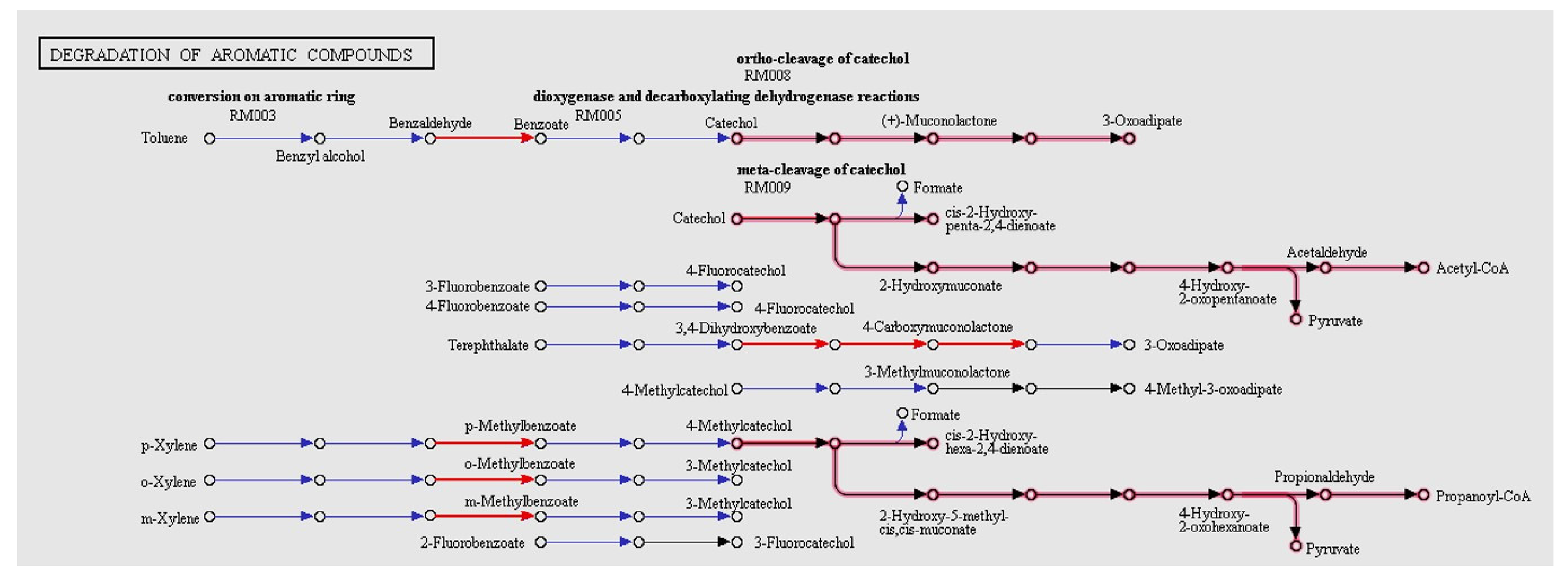
3.7. Biotechnological Potential of PF2B19
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Péwé, T.L. Permafrost. In Encyclopædia Britannica; 2006; Available online: http://search.eb.com/eb/article-9108442 (accessed on 9 April 2008).
- Zhang, T.; Barry, R.G.; Knowles, K.; Heginbottom, J.A.; Brown, J. Statistics and characteristics of permafrost and ground-ice distribution in the Northern Hemisphere 1. Polar Geogr. 1999, 23, 132–154. [Google Scholar] [CrossRef]
- Aislabie, J.M.; Chhour, K.L.; Saul, D.J.; Miyauchi, S.; Ayton, J.; Paetzold, R.F.; Balks, M.R. Dominant bacteria in soils of Marble point and Wright valley, Victoria land, Antarctica. Soil Biol. Biochem. 2006, 38, 3041–3056. [Google Scholar] [CrossRef]
- Chan, Y.; Van Nostrand, J.D.; Zhou, J.; Pointing, S.B.; Farrell, R.L. Functional ecology of an Antarctic dry valley. Proc. Natl. Acad. Sci. USA 2013, 110, 8990–8995. [Google Scholar] [CrossRef] [PubMed]
- Humlum, O.; Elberling, B.; Hormes, A.; Fjordheim, K.; Hansen, O.H.; Heinemeier, J. Late-Holocene glacier growth in Svalbard, documented by subglacial relict vegetation and living soil microbes. Holocene 2005, 15, 396–407. [Google Scholar] [CrossRef]
- Singh, S.M.; Sharma, J.; Gawas-Sakhalkar, P.; Upadhyay, A.K.; Naik, S.; Bande, D.; Ravindra, R. Chemical and bacteriological analysis of soil from the Middle and Late Weichselian from Western Spitsbergen, Arctic. Quat. Int. 2012, 271, 98–105. [Google Scholar] [CrossRef]
- Friedmann, E.I. Permafrost as microbial habitat. Viable Microorganisms in Permafrost 1994, 21–26,–26. [Google Scholar] [CrossRef]
- Piette, F.; D’Amico, S.; Mazzucchelli, G.; Danchin, A.; Leprince, P.; Feller, G. Life in the cold: A proteomic study of cold-repressed proteins in the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Appl. Environ. Microbiol. 2011, 77, 3881–3883. [Google Scholar] [CrossRef]
- Mesbah, N.M.; Wiegel, J. Life at extreme limits. Ann. N. Y. Acad. Sci. 2008, 1125, 44–57. [Google Scholar] [CrossRef]
- De Maayer, P.; Anderson, D.; Cary, C.; Cowan, D.A. Some like it cold: Understanding the survival strategies of psychrophiles. EMBO Rep. 2014, 15, 508–517. [Google Scholar] [CrossRef]
- Willerslev, E.; Hansen, A.J.; Poinar, H.N. Isolation of nucleic acids and cultures from fossil ice and permafrost. Trends Ecol. Evol. 2004, 19, 141–147. [Google Scholar] [CrossRef]
- Margesin, R.; Dieplinger, H.; Hofmann, J.; Sarg, B.; Lindner, H. A cold-active extracellular metalloprotease from Pedobacter cryoconitis—production and properties. Res. Microbiol. 2005, 156, 499–505. [Google Scholar] [CrossRef]
- Kuddus, M.; Ramteke, P.W. A cold-active extracellular metalloprotease from Curtobacterium luteum (MTCC 7529): Enzyme production and characterization. J. Gen. Appl. Microbiol. 2008, 54, 385–392. [Google Scholar] [CrossRef]
- Goordial, J.; Lamarche-Gagnon, G.; Lay, C.Y.; Whyte, L. Left out in the cold: Life in cryoenvironments. In Polyextremophiles; Springer: Dordrecht, The Netherlands, 2013; pp. 335–363. [Google Scholar]
- Hansen, A.A.; Herbert, R.A.; Mikkelsen, K.; Jensen, L.L.; Kristoffersen, T.; Tiedje, J.M.; Finster, K.W. Viability, diversity and composition of the bacterial community in a high Arctic permafrost soil from Spitsbergen, Northern Norway. Environ. Microbiol. 2007, 9, 2870–2884. [Google Scholar] [CrossRef]
- Schostag, M.; Stibal, M.; Jacobsen, C.S.; Bælum, J.; Taş, N.; Elberling, B.; Jansson, J.K.; Semenchuk, P.; Priemé, A. Distinct summer and winter bacterial communities in the active layer of Svalbard permafrost revealed by DNA-and RNA-based analyses. Front. Microbiol. 2015, 6, 399. [Google Scholar] [CrossRef]
- Bakermans, C.; Bergholz, P.W.; Rodrigues, D.F.; Vishnivetskaya, T.A.; Ayala-del-Río, H.L.; Tiedje, J.M. Genomic and expression analyses of cold-adapted microorganisms. In Polar Microbiology: Life in a Deep Freeze; Miller, R.V., Whyte, L.G., Eds.; ASM Press: Washington, DC, USA, 2012; pp. 126–155. [Google Scholar]
- Stackebrandt, E.; Koch, C.; Gvozdiak, O.; Schumann, P. Taxonomic Dissection of the Genus Micrococcus: Kocuria gen. nov., Nesterenkonia gen. nov., Kytococcus gen. nov., Dermacoccus gen. nov., and Micrococcus Cohn 1872 gen. emend. Int. J. Syst. Evol. Microbiol. 1995, 45, 682–692. [Google Scholar] [CrossRef]
- Li, W.J.; Chen, H.H.; Kim, C.J.; Zhang, Y.Q.; Park, D.J.; Lee, J.C.; Jiang, C.L. Nesterenkonia sandarakina sp. nov. and Nesterenkonia lutea sp. nov., novel actinobacteria, and emended description of the genus Nesterenkonia. Int. J. Syst. Evol. Microbiol. 2005, 55, 463–466. [Google Scholar] [CrossRef]
- Shafiei, M.; Ziaee, A.A.; Amoozegar, M.A. Purification and characterization of a halophilic α-amylase with increased activity in the presence of organic solvents from the moderately halophilic Nesterenkonia sp. strain F. Extremophiles 2012, 16, 627–635. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Pevzner, P.A. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Auch, A.F.; Jan, M.; Klenk, H.P.; Göker, M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genom. Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.F.; Petty, N.K.; Zakour, N.L.B.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Moe, L.A.; Rodbumrer, J.; Gaarder, A.; Handelsman, J. Functional metagenomics reveals diverse β-lactamases in a remote Alaskan soil. ISME J. 2009, 3, 243–251. [Google Scholar] [CrossRef]
- Dib, J.R.; Weiss, A.; Neumann, A.; Ordoñez, O.; Estévez, M.C.; Farías, M.E. Isolation of bacteria from remote high altitude Andean lakes able to grow in the presence of antibiotics. Recent Pat. Anti-Infect. Drug Discov. 2009, 4, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Thaller, M.C.; Migliore, L.; Marquez, C.; Tapia, W.; Cedeño, V.; Rossolini, G.M.; Gentile, G. Tracking acquired antibiotic resistance in commensal bacteria of Galapagos land iguanas: No man, no resistance. PLoS ONE 2010, 5, e8989. [Google Scholar] [CrossRef]
- Ushida, K.; Segawa, T.; Kohshima, S.; Takeuchi, N.; Fukui, K.; Li, Z.; Kanda, H. Application of real-time PCR array to the multiple detection of antibiotic resistant genes in glacier ice samples. J. Gen. Appl. Microbiol. 2010, 56, 43–52. [Google Scholar] [CrossRef]
- Vincent, W.F. Cold tolerance in cyanobacteria and life in the cryosphere. In Algae and Cyanobacteria in Extreme Environments; Springer: Dordrecht, The Netherlands, 2007; pp. 287–301. [Google Scholar]
- Mueller, D.R.; Vincent, W.F.; Bonilla, S.; Laurion, I. Extremotrophs, extremophiles and broadband pigmentation strategies in a high arctic ice shelf ecosystem. FEMS Microbiol. Ecol. 2005, 53, 73–87. [Google Scholar] [CrossRef]
- Berger, F.; Morellet, N.; Menu, F.; Potier, P. Cold shock and cold acclimation proteins in the psychrotrophic bacterium Arthrobacter globiformis SI55. J. Bacteriol. 1996, 178, 2999–3007. [Google Scholar] [CrossRef] [PubMed]
- Médigue, C.; Krin, E.; Pascal, G.; Barbe, V.; Bernsel, A.; Bertin, P.N.; Fang, G. Coping with cold: The genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 2005, 15, 1325–1335. [Google Scholar] [CrossRef] [PubMed]
- Methé, B.A.; Nelson, K.E.; Deming, J.W.; Momen, B.; Melamud, E.; Zhang, X.; Moult, J.; Madupu, R.; Nelson, W.C.; Dodson, R.J.; et al. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc. Natl. Acad. Sci. USA 2005, 102, 10913–10918. [Google Scholar] [CrossRef]
- Gao, H.; Yang, Z.K.; Wu, L.; Thompson, D.K.; Zhou, J. Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J. Bacteriol. 2006, 188, 4560–4569. [Google Scholar] [CrossRef]
- Riley, M.; Staley, J.T.; Danchin, A.; Wang, T.Z.; Brettin, T.S.; Hauser, L.J.; Land, M.L.; Thompson, L.S. Genomics of an extreme psychrophile, Psychromonas ingrahamii. BMC Genom. 2008, 9, 210. [Google Scholar] [CrossRef]
- Phadtare, S.; Alsina, J.; Inouye, M. Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 1999, 2, 175–180. [Google Scholar] [CrossRef]
- Rabus, R.; Ruepp, A.; Frickey, T.; Rattei, T.; Fartmann, B.; Stark, M.; Amann, J. The genome of Desulfotalea psychrophila, a sulfate-reducing bacterium from permanently cold Arctic sediments. Environ. Microbiol. 2004, 6, 887–902. [Google Scholar] [CrossRef]
- Chaikam, V.; Karlson, D.T. Comparison of structure, function and regulation of plant cold shock domain proteins to bacterial and animal cold shock domain proteins. BMB Rep. 2010, 43, 1–8. [Google Scholar] [CrossRef]
- Goordial, J.; Raymond-Bouchard, I.; Zolotarov, Y.; de Bethencourt, L.; Ronholm, J.; Shapiro, N.; Whyte, L. Cold adaptive traits revealed by comparative genomic analysis of the eurypsychrophile Rhodococcus sp. JG3 isolated from high elevation McMurdo Dry Valley permafrost, Antarctica. FEMS Microbiol. Ecol. 2016, 92, 154. [Google Scholar]
- Bae, W.; Xia, B.; Inouye, M.; Severinov, K. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl. Acad. Sci. USA 2000, 97, 7784–7789. [Google Scholar] [CrossRef]
- Feller, G. Molecular adaptations to cold in psychrophilic enzymes. Cell. Mol. Life Sci. CMLS 2003, 60, 648–662. [Google Scholar] [CrossRef]
- Feller, G.; Gerday, C. Psychrophilic enzymes: Hot topics in cold adaptation. Nat. Rev. Microbiol. 2003, 1, 200–208. [Google Scholar] [CrossRef]
- D’amico, S.; Collins, T.; Marx, J.C.; Feller, G.; Gerday, C. Psychrophilic microorganisms: Challenges for life. EMBO Rep. 2006, 7, 385–389. [Google Scholar] [CrossRef]
- Hoang, T.T.; Sullivan, S.A.; Cusick, J.K.; Schweizer, H.P. β-Ketoacyl acyl carrier protein reductase (FabG) activity of the fatty acid biosynthetic pathway is a determining factor of 3-oxo-homoserine lactone acyl chain lengths. Microbiology 2002, 148, 3849–3856. [Google Scholar] [CrossRef]
- Chintalapati, S.; Kiran, M.D.; Shivaji, S. Role of membrane lipid fatty acids in cold adaptation. Cell. Mol. Biol. 2004, 50, 631–642. [Google Scholar]
- Morgan-Kiss, R.M.; Priscu, J.C.; Pocock, T.; Gudynaite-Savitch, L.; Huner, N.P. Adaptation and acclimation of photosynthetic microorganisms to permanently cold environments. Microbiol. Mol. Biol. Rev. 2006, 70, 222–252. [Google Scholar] [CrossRef]
- Dieser, M.; Greenwood, M.; Foreman, C.M. Carotenoid pigmentation in Antarctic heterotrophic bacteria as a strategy to withstand environmental stresses. Arct. Antarct. Alp. Res. 2010, 42, 396–405. [Google Scholar] [CrossRef]
- Chattopadhyay, M.K. Mechanism of bacterial adaptation to low temperature. J. Biosci. 2006, 31, 157–165. [Google Scholar] [CrossRef]
- Ballal, A.; Manna, A.C. Control of thioredoxin reductase gene (trxB) transcription by SarA in Staphylococcus aureus. J. Bacteriol. 2010, 192, 336–345. [Google Scholar] [CrossRef]
- Welsh, D.T. Ecological significance of compatible solute accumulation by micro-organisms: From single cells to global climate. FEMS Microbiol. Rev. 2000, 24, 263–290. [Google Scholar] [CrossRef]
- Le Rudulier, D.; Strom, A.R.; Dandekar, A.M.; Smith, L.T.; Valentine, R.C. Molecular biology of osmoregulation. Science 1984, 224, 1064–1069. [Google Scholar] [CrossRef]
- Garnier, M.; Matamoros, S.; Chevret, D.; Pilet, M.F.; Leroi, F.; Tresse, O. Adaptation to cold and proteomic responses of the psychrotrophic biopreservative Lactococcus piscium strain CNCM I-4031. Appl. Environ. Microbiol. 2010, 76, 8011–8018. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Charlton, T.; Ertan, H.; Omar, S.M.; Siddiqui, K.S.; Williams, T.J. Biotechnological uses of enzymes from psychrophiles. Microb. Biotechnol. 2011, 4, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.R. The biodegradation of aromatic hydrocarbons by bacteria. In Physiology of Biodegradative Microorganisms; Springer: Wageningen, The Netherlands, 1991; pp. 191–206. [Google Scholar]
- Ryu, C.M.; Farag, M.A.; Hu, C.H.; Reddy, M.S.; Wei, H.X.; Paré, P.W.; Kloepper, J.W. Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 100, 4927–4932. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef] [PubMed]

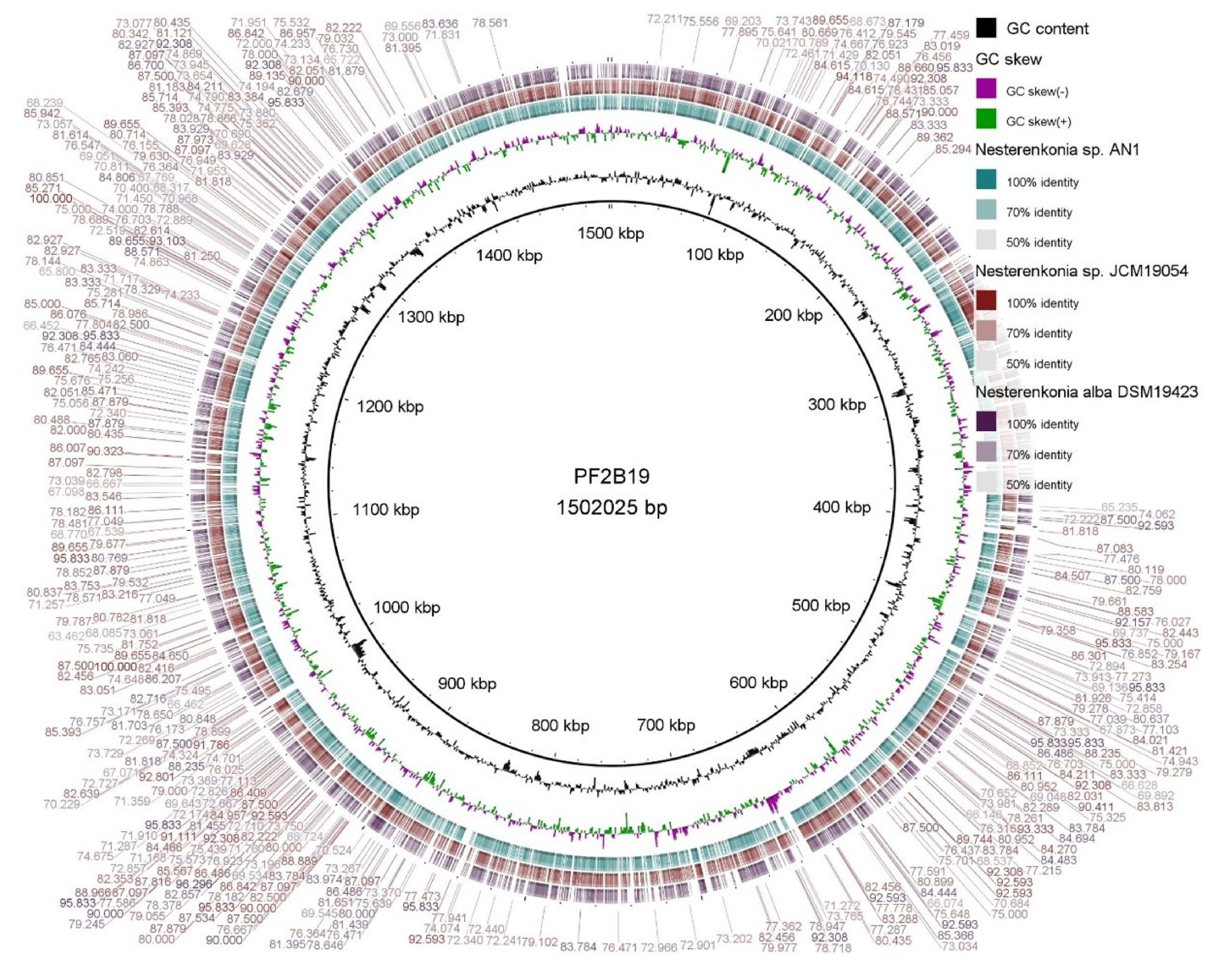
| Attributes | Strains of the Genus Nesterenkonia * | ||||||
|---|---|---|---|---|---|---|---|
| PF2B19 | CD08_7 | AN1 | F | JCM 19054 | NP1 | DSM 19423 | |
| Accession no. | MDSS00000000 | LQBM00000000 | JEMO00000000 | AFRW00000000 | BAXI00000000 | CBLL00000000 | ATXP00000000 |
| Isolation source | Permafrost soil Svalbard, Arctic | Duodenal mucosa of CD patient | Salt Lake, Iran | Antarctic soil | Sea snail Nassarius glans | Feces of AIDS patient | Black liquor treatment system of a cotton pulp mill |
| Growth temp | 15 °C | 37 °C | 21 °C | 32 °C | 28 °C | 37 °C | 42 °C |
| Size | 3.6 Mb | 2.9 Mb | 3.0 Mb | 2.8 Mb | 2.5 Mb | 2.6 Mb | 2.5 Mb |
| Contigs | 135 | 8 | 42 | 138 | 1086 | 175 | 36 |
| G+C (%) | 69.5 | 67.6 | 67.4 | 71.5 | 67.1 | 62.9 | 63.7 |
| No. of RNAs | 55 | 52 | 52 | 50 | 48 | 49 | 51 |
| No. of subsystem | 394 | 379 | 374 | 347 | 292 | 355 | 343 |
| Coding sequences | 3708 | 2531 | 2846 | 2480 | 3901 | 2435 | 2295 |
| Genome Used for Comparison | No. of Unique Genes Detected in PF2B19 on Comparison | Type of Distinct Genes Detected in Relation to Psychrophilic Lifestyle of PF2B19 | Role |
|---|---|---|---|
| Nesterenkonia alba DSM 19423(T) | 323 |
| Counteract against cold-induced osmotic stress |
| Counteract against cold-induced oxidative stress | ||
| Modulate membrane fluidity at low temperatures | ||
| Nesterenkonia massilensis NP1 | 310 |
| Carbon Starvation |
| Counteract against cold-induced osmotic stress | ||
| Counteract against cold-induced oxidative stress | ||
| Nesterenkonia sp. F | 215 |
| Counteract against cold-induced osmotic stress |
| Counteract against cold-induced oxidative stress | ||
| Carbon starvation | ||
| Modulate membrane fluidity at low temperatures | ||
| Nesterenkonia jeotgali CD08_7 | 218 |
| Counteract against cold-induced osmotic stress |
| Counteract against cold-induced oxidative stress | ||
| Carbon starvation | ||
| Nesterenkonia sp. AN1 | 202 |
| Counteract against cold-induced osmotic stress |
| Counteract against cold-induced oxidative stress | ||
| Carbon starvation | ||
| Nesterenkonia sp. JCM 19054 | 345 |
| Cold shock response |
| Counteract against cold-induced osmotic stress | ||
| Counteract against cold-induced oxidative stress | ||
| Carbon starvation |
| Gene Name | Gene Products | Function |
|---|---|---|
| cshA | Putative ATP-dependent RNA helicase | Cold stress |
| cspC | Cold shock protein C | |
| cspA | Cold shock protein A | |
| infB | Translation initiation factor 1 | |
| deaD | DEAD-box ATP-dependent RNA helicase CshA | |
| Pnp | Polyribonucleotide nucleotidyl transferase | |
| infB | Translation initiation factor 2 | |
| rbfA | Ribosome-binding factor A | |
| nusA | Transcription termination protein | |
| dnaJ | Chaperone protein | |
| dnaK | Chaperone protein | |
| grpE | Heat shock protein | |
| hrpA | ATP-dependent helicase | |
| ygcA | RNA methyltransferase, TrmA family | |
| cstA | Carbon starvation protein A | |
| hrpA | ATP-dependent helicase | |
| recA | Recombinase | DNA repair |
| recN | DNA repair protein | |
| recR | Recombination protein | |
| uvrA | Excinuclease ABC subunit A paralog of unknown function | |
| xthA | Exodeoxyribonuclease III | |
| mutM | Formamidopyrimidine-DNA glycosylase | |
| mutY | A/G-specific adenine glycosylase | |
| recA | RecA protein | |
| recX | Regulatory protein | |
| uvrC | Excinuclease ABC subunit C | |
| uvrB | Excinuclease ABC subunit B | |
| uvrA | Excinuclease ABC subunit A | |
| ruvA | Holliday junction DNA helicase | |
| ruvB | Holliday junction DNA helicase | |
| ruvC | Crossover junction endodeoxyribonuclease | |
| recO | DNA recombination and repair protein | |
| Pdg | Endonuclease III | |
| -- | Phytoene dehydrogenase and related proteins | Membrane fluidity |
| -- | Fatty acid desaturase | |
| hepT | Octaprenyl diphosphate synthase | |
| fabG | 3-oxoacyl-[acyl-carrier protein] reductase | |
| CrtEb | Lycopene elongase | |
| crtB | Phytoene synthase | |
| Idi | Isopentenyl-diphosphate delta-isomerase | |
| fabG | short-chain dehydrogenase/reductase SDR | |
| aas | 1-acyl-sn-glycerol-3-phosphate acyltransferase | |
| Gds | Geranylgeranyl diphosphate synthase | |
| fabH | 3-oxoacyl-[ACP] synthase III in alkane synthesis cluster | |
| fabF | 3-oxoacyl-[acyl-carrier-protein] synthase, KASII | |
| plsC | 1-acyl-sn-glycerol-3-phosphate acyltransferase | |
| pcaH | Protocatechuate 3,4-dioxygenase beta chain | Oxidative stress |
| pcaG | Protocatechuate 3,4-dioxygenase alpha chain | |
| trxC | Thiosulfate sulfurtransferase, rhodanese | |
| ntcA | Transcriptional regulator, Crp/Fnr family | |
| Lactoylglutathione lyase | ||
| yrkH | Hydroxyacylglutathione hydrolase | |
| sodC | Superoxide dismutase [Cu-Zn] precursor | |
| Cob | NAD-dependent protein deacetylase of SIR2 family | |
| --- | Glutathione S-transferase domain protein | |
| hcaC | Ferredoxin, 2Fe-2S | |
| soda | Superoxide dismutase [Mn] | |
| Fur | Zinc uptake regulation protein ZUR | |
| Gap | NADPH-dependent glyceraldehyde-3-phosphate dehydrogenase | |
| kata | Catalase | |
| nrdH | Glutaredoxin-like protein NrdH, required for reduction of Ribonucleotide reductase class Ib | |
| trxA | Thioredoxin | |
| trxB | Thioredoxin reductase | |
| capD | Gamma-glutamyltranspeptidase | |
| -- | Lactoylglutathione lyase and related lyases | |
| msrA | Peptide methionine sulfoxide reductase | |
| Dps | Ferroxidase | |
| yeaX | Vanillate O-demethylase oxidoreductase | |
| Line | Glyoxalase family protein | |
| Ohr | Organic hydroperoxide resistance protein | |
| rsmE | Ribosomal RNA small subunit methyltransferase E | |
| ywrD | Gamma-glutamyltranspeptidase | |
| ahpC | Alkyl hydroperoxide reductase subunit C-like protein | |
| Bcp | Thiol peroxidase, Bcp-type | |
| trxB | Thioredoxin reductase | |
| pncB1 | Nicotinate phosphoribosyltransferase | |
| Fur | Transcriptional regulator, FUR family | |
| hcaC | 3-phenylpropionate dioxygenase ferredoxin subunit | |
| bphG | Ferredoxin reductase | |
| pcaR | Transcriptional regulator, IclR family | |
| cobB1 | NAD-dependent protein deacetylase of SIR2 family | |
| ntcA | Transcriptional regulator, Crp/Fnr family | |
| bphC | Catechol 2,3-dioxygenase | |
| bphG | 3-phenylpropionate dioxygenase ferredoxin subunit | |
| Nicotinamidase | ||
| betA | Choline dehydrogenase | Osmo-protection |
| betP | High-affinity choline uptake protein | |
| gltB | Glutamate synthase [NADPH] large chain | |
| betC | Choline-sulfatase | |
| opuD | Glycine betaine transporter | |
| opuCA | L-proline glycine betaine ABC transport system permease protein ProV | |
| otsB | Trehalose-6-phosphate phosphatase | |
| proW | L-proline glycine betaine ABC transport system permease protein | |
| tcrY | Osmosensitive K+ channel histidine kinase KdpD | |
| otsA | Alpha, alpha-trehalose-phosphate synthase [UDP-forming] | |
| - | Na(+) H(+) antiporter subunit G | |
| - | Na(+) H(+) antiporter subunit F | |
| mrpD | Na(+) H(+) antiporter subunit D | |
| mrpE | Na(+) H(+) antiporter subunit E | |
| mnhC1 | Na(+) H(+) antiporter subunit C | |
| mrpA | Na(+) H(+) antiporter subunit A; Na(+) H(+) antiporter subunit B | |
| opuCB | Glycine betaine ABC transport system permease protein | |
| mrpG | Na(+) H(+) antiporter subunit G | |
| mrpC | Na(+) H(+) antiporter subunit C | |
| - | FIG152265: Sodium:solute symporter associated protein | |
| - | Na(+) H(+) antiporter subunit F | |
| - | Na(+) H(+) antiporter subunit E | |
| mrpD | Na(+) H(+) antiporter subunit D | |
| betT | High-affinity choline uptake protein | |
| gltB | Glutamate synthase [NADPH] small chain | |
| ectA | L-2,4-diaminobutyric acid acetyltransferase | |
| gbsA | Betaine aldehyde dehydrogenase | |
| betA | Choline dehydrogenase | |
| baeS | Osmosensitive K+ channel histidine kinase KdpD | |
| - | Glutamate synthase [NADPH] large chain | |
| gltB | Glutamate synthase [NADPH] small chain | |
| opuBB | Glycine betaine ABC transport system permease protein | |
| putA | Proline dehydrogenase (Proline oxidase) | |
| ectC | L-ectoine synthase | |
| ectB | Diaminobutyrate-pyruvate aminotransferase | |
| panF | Sodium:solute symporter, putative | |
| treS | Trehalose synthase | |
| osmF | L-proline glycine betaine binding ABC transporter protein ProX | |
| - | Universal stress protein | General stress |
| - | Serine phosphatase RsbU, regulator of sigma subunit | |
| glbO | Hemoglobin-like protein HbO | |
| rpoE | RNA polymerase sigma-70 factor, ECF subfamily |
| Cold-Active Enzymes Detected in PF2B19 Genome | Applications |
|---|---|
| Lipase, protease, phytase, xylanase | Improves digestibility and assimilation of animal feed |
| Chitinase, Protease | Meat tenderizing |
| α-amylase, xylanase | Textile industry |
| Esterase | Chiral resolution of drugs to escalate effectiveness and range |
| β-lactamase | Antibiotic degradation |
| Lipase | Cosmetics, detergents |
| Chitinase | Additive for anti-fungal creams and lotions, Anti-fungal drug |
| β-galactosidase | Bioethanol production from dairy waste, improves the digestibility of dairy products for lactose-intolerant consumers |
| β-glucosidase | Wine industry |
| Xylanase | Biobleaching in paper and pulp industry |
| Lipase | Biodiesel production by trans-esterification of oils and alcohols |
| Alkaline phosphatase | Cloning experiments in molecular biology |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, P.; Kapse, N.; Gowdaman, V.; Tsuji, M.; Singh, S.M.; Dhakephalkar, P.K. Comparative Genomic Analysis of Arctic Permafrost Bacterium Nesterenkonia sp. PF2B19 to Gain Insights into Its Cold Adaptation Tactic and Diverse Biotechnological Potential. Sustainability 2021, 13, 4590. https://doi.org/10.3390/su13084590
Singh P, Kapse N, Gowdaman V, Tsuji M, Singh SM, Dhakephalkar PK. Comparative Genomic Analysis of Arctic Permafrost Bacterium Nesterenkonia sp. PF2B19 to Gain Insights into Its Cold Adaptation Tactic and Diverse Biotechnological Potential. Sustainability. 2021; 13(8):4590. https://doi.org/10.3390/su13084590
Chicago/Turabian StyleSingh, Purnima, Neelam Kapse, Vasudevan Gowdaman, Masaharu Tsuji, Shiv Mohan Singh, and Prashant K. Dhakephalkar. 2021. "Comparative Genomic Analysis of Arctic Permafrost Bacterium Nesterenkonia sp. PF2B19 to Gain Insights into Its Cold Adaptation Tactic and Diverse Biotechnological Potential" Sustainability 13, no. 8: 4590. https://doi.org/10.3390/su13084590
APA StyleSingh, P., Kapse, N., Gowdaman, V., Tsuji, M., Singh, S. M., & Dhakephalkar, P. K. (2021). Comparative Genomic Analysis of Arctic Permafrost Bacterium Nesterenkonia sp. PF2B19 to Gain Insights into Its Cold Adaptation Tactic and Diverse Biotechnological Potential. Sustainability, 13(8), 4590. https://doi.org/10.3390/su13084590








