Transcriptome Analysis Revealed Ameliorative Effects of Bacillus Based Probiotic on Immunity, Gut Barrier System, and Metabolism of Chicken under an Experimentally Induced Eimeria tenella Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. E. tenella and Probiotic
2.2. Ethical Statement
2.3. Experimental Design
2.4. Evolution of Clinical Parameters and Sample Collection for RNA-Sequencing
2.5. RNA Extraction
2.6. Library Construction for Illumina Sequencing
2.7. Clustering and Sequencing
2.8. Raw Data Processing and Analysis
2.9. Screening of Differentially Expressed Genes
2.10. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Enrichment Analysis of Differentially Expressed Genes
2.11. Validation of DEGs by qRT-PCR
2.12. Statistical Analysis
3. Results
3.1. Clinical Parameters
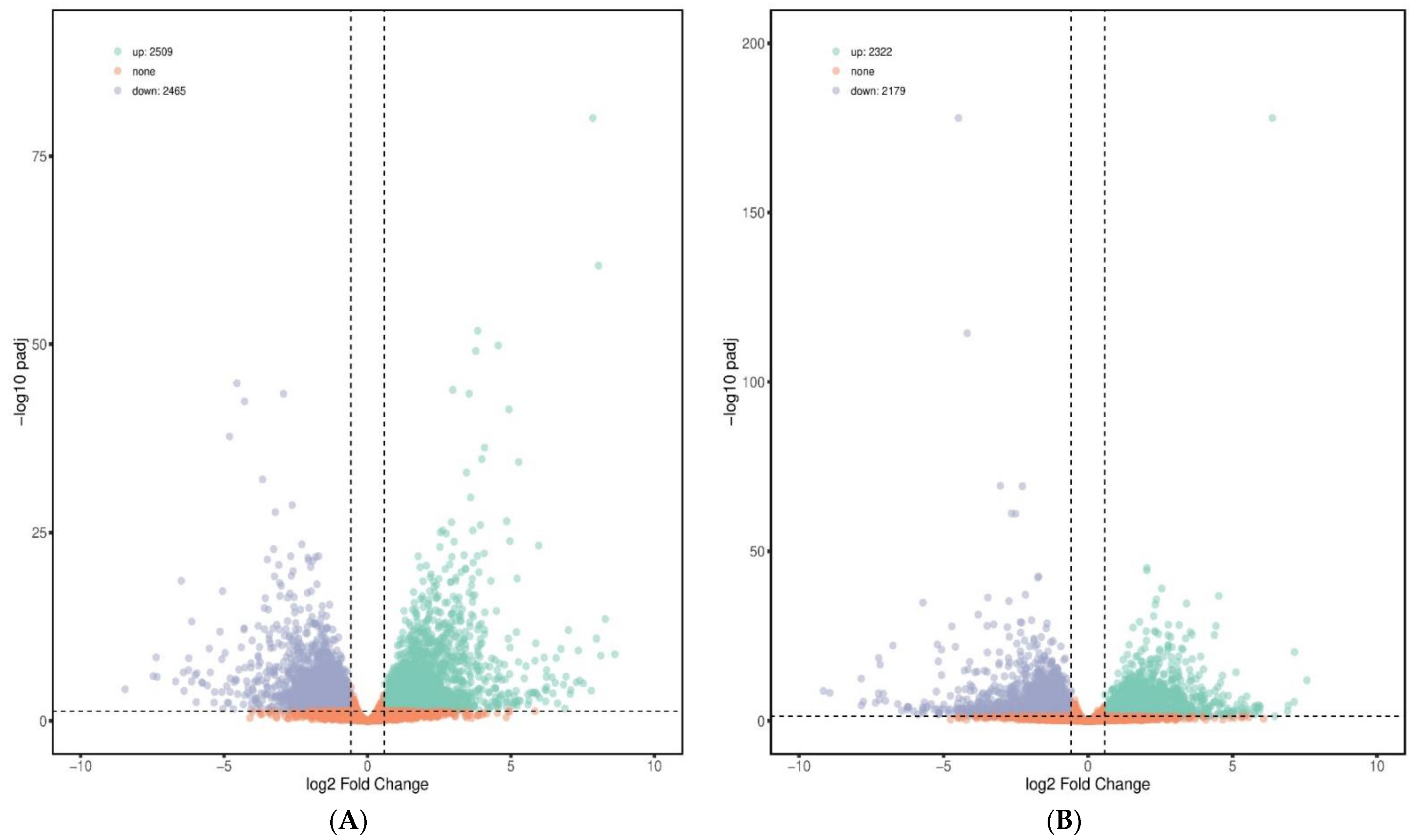
3.2. Data Analysis of Transcriptome
3.3. Differentially Expressed Genes Regulated by the Exposure of E. tenella and Probiotic
3.4. Gene Ontology Enrichment Analysis for Unique Differentially Expressed Genes
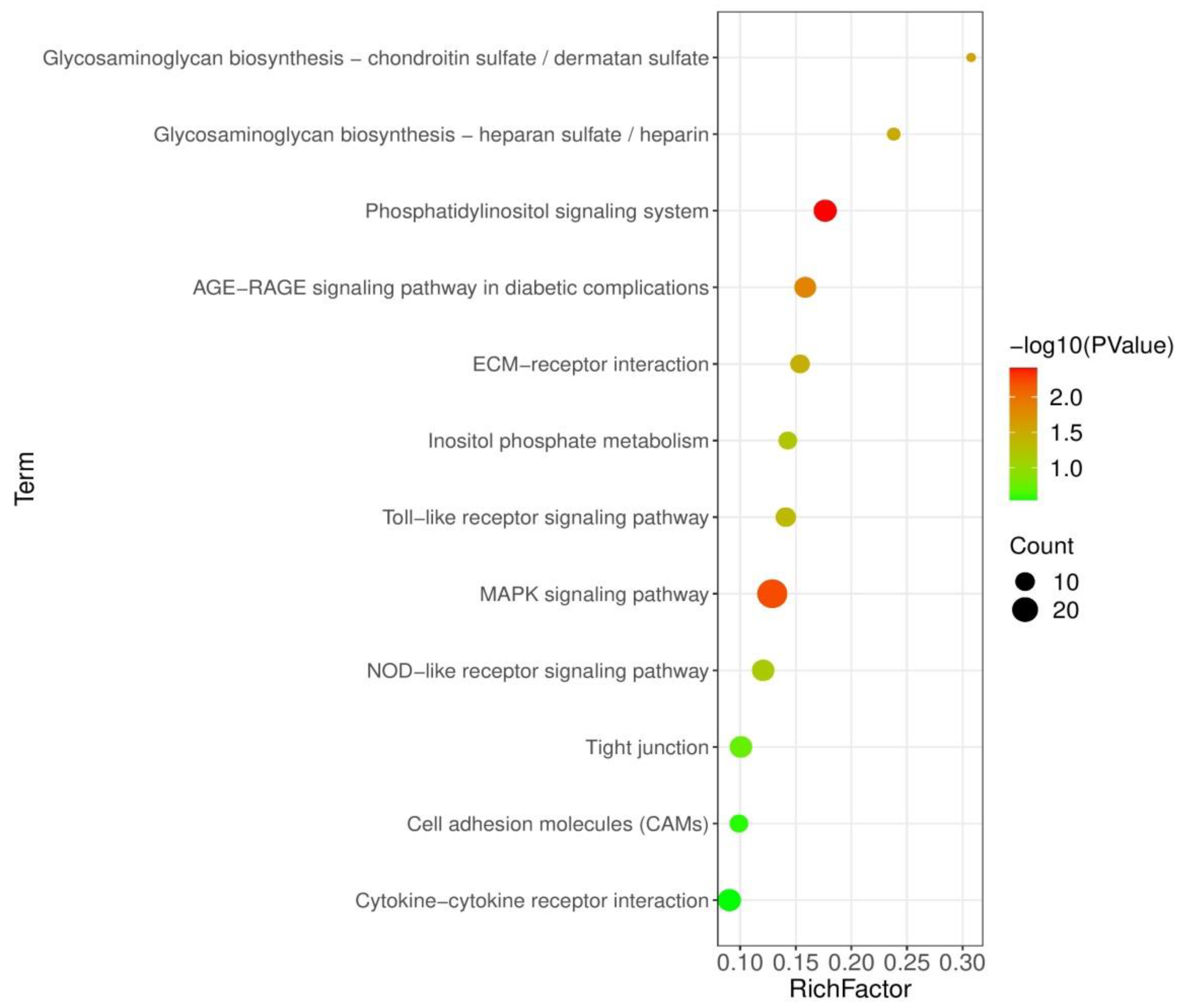
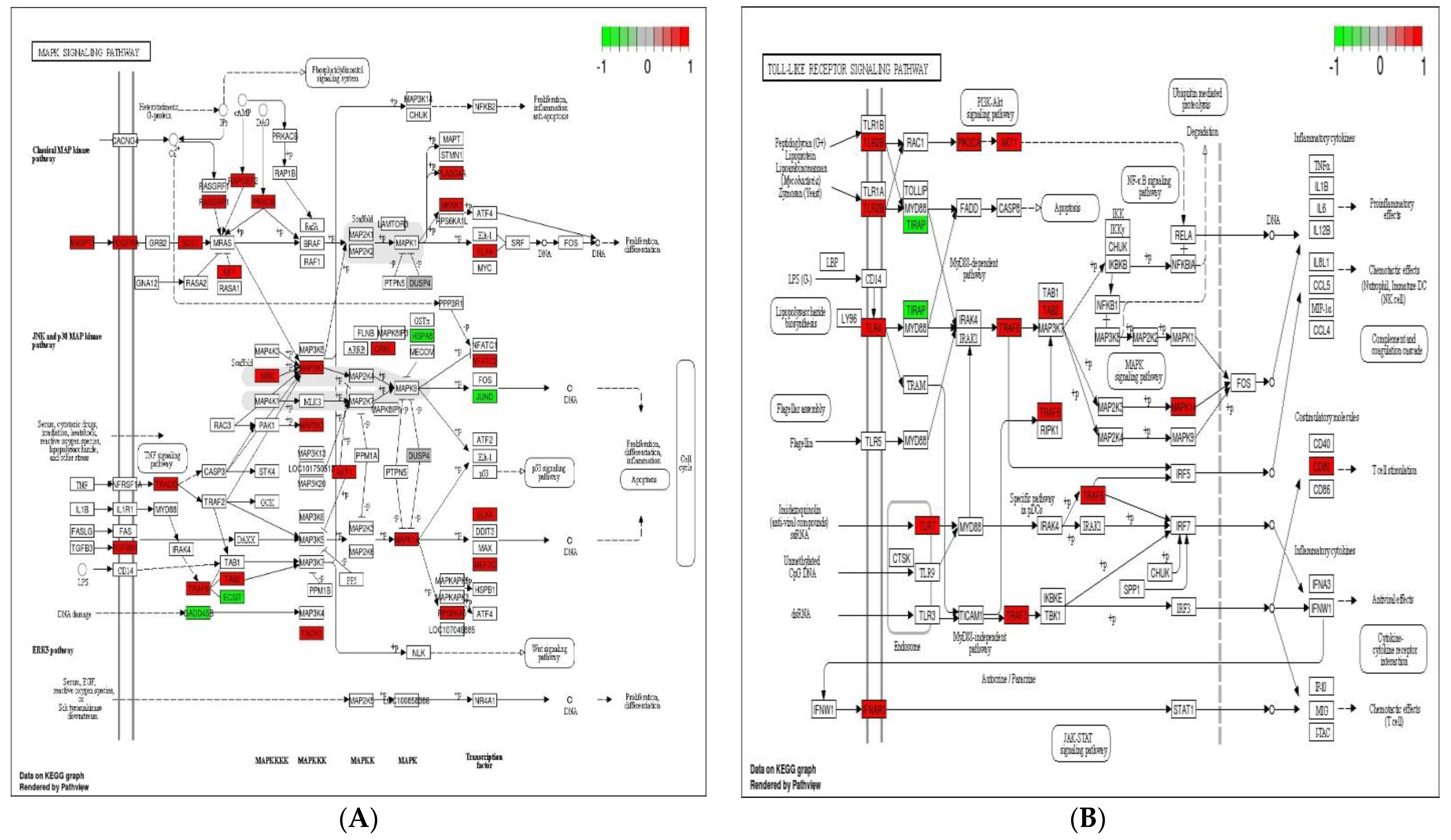
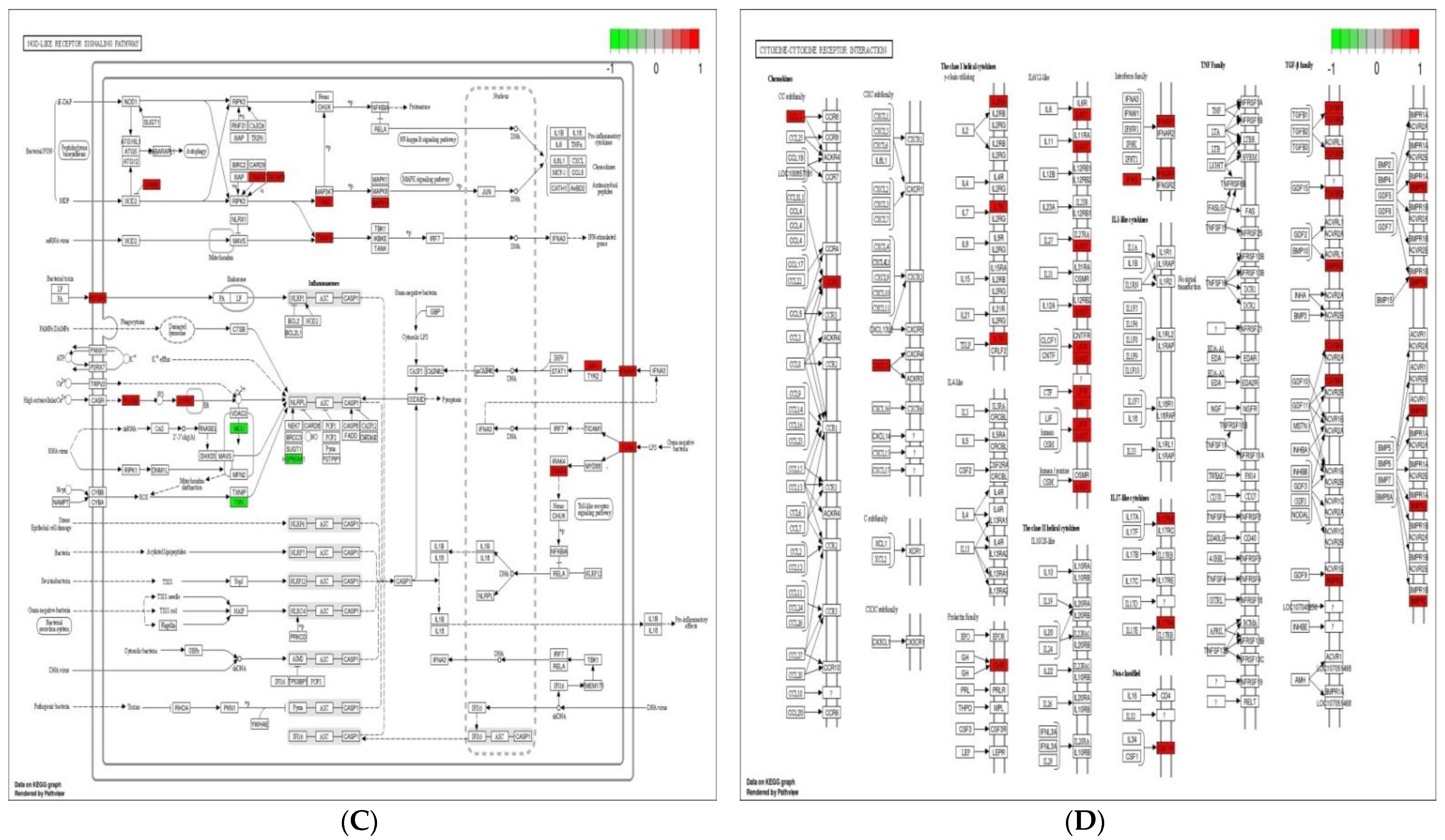
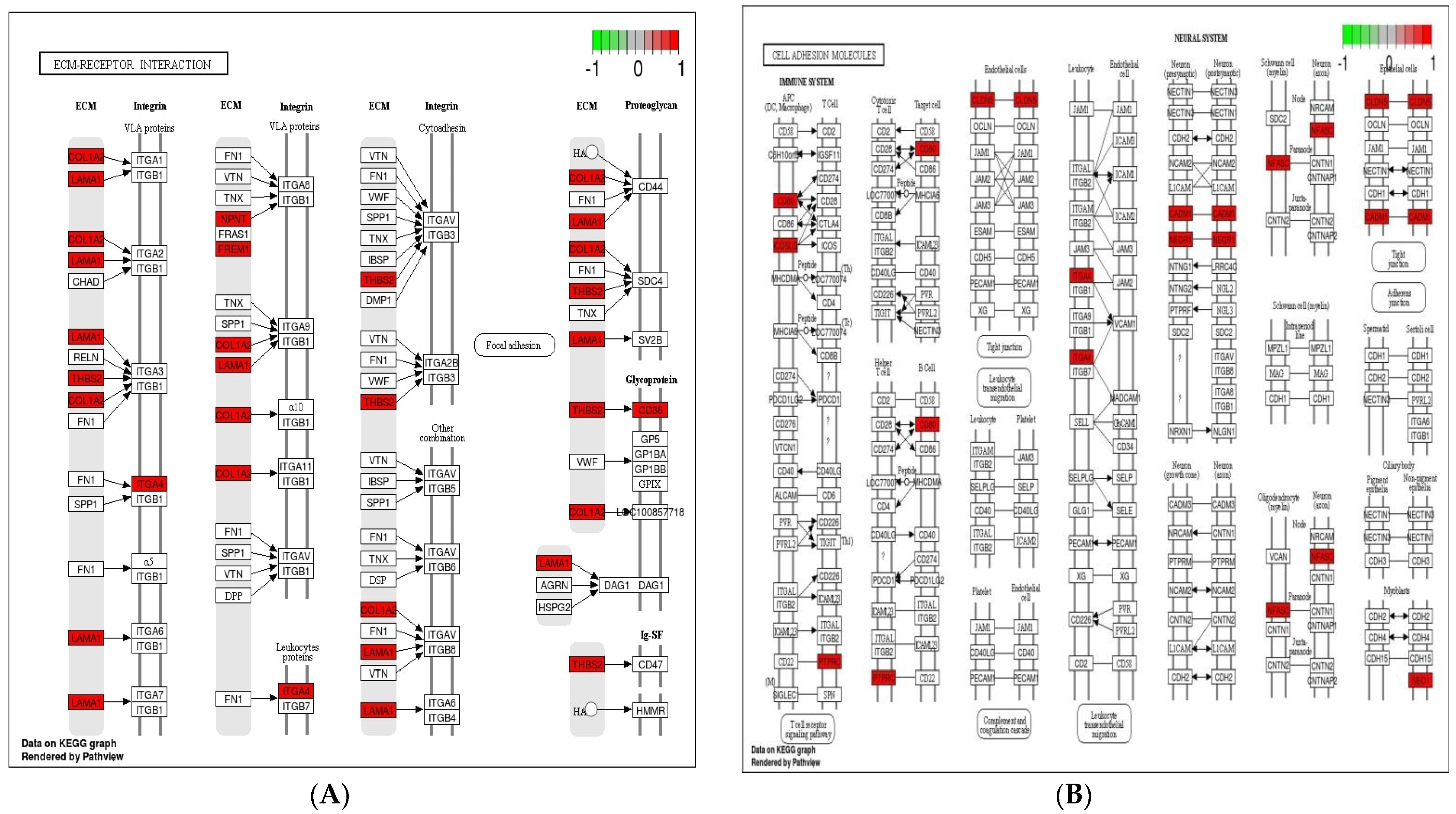
3.5. Analysis of KEGG Pathways
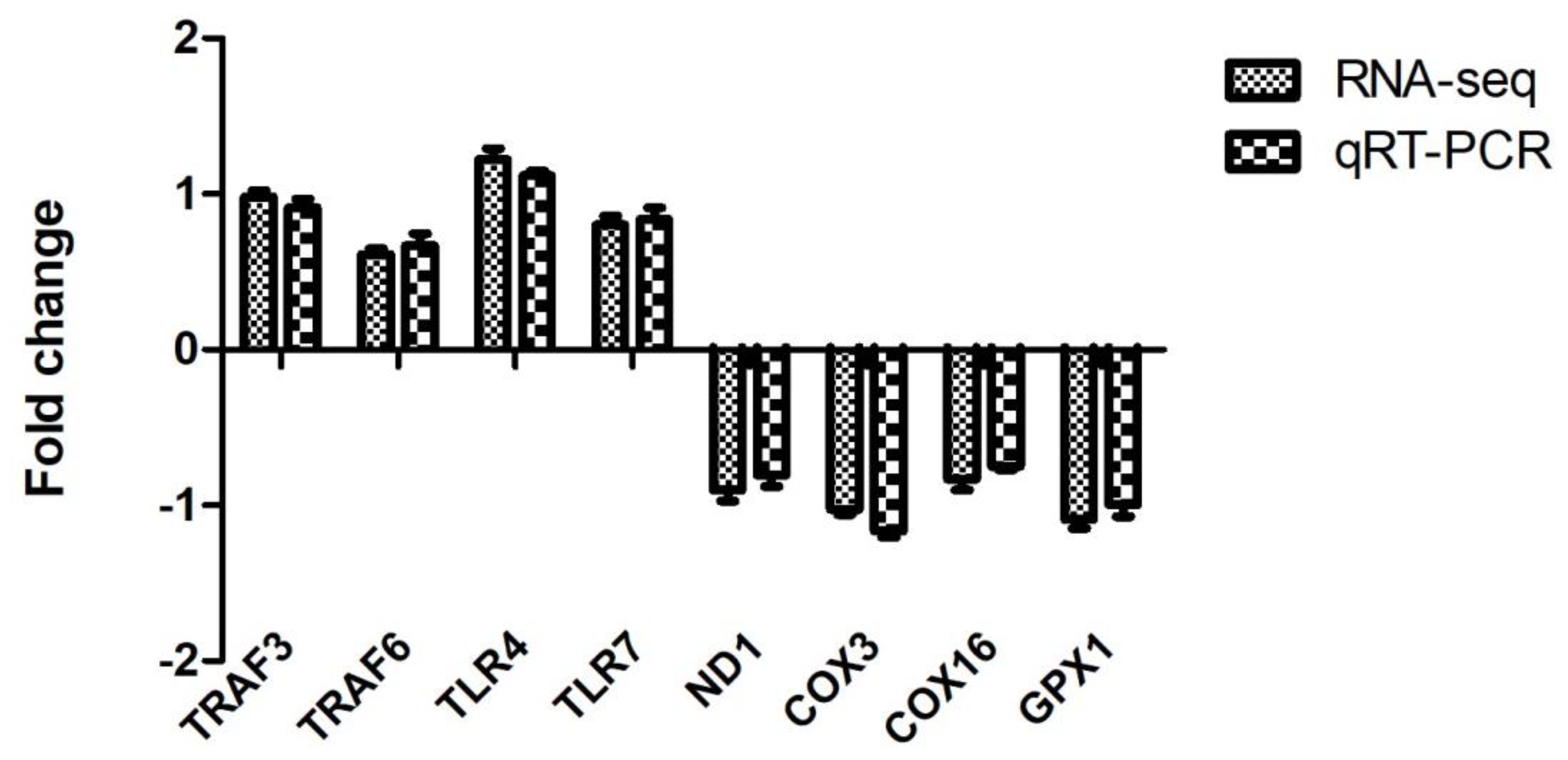
3.6. Validation of DEGs by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blake, D.P.; Tomley, F.M. Securing poultry production from the ever-present Eimeria challenge. Trends Parasitol. 2014, 30, 12–19. [Google Scholar] [CrossRef]
- Vermeulen, A.; Schaap, D.; Schetters, T.P. Control of coccidiosis in chickens by vaccination. Vet. Parasitol. 2001, 100, 13–20. [Google Scholar] [CrossRef]
- Chapman, H.; Jeffers, T.; Williams, R. Forty years of monensin for the control of coccidiosis in poultry. Poult. Sci. 2010, 89, 1788–1801. [Google Scholar] [CrossRef]
- Liu, Y.; Zheng, J.; Li, J.; Gong, P.; Zhang, X. Protective immunity induced by a DNA vaccine encoding Eimeria tenella rhomboid against homologous challenge. Parasitol. Res. 2013, 112, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, Q.; Huang, Z.; Lv, L.; Liu, X.; Yin, C.; Yan, H.; Yuan, J. Effect of Bacillus subtilis CGMCC 1.1086 on the growth performance and intestinal microbiota of broilers. J. Appl. Microbiol. 2016, 120, 195–204. [Google Scholar]
- Bozkurt, M.; Aysul, N.; Küçükyilmaz, K.; Aypak, S.; Ege, G.; Catli, A.; Akşit, H.; Çöven, F.; Seyrek, K.; Çınar, M. Efficacy of in-feed preparations of an anticoccidial, multienzyme, prebiotic, probiotic, and herbal essential oil mixture in healthy and Eimeria spp.-infected broilers. Poult. Sci. 2014, 93, 389–399. [Google Scholar] [CrossRef]
- Salim, H.; Kang, H.; Akter, N.; Kim, D.; Kim, J.; Kim, M.; Na, J.; Jong, H.; Choi, H.; Suh, O. Supplementation of direct-fed microbials as an alternative to antibiotic on growth performance, immune response, cecal microbial population, and ileal morphology of broiler chickens. Poult. Sci. 2013, 92, 2084–2090. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Farnell, Y.Z.; Kiess, A.S.; Peebles, E.D.; Wamsley, K.G.; Zhai, W. Effects of Bacillus subtilis and coccidial vaccination on cecal microbial diversity and composition of Eimeria-challenged male broilers. Poult. Sci. 2019, 98, 3839–3849. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, W. Roles of Bacillus endospores in the environment. Cell. Mol. Life Sci. CMLS 2002, 59, 410–416. [Google Scholar] [CrossRef]
- Rajput, I.R.; Ying, H.; Yajing, S.; Arain, M.A.; Weifen, L.; Ping, L.; Bloch, D.M.; Wenhua, L. Saccharomyces boulardii and Bacillus subtilis B10 modulate TLRs and cytokines expression patterns in jejunum and ileum of broilers. PLoS ONE 2017, 12, e0173917. [Google Scholar]
- Sikandar, A.; Zaneb, H.; Younus, M.; Masood, S.; Aslam, A.; Shah, M.; Rehman, H. Growth performance, immune status and organ morphometry in broilers fed Bacillus subtilis-supplemented diet. S. Afr. J. Anim. Sci. 2017, 47, 378–388. [Google Scholar]
- Yang, X.; Kui, L.; Tang, M.; Li, D.; Wei, K.; Chen, W.; Miao, J.; Dong, Y. High-throughput transcriptome profiling in drug and biomarker discovery. Front. Genet. 2020, 11, 19. [Google Scholar] [CrossRef] [PubMed]
- Mack, D.R.; Michail, S.; Wei, S.; McDougall, L.; Hollingsworth, M.A. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am. J. Physiol. Gastrointest. Liver Physiol. 1999, 276, G941–G950. [Google Scholar] [CrossRef]
- Jiang, S.; Mohammed, A.; Jacobs, J.; Cramer, T.; Cheng, H. Effect of synbiotics on thyroid hormones, intestinal histomorphology, and heat shock protein 70 expression in broiler chickens reared under cyclic heat stress. Poult. Sci. 2020, 99, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Kukurba, K.R.; Montgomery, S.B. RNA sequencing and analysis. Cold Spring Harb. Protoc. 2015. [Google Scholar] [CrossRef] [PubMed]
- Memon, F.Q.; Yang, Y.; Lv, F.; Soliman, A.; Chen, Y.; Sun, J.; Wang, Y.; Zhang, G.; Li, Z.; Xu, B. Effects of probiotic and Bidens pilosa on the performance and gut health of chicken during induced Eimeria tenella infection. J. Appl. Microbiol. 2020. [Google Scholar] [CrossRef]
- Morehouse, N.F.; Baron, R.R. Coccidiosis: Evaluation of coccidiostats by mortality, weight gains, and fecal scores. Exp. Parasitol. 1970, 28, 25–29. [Google Scholar] [CrossRef]
- Hodgson, J. Coccidiosis: Oocyst counting technique for coccidiostat evaluation. Exp. Parasitol. 1970, 28, 99–102. [Google Scholar] [CrossRef]
- Johnson, J.; Reid, W.M. Anticoccidial drugs: Lesion scoring techniques in battery and floor-pen experiments with chickens. Exp. Parasitol. 1970, 28, 30–36. [Google Scholar] [CrossRef]
- Zhao, P.; Zhang, K.; Guo, G.; Sun, X.; Chai, H.; Zhang, W.; Xing, M. Heat shock protein alteration in the gastrointestinal tract tissues of chickens exposed to arsenic trioxide. Biol. Trace Elem. Res. 2016, 170, 224–236. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Meth. Nat. Res. 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Anders, S.; Kim, V.; Huber, W. RNA-Seq workflow: Gene-level exploratory analysis and differential expression. F1000Research 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, 1–12. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Giannenas, I.; Papadopoulos, E.; Tsalie, E.; Triantafillou, E.; Henikl, S.; Teichmann, K.; Tontis, D. Assessment of dietary supplementation with probiotics on performance, intestinal morphology and microflora of chickens infected with Eimeria tenella. Vet. Parasitol. 2012, 188, 31–40. [Google Scholar] [CrossRef]
- Lee, K.-W.; Lillehoj, H.S.; Jang, S.I.; Li, G.; Lee, S.-H.; Lillehoj, E.P.; Siragusa, G.R. Effect of Bacillus-based direct-fed microbials on Eimeria maxima infection in broiler chickens. Comp. Immunol. Microbiol. Infect. Dis. 2010, 33, e105–e110. [Google Scholar] [CrossRef]
- Lillehoj, H.S.; Lillehoj, E.P. Avian coccidiosis. A review of acquired intestinal immunity and vaccination strategies. Avian Dis. 2000, 44, 408–425. [Google Scholar]
- Adams, C.; Vahl, H.; Veldman, A. Interaction between nutrition and Eimeria acervulina infection in broiler chickens: Development of an experimental infection model. Br. J. Nutr. 1996, 75, 867–873. [Google Scholar] [CrossRef]
- Sanders, M.E.; Guarner, F.; Guerrant, R.; Holt, P.R.; Quigley, E.M.; Sartor, R.B.; Sherman, P.M.; Mayer, E.A. An update on the use and investigation of probiotics in health and disease. Gut 2013, 62, 787–796. [Google Scholar] [CrossRef]
- Cox, C.; Dalloul, R. Immunomodulatory role of probiotics in poultry and potential in ovo application. Benef. Microbes 2015, 6, 45–52. [Google Scholar] [CrossRef]
- Dunislawska, A.; Slawinska, A.; Bednarczyk, M.; Siwek, M. Transcriptome modulation by in ovo delivered Lactobacillus synbiotics in a range of chicken tissues. Gene 2019, 698, 27–33. [Google Scholar] [CrossRef]
- Kumar, A.; Vlasova, A.N.; Liu, Z.; Chattha, K.S.; Kandasamy, S.; Esseili, M.; Zhang, X.; Rajashekara, G.; Saif, L.J. In vivo gut transcriptome responses to Lactobacillus rhamnosus GG and Lactobacillus acidophilus in neonatal gnotobiotic piglets. Gut Microbes 2014, 5, 152–164. [Google Scholar] [CrossRef]
- Modak, T.H.; Gomez-Chiarri, M. Contrasting Immunomodulatory Effects of Probiotic and Pathogenic Bacteria on Eastern Oyster, Crassostrea Virginica, Larvae. Vaccines 2020, 8, 588. [Google Scholar] [CrossRef]
- Marcobal, A.; Yusufaly, T.; Higginbottom, S.; Snyder, M.; Sonnenburg, J.L.; Mias, G.I. Metabolome progression during early gut microbial colonization of gnotobiotic mice. Sci. Rep. 2015, 5, 1–13. [Google Scholar] [CrossRef]
- Gensollen, T.; Blumberg, R.S. Correlation between early-life regulation of the immune system by microbiota and allergy development. J. Allergy Clin. Immunol. 2017, 139, 1084–1091. [Google Scholar] [CrossRef]
- Fukata, M.; Vamadevan, A.S.; Abreu, M.T. Toll-like receptors (TLRs) and Nod-like receptors (NLRs) in inflammatory disorders. Semin. Immunol. 2009, 21, 242–253. [Google Scholar] [CrossRef]
- Symons, A.; Beinke, S.; Ley, S.C. MAP kinase kinase kinases and innate immunity. Trends Immunol. 2006, 27, 40–48. [Google Scholar] [CrossRef]
- Lai, Y.; Gallo, R.L. Toll-like receptors in skin infections and inflammatory diseases. Infect. Disord. Drug Targets 2008, 8, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Petruzzelli, L.; Takami, M.; Humes, H.D. Structure and function of cell adhesion molecules. Am. J. Med. 1999, 106, 467–476. [Google Scholar] [CrossRef]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef] [PubMed]
- Bonazzi, M.; Cossart, P. Impenetrable barriers or entry portals? The role of cell–cell adhesion during infection. J. Cell Biol. 2011, 195, 349–358. [Google Scholar] [PubMed]



| Parameters | Groups | ||
|---|---|---|---|
| NC | PC | BS + ET | |
| Lesion scores (Grade) | |||
| Day 7 post-infection | 0.00 ± 0.00 c | 3.00 ± 0.00 a | 1.66 ± 0.33 b |
| Bloody diarrhea scores | |||
| Day 5 post-infection | 0.00 ± 0.00 c | 2.33 ± 0.33 a | 1.33 ± 0.33 b |
| Day 6 post-infection | 0.00 ± 0.00 c | 3.33 ± 0.33 a | 1.66 ± 0.33 b |
| Day 7 post-infection | 0.00 ± 0.00 c | 3.00 ± 0.57 a | 1.33 ± 0.33 b |
| Oocyst counting (OPG/g × 103) | |||
| Day 5 post-infection | 0.00 ± 0.00 c | 34.00 ± 2.51 a | 20.33 ± 1.20 b |
| Day 6 post-infection | 0.00 ± 0.00 c | 51.33 ± 4.05 a | 28.66 ± 2.18 b |
| Day 7 post-infection | 0.00 ± 0.00 c | 77.66 ± 2.90 a | 37.33 ± 3.17 b |
| Sample | Raw Reads | Raw Bases (G) | Clean Reads | Clean Bases (G) | Q20 (%) | GC Content (%) |
|---|---|---|---|---|---|---|
| NC-1 | 20,395,539 | 6.12 | 19,319,443 | 5.80 | 97.28 | 46.94 |
| NC-2 | 19,948,996 | 5.98 | 18,807,009 | 5.64 | 97.73 | 47.16 |
| NC-3 | 20,615,240 | 6.18 | 19,233,599 | 5.77 | 97.49 | 48.18 |
| PC-1 | 20,963,369 | 6.29 | 19,772,595 | 5.93 | 96.96 | 51.25 |
| PC-2 | 19,983,927 | 6.00 | 19,174,592 | 5.75 | 96.94 | 50.95 |
| PC-3 | 20,852,755 | 6.26 | 20,110,347 | 6.03 | 96.86 | 51.46 |
| BS + ET-1 | 20,851,951 | 6.26 | 20,048,311 | 6.01 | 96.79 | 51.06 |
| BS + ET-2 | 19,842,850 | 5.95 | 18,763,598 | 5.63 | 97 | 51.19 |
| BS + ET-3 | 20,570,711 | 6.17 | 19,463,192 | 5.84 | 96.92 | 50.16 |
| Sum | 184,025,338 | 55.51 | 174,692,686 | 52.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Memon, F.U.; Yang, Y.; Leghari, I.H.; Lv, F.; Soliman, A.M.; Zhang, W.; Si, H. Transcriptome Analysis Revealed Ameliorative Effects of Bacillus Based Probiotic on Immunity, Gut Barrier System, and Metabolism of Chicken under an Experimentally Induced Eimeria tenella Infection. Genes 2021, 12, 536. https://doi.org/10.3390/genes12040536
Memon FU, Yang Y, Leghari IH, Lv F, Soliman AM, Zhang W, Si H. Transcriptome Analysis Revealed Ameliorative Effects of Bacillus Based Probiotic on Immunity, Gut Barrier System, and Metabolism of Chicken under an Experimentally Induced Eimeria tenella Infection. Genes. 2021; 12(4):536. https://doi.org/10.3390/genes12040536
Chicago/Turabian StyleMemon, Fareed Uddin, Yunqiao Yang, Imdad Hussain Leghari, Feifei Lv, Ahmed M. Soliman, Weiyu Zhang, and Hongbin Si. 2021. "Transcriptome Analysis Revealed Ameliorative Effects of Bacillus Based Probiotic on Immunity, Gut Barrier System, and Metabolism of Chicken under an Experimentally Induced Eimeria tenella Infection" Genes 12, no. 4: 536. https://doi.org/10.3390/genes12040536
APA StyleMemon, F. U., Yang, Y., Leghari, I. H., Lv, F., Soliman, A. M., Zhang, W., & Si, H. (2021). Transcriptome Analysis Revealed Ameliorative Effects of Bacillus Based Probiotic on Immunity, Gut Barrier System, and Metabolism of Chicken under an Experimentally Induced Eimeria tenella Infection. Genes, 12(4), 536. https://doi.org/10.3390/genes12040536






