Unlocking the Patterns of the Tunisian Durum Wheat Landraces Genetic Structure Based on Phenotypic Characterization in Relation to Farmer’s Vernacular Name
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Field Experimental Trials
2.3. Phenotypic Characterization
2.4. Genetic Landraces Patterns Analysis
2.5. Statistical Analysis
3. Results
3.1. Heritability and Variability of Agro-Morphological and Phenological Traits in Tunisian Durum Wheat Germplasm Collection
3.2. Analysis of Variance
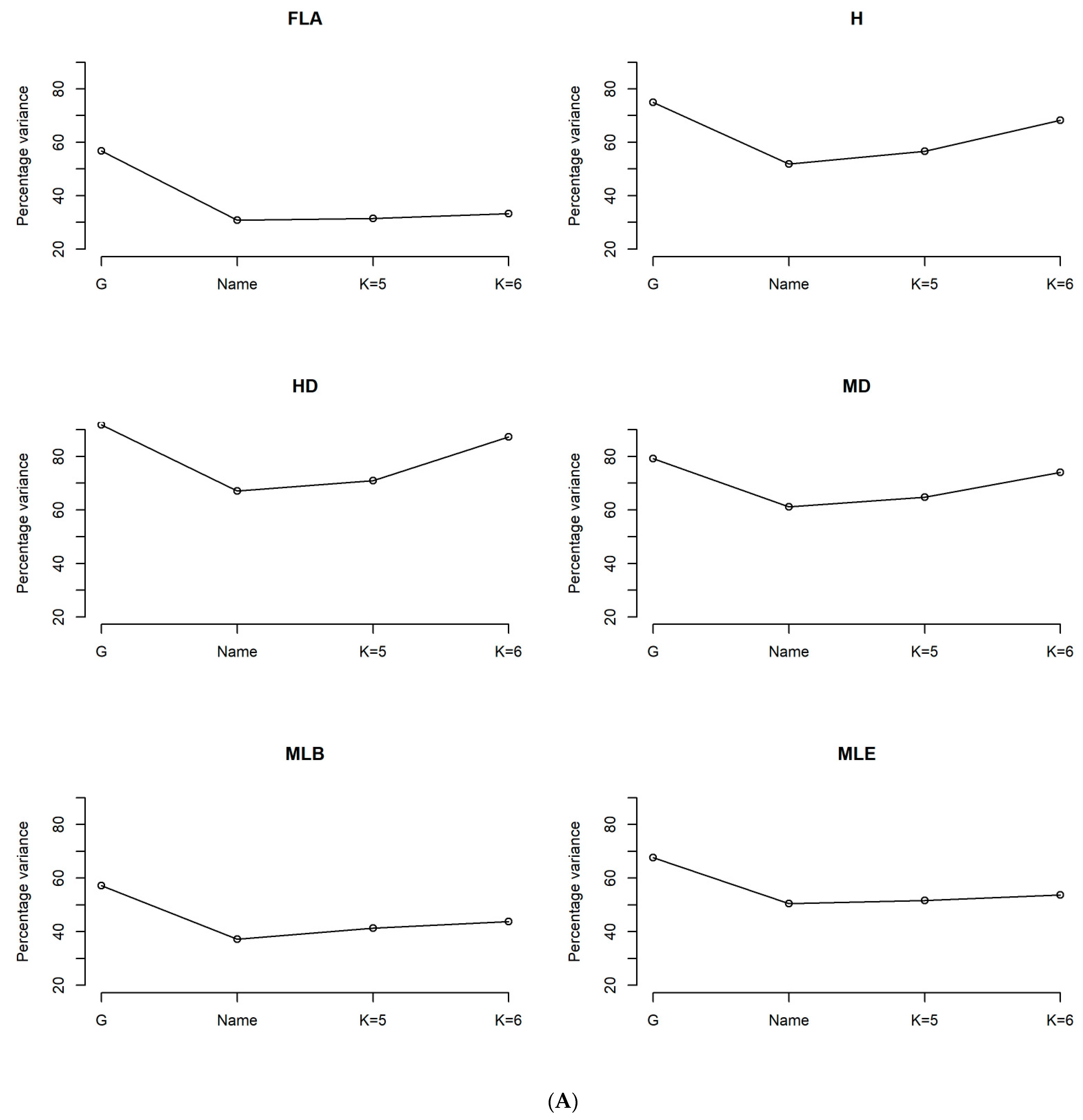
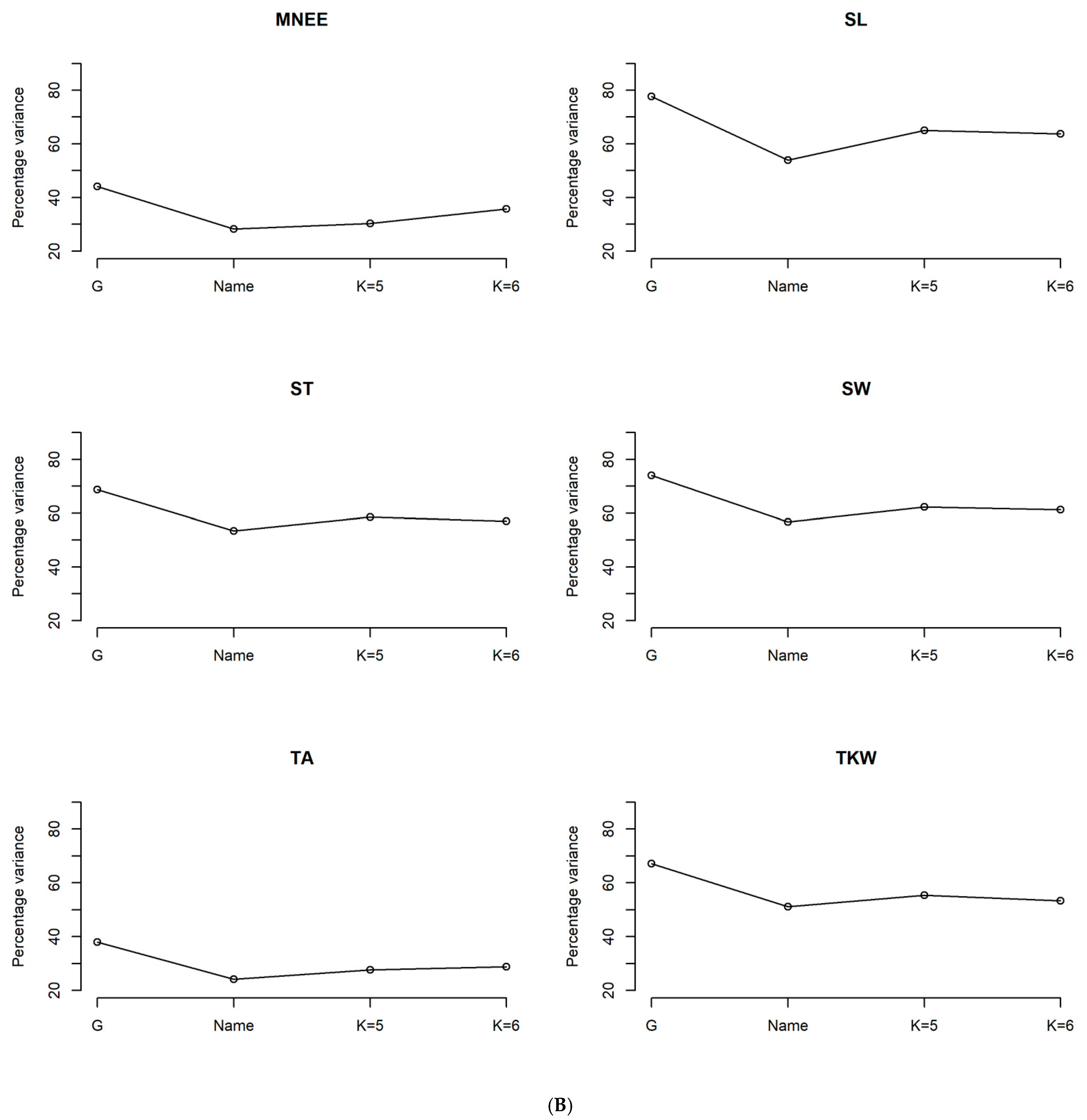
3.3. Phenotypic Characterization Based on the Genetic Structure Using the Optimal Number of Sub-Population (K = 6)
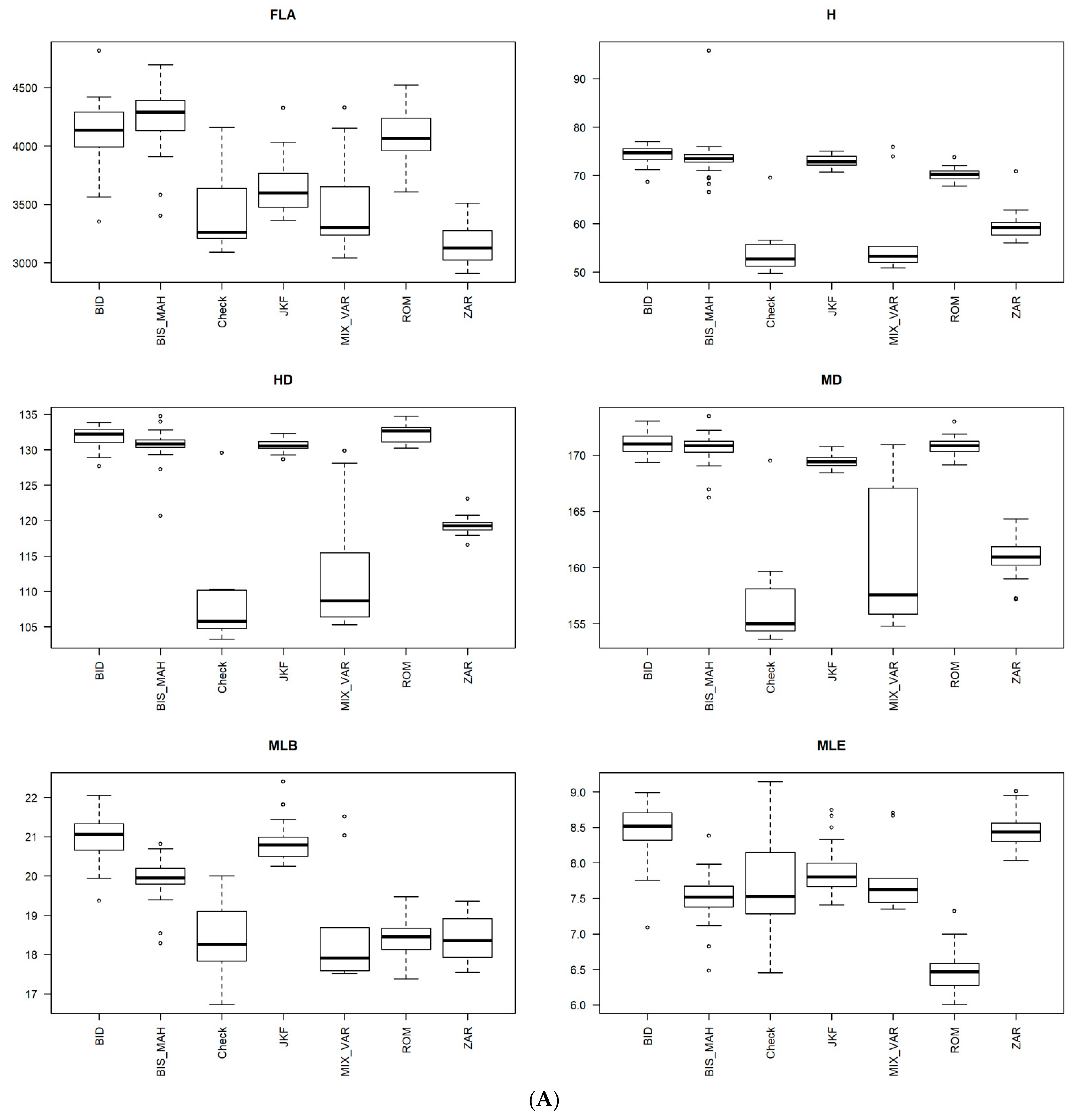
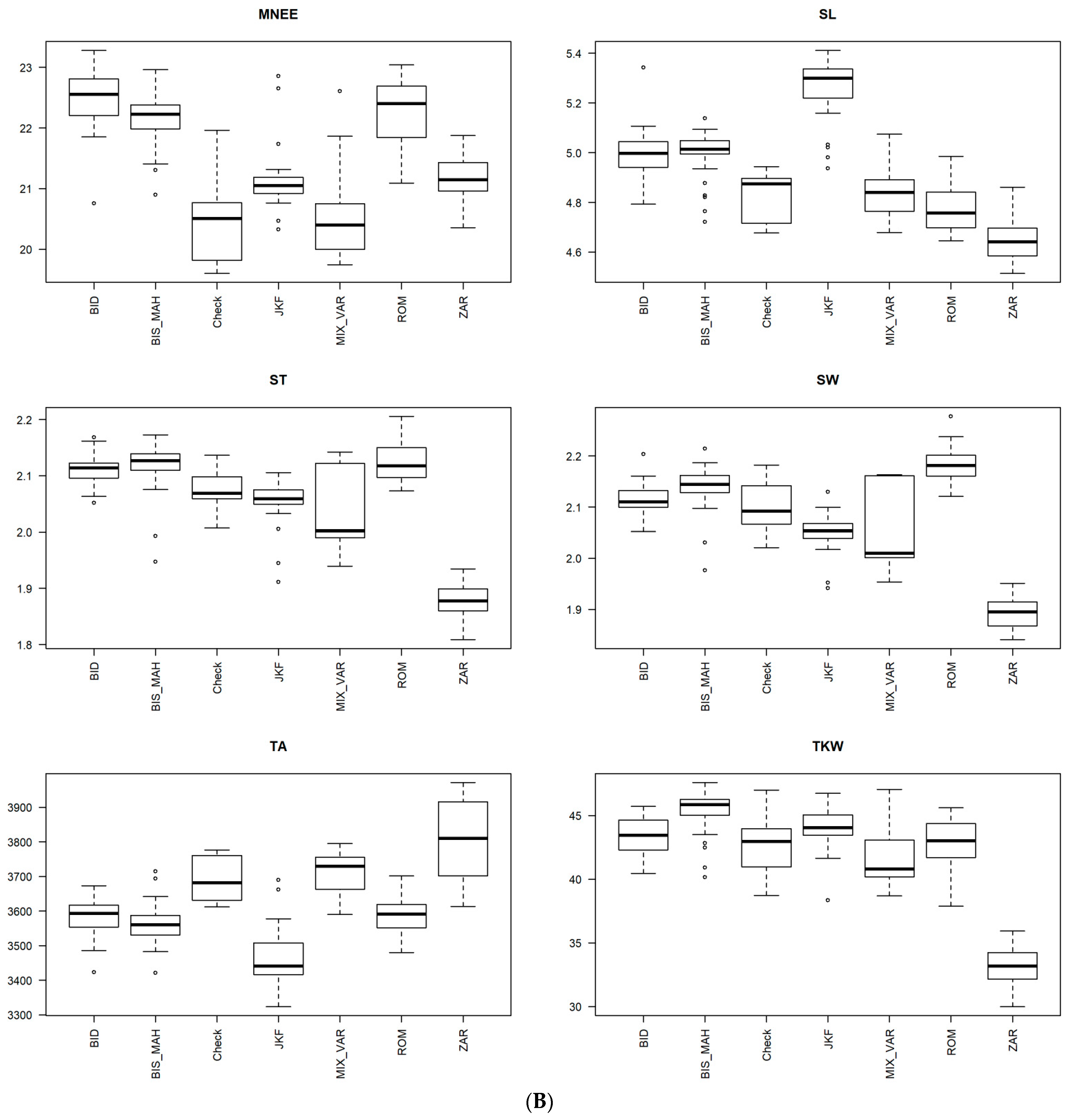
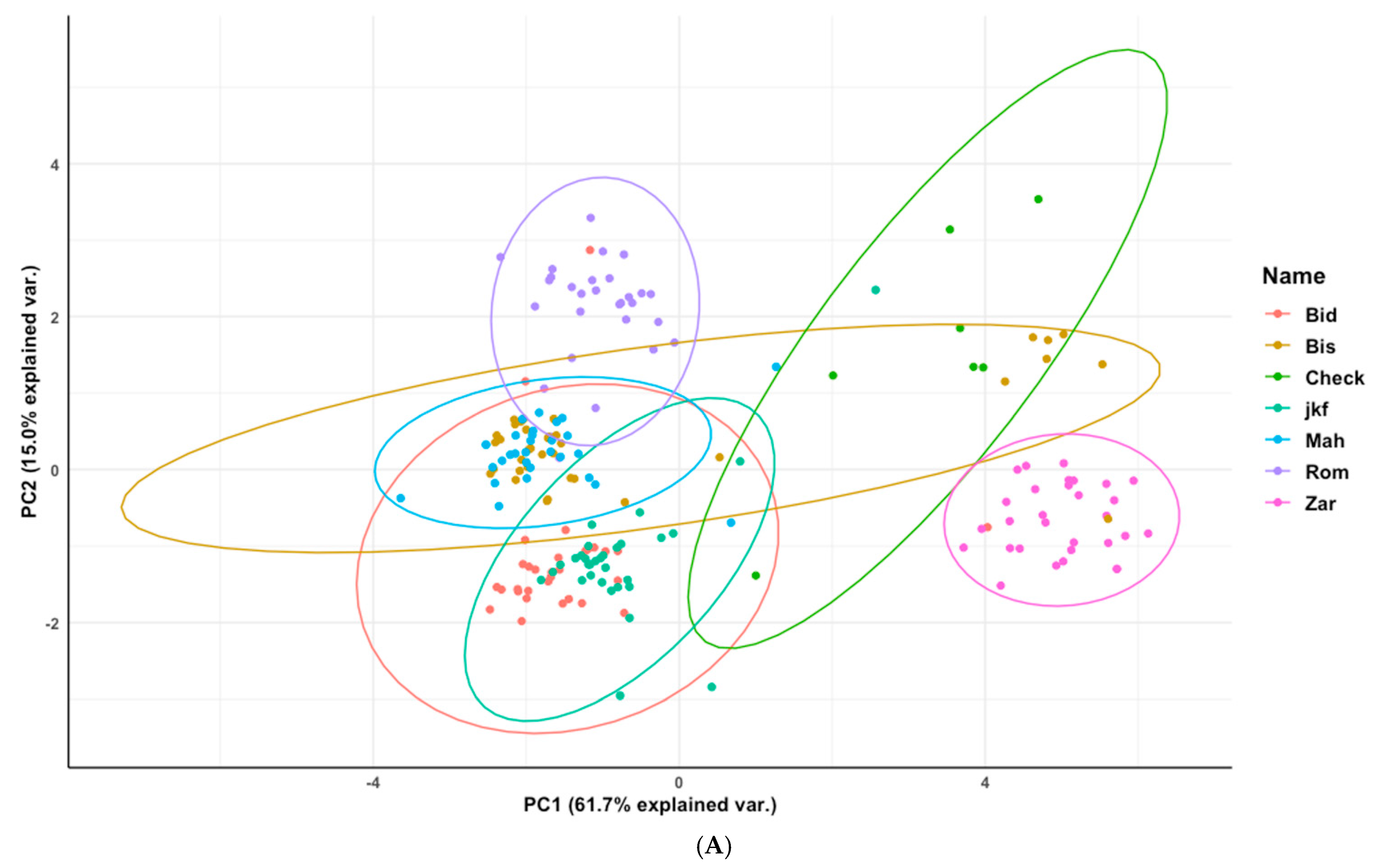
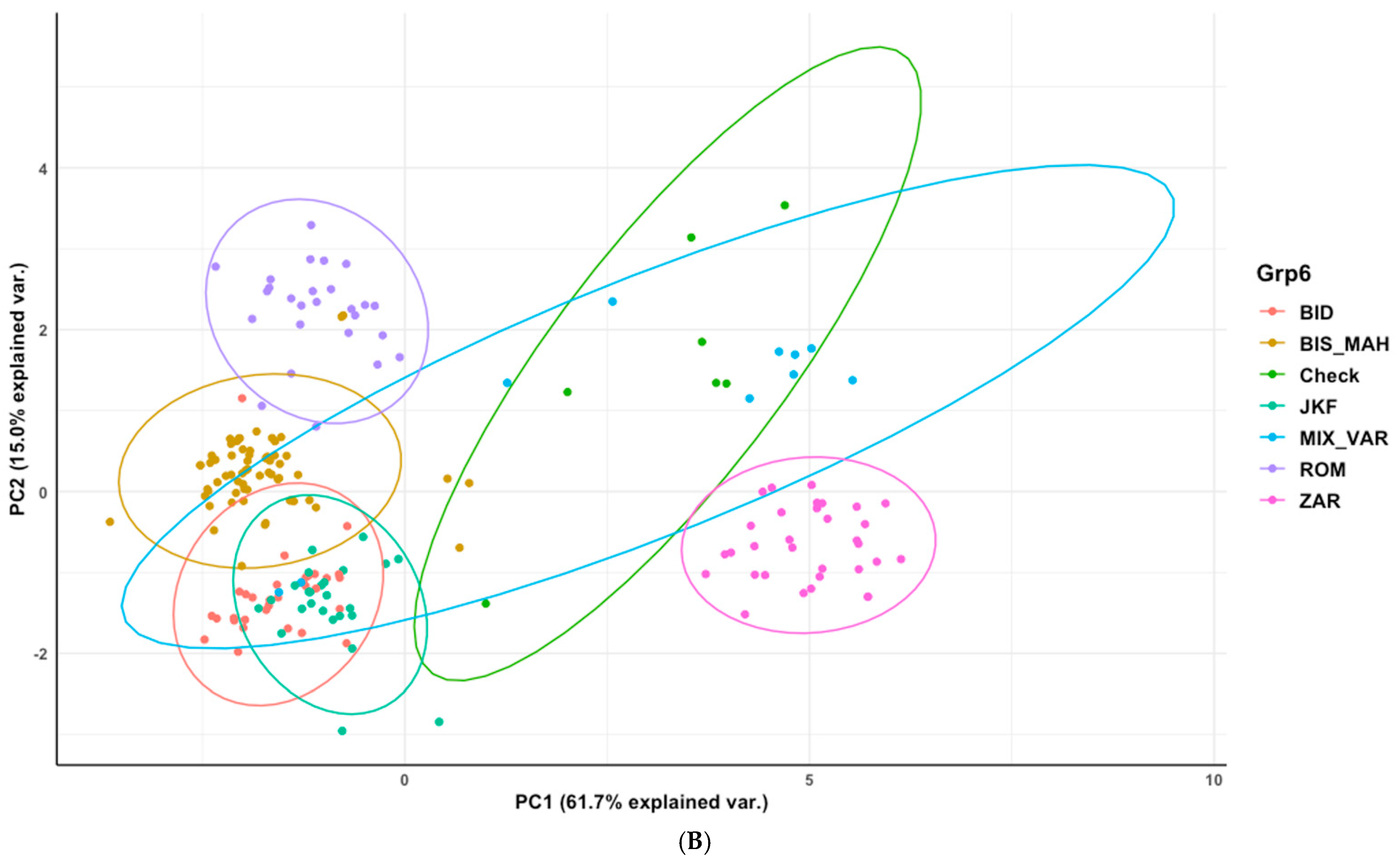
3.4. Phenotypic Structure and Relationships among the Tunisian Germplasm
4. Discussion
4.1. Strong Genetic Effect of the Agro-Morphological and Phenological Traits in the Tunisian Durum Wheat Collection
4.2. Phenotypic Variability Inferred from Genetic Patterns and Importance of the Population Jenah Zarzoura
4.3. Phenotypic Structure and Relationships between the Tunisian Durum Wheat Landraces
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feldman, M.; Levy, A. Allopolyploidy—A shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 2005, 109, 250–258. [Google Scholar] [CrossRef]
- Zohary, D.; Hopf, M. Domestication of Plants in the Old World: The Origin and Spread of Cultivated Plants in West Asia, Europe, and the Nile Valley, 3rd ed.; Oxford University Press: Oxford, UK, 2000; ISBN 9780198503576. [Google Scholar]
- Tanno, K.-I. How Fast Was Wild Wheat Domesticated? Science 2006, 311, 1886. [Google Scholar] [CrossRef] [Green Version]
- Dvorak, J.; Akhunov, E.D. Tempos of Gene Locus Deletions and Duplications and Their Relationship to Recombination Rate During Diploid and Polyploid Evolution in the Aegilops-Triticum Alliance. Genetics 2005, 171, 323–332. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.H.; Sun, D.; Nevo, E. Domestication evolution, genetics and genomics in wheat. Mol. Breed. 2011, 28, 281–301. [Google Scholar] [CrossRef]
- Avni, R.; Nave, M.; Barad, O.; Baruch, K.; Twardziok, S.O.; Gundlach, H.; Hale, I.; Mascher, M.; Spannagl, M.; Wiebe, K.; et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 2017, 357, 93–97. [Google Scholar] [CrossRef] [Green Version]
- Hughes, N.; Oliveira, H.R.; Fradgley, N.; Corke, F.M.K.; Cockram, J.; Doonan, J.H.; Nibau, C. µ CT trait analysis reveals morphometric differences between domesticated temperate small grain cereals and their wild relatives. Plant J. 2019, 99, 98–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonjean, A.P.; Angus, W.J.; Van Ginkel, M. The World Wheat Book: A History of Wheat Breeding; Lavoisier: Paris, France, 2016; Volume 3, ISBN 9782743020910. [Google Scholar]
- Feldman, M. Origin of Cultivated Wheat. In The World Wheat Book: A History of Wheat Breeding, 1st ed.; Bonjean, A.P., Angus, W.J., Van Ginkel, M., Eds.; Lavoisier Publishing: Paris, France, 2001; Volume 1, pp. 1–56. [Google Scholar]
- Lopes, M.S.; El-Basyoni, I.; Baenziger, P.S.; Singh, S.; Royo, C.; Ozbek, K.; Aktas, H.; Ozer, E.; Ozdemir, F.; Manickavelu, A.; et al. Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J. Exp. Bot. 2015, 66, 3477–3486. [Google Scholar] [CrossRef]
- Mengistu, D.K.; Kidane, Y.G.; Fadda, C.; Pè, M.E. Genetic diversity in Ethiopian Durum Wheat (Triticum turgidum var. durum) inferred from phenotypic variations. Plant. Genet. Resour. 2016, 16, 39–49. [Google Scholar] [CrossRef] [Green Version]
- Soriano, J.M.; Villegas, L.; Aranzana, M.J.; Del Moral, L.F.G.; Royo, C. Genetic Structure of Modern Durum Wheat Cultivars and Mediterranean Landraces Matches with Their Agronomic Performance. PLoS ONE 2016, 11, e0160983. [Google Scholar] [CrossRef] [Green Version]
- Mangini, G.; Margiotta, B.; Marcotuli, I.; Signorile, M.A.; Gadaleta, A.; Blanco, A. Genetic diversity and phenetic analysis in wheat (Triticum turgidum subsp. durum and Triticum aestivum subsp. aestivum) landraces based on SNP markers. Genet. Resour. Crop. Evol. 2016, 64, 1269–1280. [Google Scholar] [CrossRef]
- Moragues, M.; Zarco-Hernández, J.; Moralejo, M.A.; Royo, C. Genetic Diversity of Glutenin Protein Subunits Composition in Durum Wheat Landraces [Triticum turgidum ssp. turgidum Convar. durum (Desf.) MacKey] from the Mediterranean Basin. Genet. Resour. Crop. Evol. 2005, 53, 993–1002. [Google Scholar] [CrossRef]
- Nazco, R.; Villegas, D.; Ammar, K.; Peña, R.J.; Moragues, M.; Royo, C. Can Mediterranean durum wheat landraces contribute to improved grain quality attributes in modern cultivars? Euphytica 2011, 185, 1–17. [Google Scholar] [CrossRef]
- Roselló, M.; Royo, C.; Sanchez-Garcia, M.; Soriano, J.M. Genetic Dissection of the Seminal Root System Architecture in Mediterranean Durum Wheat Landraces by Genome-Wide Association Study. Agronomy 2019, 9, 364. [Google Scholar] [CrossRef] [Green Version]
- Oumata, S.; David, J.; Mekliche-Hanifi, L.; Kharsi, M.; Zaharieva, M.; Monneveux, P. Oasis wheats of the South of Algeria: Landraces, cultural practices and utilization. Genet. Resour. Crop. Evol. 2020, 67, 325–337. [Google Scholar] [CrossRef]
- Ouaja, M.; Aouini, L.; Bahri, B.; Ferjaoui, S.; Medini, M.; Marcel, T.C.; Hamza, S. Identification of valuable sources of resistance to Zymoseptoria tritici in the Tunisian durum wheat landraces. Eur. J. Plant. Pathol. 2020, 156, 647–661. [Google Scholar] [CrossRef]
- Yacoubi, I.; Nigro, D.; Sayar, R.; Masmoudi, K.; Seo, Y.W.; Brini, F.; Giove, S.L.; Mangini, G.; Giancaspro, A.; Marcotuli, I.; et al. New insight into the North-African durum wheat biodiversity: Phenotypic variations for adaptive and agronomic traits. Genet. Resour. Crop. Evol. 2019, 67, 445–455. [Google Scholar] [CrossRef]
- Daaloul, A.; Harrabi, M.; Amara, H. Evaluation de la Collection Nationale de Blé Dur. In Revure de l’INAT; numéro spécial; INAT: Tunis, Tunisia, 1998; pp. 337–358. (In French) [Google Scholar]
- Jaradat, A.A.; Shahid, M. How diverse a farmer-managed wheat landrace can be? Emir. J. Food Agric. 2014, 26, 93–118. [Google Scholar] [CrossRef] [Green Version]
- Pingali, P. The Green Revolution and Crop Diversity (Chapter 12). In Handbook of Agricultural Biodiversity; Hunter, D., Guarino, L., Spillane, C., McKeown, P., Eds.; Routledge: New York, NY, USA, 2017; pp. 213–223. [Google Scholar]
- Boeuf, F. Le blé en Tunisie: La Plante. Le Milieu Physico-Chimique; Société Anonyme de L’Imprimerie Rapide de Tunis: Tunis, Tunisia, 1932; Volume 1. (In French) [Google Scholar]
- Deghaïs, M.; Kouki, M.M.; Gharbi, M.M.; El Felah, M. Les Variétés de Céréales Cultivées en Tunisie (blé dur, blé Tendre, orge et Triticale; INRAT: Tunis, Tunisia, 2007. (In French) [Google Scholar]
- Vavilov, N.I. The Origin, Variation, Immunity and Breeding of Cultivated Plants. Soil Sci. 1951, 72, 482. [Google Scholar] [CrossRef]
- Sourour, A.; Chahine, K.; Youssef, T.; Olfa, S.; Hajer, S. Phenotypic diversity of Tunisian durum wheat landraces. Afr. Crop. Sci. J. 2010, 18, 18. [Google Scholar] [CrossRef] [Green Version]
- Belhadj, H.; Medini, M.; Bouhaouel, I.; Amara, H. Analyse de la diversitéphénotypique de quelques accessions autochtones de blé dur (Triticum turgidum ssp. durum Desf.) du sud tunisien. J. New Sci. Agric. Biotechnol. 2015, 24, 1115–1125. [Google Scholar]
- Baloch, F.S.; Alsaleh, A.; Shahid, M.Q.; Çiftçi, V.; De Miera, L.E.S.; Aasim, M.; Nadeem, M.A.; Aktaş, H.; Özkan, H.; Hatipoğlu, R. A Whole Genome DArTseq and SNP Analysis for Genetic Diversity Assessment in Durum Wheat from Central Fertile Crescent. PLoS ONE 2017, 12, e0167821. [Google Scholar] [CrossRef] [Green Version]
- Soriano, J.M.; Villegas, D.; Sorrells, M.E.; Royo, C. Durum Wheat Landraces from East and West Regions of the Mediterranean Basin Are Genetically Distinct for Yield Components and Phenology. Front. Plant. Sci. 2018, 9, 80. [Google Scholar] [CrossRef] [Green Version]
- Kabbaj, H.; Sall, A.T.; Al-Abdallat, A.; Geleta, M.; Amri, A.; Filali-Maltouf, A.; Belkadi, B.; Ortiz, R.; Bassi, F.M. Genetic Diversity within a Global Panel of Durum Wheat (Triticum durum) Landraces and Modern Germplasm Reveals the History of Alleles Exchange. Front. Plant. Sci. 2017, 8, 1277. [Google Scholar] [CrossRef] [Green Version]
- Robbana, C.; Kehel, Z.; Ben Naceur, M.; Sansaloni, C.; Bassi, F.; Amri, A. Genome-Wide Genetic Diversity and Population Structure of Tunisian Durum Wheat Landraces Based on DArTseq Technology. Int. J. Mol. Sci. 2019, 20, 1352. [Google Scholar] [CrossRef] [Green Version]
- Slim, A.; Piarulli, L.; Kourda, H.C.; Rouaissi, M.; Robbana, C.; Chaabane, R.; Pignone, D.; Montemurro, C.; Mangini, G. Genetic Structure Analysis of a Collection of Tunisian Durum Wheat Germplasm. Int. J. Mol. Sci. 2019, 20, 3362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kehel, Z.; Garcia-Ferrer, A.; Nachit, M.M. Using Bayesian and Eigen approaches to study spatial genetic structure of Moroccan and Syrian durum wheat landraces. Am. J. Mol. Biol. 2013, 3, 17–31. [Google Scholar] [CrossRef] [Green Version]
- Falconer, D.S. Introduction to Quantitative Genetics; Longmans, Green & Co publishers: Harlow, UK, 1989; pp. 65–71. [Google Scholar]
- Butler, D.G.; Cullis, B.R.; Gilmour, A.R.; Gogel, B.G.; Thompson, R. ASReml-R Reference Manual Version 4; VSN International Ltd.: Hemel Hempstead, UK, 2017; Available online: https://asreml.kb.vsni.co.uk/wp-content/uploads/sites/3/2018/02/ASReml-R-Reference-Manual-4.pdf (accessed on 2 February 2018).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org (accessed on 3 November 2018).
- Stacklies, W.; Redestig, H.; Scholz, M.; Walther, D.; Selbig, J. pcaMethods a bioconductor package providing PCA methods for incomplete data. Bioinformatics 2007, 23, 1164–1167. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Wagner, H. Vegan: Community Ecology Package. 2019. Available online: https://CRAN.R-project.org/package=vegan (accessed on 28 November 2020).
- Khan, M.K.; Pandey, A.; Choudhary, S.; Hakki, E.E.; Akkaya, M.S.; Thomas, G. From RFLP to DArT: Molecular tools for wheat (Triticum spp.) diversity analysis. Genet. Resour. Crop. Evol. 2014, 61, 1001–1032. [Google Scholar] [CrossRef]
- Taranto, F.; D’Agostino, N.; Rodriguez, M.; Pavan, S.; Minervini, A.P.; Pecchioni, N.; Papa, R.; De Vita, P. Whole Genome Scan Reveals Molecular Signatures of Divergence and Selection Related to Important Traits in Durum Wheat Germplasm. Front. Genet. 2020, 11, 217. [Google Scholar] [CrossRef] [Green Version]
- Royo, C.; Maccaferri, M.; Álvaro, F.; Moragues, M.; Sanguineti, M.C.; Tuberosa, R.; Maalouf, F.; Del Moral, L.F.G.; Demontis, A.; Rhouma, S.; et al. Understanding the relationships between genetic and phenotypic structures of a collection of elite durum wheat accessions. Field Crop. Res. 2010, 119, 91–105. [Google Scholar] [CrossRef]
- Allard, R.W. Principles of Plant Breeding. Soil Sci. 1961, 91, 414. [Google Scholar] [CrossRef]
- Johnson, H.W.; Robinson, H.F.; Comstock, R.E. Estimates of Genetic and Environmental Variability in Soybeans1. Agron. J. 1955, 47, 314–318. [Google Scholar] [CrossRef]
- Liu, Y.; Lin, Y.; Gao, S.; Li, Z.; Ma, J.; Deng, M.; Chen, G.; Wei, Y.; Zheng, Y. A genome-wide association study of 23 agronomic traits in Chinese wheat landraces. Plant. J. 2017, 91, 861–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, B.D. Plant Breeding: Principles and Methods; Kalyani Publishers: New Delhi, India, 2001. [Google Scholar]
- Sahri, A.; Chentoufi, L.; Arbaoui, M.; Ardisson, M.; Belqadi, L.; Birouk, A.; Roumet, P.; Muller, M.-H. Towards a comprehensive characterization of durum wheat landraces in Moroccan traditional agrosystems: Analysing genetic diversity in the light of geography, farmers’ taxonomy and tetraploid wheat domestication history. BMC Evol. Biol. 2014, 14, 264. [Google Scholar] [CrossRef] [Green Version]
- Zaharieva, M.; Bonjean, A.; Monneveux, P. Saharan wheats: Before they disappear. Genet. Resour. Crop. Evol. 2014, 61, 1065–1084. [Google Scholar] [CrossRef]
- Annicchiarico, P.; Pecetti, L.; Damania, A.B. Relationships between Phenotypic Variation and Climatic Factors at Collecting Sites in Durum Wheat Landraces. Hereditas 2004, 122, 163–167. [Google Scholar] [CrossRef]
- Fiore, M.C.; Mercati, F.; Spina, A.; Blangiforti, S.; Venora, G.; Dell’Acqua, M.; Lupini, A.; Preiti, G.; Monti, M.; Pè, M.E.; et al. High-Throughput Genotype, Morphology, and Quality Traits Evaluation for the Assessment of Genetic Diversity of Wheat Landraces from Sicily. Plants 2019, 8, 116. [Google Scholar] [CrossRef] [Green Version]
- Jaradat, A.A. Wheat Landraces: A mini review. Emir. J. Food Agric. 2013, 25, 20–29. [Google Scholar] [CrossRef]
| Landraces | Lines | Zones | Collecting Site | Province |
|---|---|---|---|---|
| Bidi | 30 | Centeral | Kairouan | Kairouan |
| Jenah Khotifa1 | 28 | Centeral | Kairouan | Kairouan |
| Jenah Khotifa2 | 4 | Southern | El Frid | Tozeur |
| Biskri1 | 30 | Southern | Djebel ouled ouhiba | Gafsa |
| Biskri2 * | 7 | Southern | Zarzis | Medenine |
| Mahmoudi | 30 | Southern | Snad | Gafsa |
| Rommani | 30 | Southern | Djebel ouled ouhiba | Gafsa |
| Jenah Zarzoura | 30 | Oases | Oasis of Mareth | Gabes |
| Total | 189 |
| Elite Line Name | Provider | Accession Identifier | Pedigree |
|---|---|---|---|
| - | ICARDA | MCHCB-102 | OmRabi3/T.urartu500651/ch5//980947/3/Otb4//Ossl1/Rf m6 |
| IcaJoudy | ICARDA | MCHCB-100 | Atlast1/961081//Icasyr1 |
| Nachit | ICARDA | DAWRYT-106 | Ameddkul1/T. dicoccoides Syrian collection//Loukos |
| Zeina4 | ICARDA | MCHCB-154 | GdoVZ512/Cit//Ruff/Fg/3/Src3 |
| Louiza | ICARDA | – | Rscn39/Til1 |
| Ammar 6 | ICARDA | ICARDA | IDYT37-5 ICAMORTA0472/Ammar7 |
| Ammar 10 | ICARDA | MCHCB-99 | Lgt3/Bicrecham1 |
| Site | KAIR IR | KAIR IR | KAIR-STR | KAIR STR | MOR RF | MOR RF | Across All Environments |
|---|---|---|---|---|---|---|---|
| Year | 2016 | 2017 | 2016 | 2017 | 2015 | 2016 | |
| H2 | H2 | H2 | H2 | H2 | H2 | H’2 | |
| HD | 0.97 | 0.98 | 0.94 | 0.98 | 0.95 | 0.94 | 0.98 |
| MD | 0.90 | 0.80 | 0.93 | 0.87 | 0.96 | 0.89 | 0.95 |
| H | 0.96 | 0.95 | 0.86 | 0.96 | 0.95 | 0.95 | 0.84 |
| FLA | 0.77 | 0.36 | 0.62 | 0.74 | 0.62 | 0.87 | 0.60 |
| MLB | 0.68 | 0.82 | 0.79 | 0.83 | 0.78 | 0.66 | 0.89 |
| MLE | 0.82 | 0.80 | 0.89 | 0.85 | 0.82 | 0.8 | 0.93 |
| SL | 0.91 | 0.95 | 0.95 | 0.98 | 0.80 | 0.88 | 0.96 |
| ST | 0.87 | 0.87 | 0.92 | 0.95 | 0.83 | 0.65 | 0.95 |
| SW | 0.91 | 0.93 | 0.94 | 0.97 | 0.88 | 0.71 | 0.97 |
| TA | 0.84 | 0.88 | - | 0.89 | 0.85 | - | 0.67 |
| NEE | 0.69 | 0.71 | 0.67 | 0.65 | 0.61 | 0.61 | 0.85 |
| TKW | 0.86 | 0.90 | 0.93 | 0.95 | 0.87 | 0.64 | 0.95 |
| Df | SumOfSqs | F | Pr (>F) | Percent Variance | |
|---|---|---|---|---|---|
| Farmer’s population name | 6 | 0.191 | 57.413 | 0.001 | 64.81 |
| Residual | 187 | 0.104 | 35.18 | ||
| Genetic group with K = 5 | 5 | 0.192 | 70.041 | 0.001 | 65.06 |
| Residual | 188 | 0.103 | 34.93 | ||
| Genetic group with K = 6 | 6 | 0.225 | 100.33 | 0.001 | 76.29 |
| Residual | 187 | 0.070 | 23.70 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robbana, C.; Kehel, Z.; Ammar, K.; Guzmán, C.; Naceur, M.B.; Amri, A. Unlocking the Patterns of the Tunisian Durum Wheat Landraces Genetic Structure Based on Phenotypic Characterization in Relation to Farmer’s Vernacular Name. Agronomy 2021, 11, 634. https://doi.org/10.3390/agronomy11040634
Robbana C, Kehel Z, Ammar K, Guzmán C, Naceur MB, Amri A. Unlocking the Patterns of the Tunisian Durum Wheat Landraces Genetic Structure Based on Phenotypic Characterization in Relation to Farmer’s Vernacular Name. Agronomy. 2021; 11(4):634. https://doi.org/10.3390/agronomy11040634
Chicago/Turabian StyleRobbana, Cyrine, Zakaria Kehel, Karim Ammar, Carlos Guzmán, M’Barek Ben Naceur, and Ahmed Amri. 2021. "Unlocking the Patterns of the Tunisian Durum Wheat Landraces Genetic Structure Based on Phenotypic Characterization in Relation to Farmer’s Vernacular Name" Agronomy 11, no. 4: 634. https://doi.org/10.3390/agronomy11040634







