Widely Targeted Metabolomics Analysis of Different Parts of Salsola collina Pall
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Sample Preparation and Extraction
2.3. UPLC Conditions
2.4. ESI-Q TRAP-MS/MS
2.5. Qualitative and Semi-Quantitative Analysis of Metabolites
2.6. Principal Component Analysis
2.7. Hierarchical Cluster Analysis and Pearson Correlation Coefficients
2.8. Differential Metabolites Selected
2.9. KEGG Annotation and Enrichment Analysis
3. Results
3.1. Metabolic Profiling of S. collina Based on LC-MS/MS
3.2. Multivariate Statistical Analysis
3.3. Differential Metabolites Between L-Sc and R-Sc
3.4. Putative Antioxidant Components Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Kong, X. Chenopodiaceae. In Flora Repubulicae Popularis Sinicae; Editorial Committee of Chinese Flora, Ed.; Science Press: Beijing, China, 1979; Volume 25, p. 176. [Google Scholar]
- Compilation Group of National Compilation of Chinese Herbal Medicine. National Compilation of Chinese Herbal Medicine; People′s Medical Publishing House: Beijing, China, 1975; pp. 796–797. [Google Scholar]
- Meng, X.P.; Liu, J.X. Antihypertensive effects of alcoholic extracts from Salsola. Mod. Food Sci. Technol. 2007, 23, 17–19. [Google Scholar]
- Liu, D.F.; Dong, Z.M.; Zhang, Z.X.; Zhu, X.W.; Wang, H.Y.; Wang, N. Development of fermented milk drink containing Salsola collina juice. Beverage Ind. 2009, 12, 24–28. [Google Scholar]
- Lee, H.J.; Pan, C.H.; Kim, E.S.; Kim, C.Y. Online high performance liquid chromatography (HPLC)-ABTS+ based assay and HPLC-electrospray ionization mass spectrometry analysis of antioxidant phenolic compounds in Salsola komarovii. J. Korean Soc. Appl. Biol. Chem. 2012, 55, 317–321. [Google Scholar] [CrossRef]
- Tundis, R.; Loizzo, M.R.; Statti, G.A.; Menichini, F. Inhibitory effects on the digestive enzyme α-amylase of three Salsola species (Chenopodiaceae) in vitro. Pharmazie 2007, 62, 473–475. [Google Scholar]
- Nanjing University of Traditional Chinese Medicine. The Dictionary of Chinese Herbal Medicine, 1st ed.; Shanghai Scientific & Technical Publishers: Shanghai, China, 2006; pp. 3070–3071. [Google Scholar]
- Wang, S.; Yan, M.; Guo, Y.; Sun, R.; Jin, H.; Gong, Y. In vivo and in vitro effects of Salsola collina on gastrointestinal motility in rats. Iran J. Basic Med. Sci. 2020, 23, 383. [Google Scholar] [PubMed]
- Zhao, X.; Wang, H.; Zhang, Z.; Jin, H.; Gong, Y. Effects of ethyl acetate extract of Salsola collina on brain-gut peptides and interstitial cells of gastric Cajal in rats with diabetic gastroparesis. Iran J. Basic Med. Sci. 2020, 23, 1218–1224. [Google Scholar]
- Zhao, X.L.; Yuan, W.; Li, Z.Z.; Jin, H.; Gong, Y.L. Salsola collina ethyl acetate extract alleviates diabetic gastroparesis possibly through oxidative stress inhibition. In IOP Conference Series: Earth and Environmental Science; IOP Publishing Ltd.: Bristol, UK, 2020; Volume 559, p. 012021. [Google Scholar]
- Oh, Y.N.; Jin, S.; Park, H.J.; Kwon, H.J.; Kim, B.W. Anti-oxidative and anti-cancer activities by cell cycle regulation of Salsola collina extract. Microbiol. Biotechnol. Lett. 2014, 42, 73–81. [Google Scholar] [CrossRef]
- Nikiforov, S.B.; Semenov, A.A.; Syrchina, A.I. Effect of an aqueous extract of Salsola collina on the course of experimental cholelithiasis in rabbits. Pharm. Chem. J. 2002, 36, 496–499. [Google Scholar] [CrossRef]
- Mayakova, T.I.; Leont’Eva, V.G.; Zharkaya, T.I.; Semenov, A.A.; Kuznetsova, E.E.; Chupin, S.P. Sterols of salsola collina. Chem. Nat. Compd. 1984, 20, 507. [Google Scholar] [CrossRef]
- Syrchina, A.I.; Vereshchagin, A.L.; Larin, M.F.; Semenov, A.A. Flavonoids of salsola collina. Chem. Nat. Compounds 1989, 25, 619–620. [Google Scholar] [CrossRef]
- Syrchina, A.I.; Chernykh, E.A.; Rafeichikova, I.V.; Zaikov, K.L.; Vereshchagin, A.L. Carbohydrates, carbohydrate ethers, and alcohols of Salsola collina. Chemistry of Natural Compounds, 1991; 27, 364. [Google Scholar]
- Zhao, Y.X.; Ding, X.B. Studies on the alkaloids from Salsola collina Pall. Acta Pharm. Sin. 2004, 39, 598–600. [Google Scholar] [PubMed]
- Xiang, Y.; Li, Y.B.; Zhang, J.; Li, P.; Yao, Y.Z. Studies on chemical constituents of Salsola collina. China J. Chin. Mater. Med. 2007, 32, 409–413. [Google Scholar] [PubMed]
- Jin, Y.S.; Du, J.L.; Yang, Y.; Jin, L.; Song, Y.; Zhang, W.; Chen, H.S. Chemical and biologically active constituents of Salsola collina. Chem. Nat. Compd. 2011, 47, 257–260. [Google Scholar] [CrossRef]
- Wang, X.J.; Zhao, Y.X.; Jia, X.H.; Ding, X.B. Studies on the chemical constituents of Salsola collina. J. Chin. Med. Mater. 2011, 34, 230–231. [Google Scholar] [PubMed]
- Ghorab, H.; Khettaf, A.; Lehbili, M.; Kabouche, A.; Magid, A.A.; Harakat, D.; Voutquenne-Nazabadioko, L.; Kabouche, Z. A new cardenolide and other compounds from Salsola tetragona. Nat. Prod. Commun. 2017, 12, 1934578X1701200102. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, N.; Wang, Z.; Qi, Z.; Zhu, H.; Zheng, B.; Li, P.; Liu, J. Nontargeted metabolomic analysis of four different parts of Platycodon grandiflorum grown in northeast China. Molecules 2017, 22, 1280. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Wu, M.; Chen, Y.; Zhang, Y.; Zhao, X.; Zheng, X. Revealing Metabolomic Variations in Cortex Moutan from Different Root Parts using HPLC–MS method. Phytochem. Anal. 2015, 26, 86–93. [Google Scholar] [CrossRef]
- Dey, P.; Chaudhuri, T.K. Metabolomic Fingerprinting of the Volatiles in Different Parts of Streptocaulon sylvestre. J. Herbs Spices Med. Plants 2017, 23, 308–319. [Google Scholar]
- Wei, G.; Dong, L.; Yang, J.; Zhang, L.; Xu, J.; Yang, F.; Cheng, R.; Xu, R.; Chen, S. Integrated metabolomic and transcriptomic analyses revealed the distribution of saponins in Panax notoginseng. Acta Pharm. Sin. B 2018, 8, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lv, Q.; Ma, C.; Qu, J.; Cai, F.; Deng, J.; Huang, J.; Ran, P.; Shi, T.; Chen, Q. Metabolite profiling and transcriptome analyses provide insights into the flavonoid biosynthesis in the developing seed of tartary buckwheat (Fagopyrum tataricum). J. Agric. Food Chem. 2019, 67, 11262–11276. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Wang, B.; He, G.; Wang, Y.; Tang, X.; Wang, S.; Ma, Y.; Fu, C.; Chai, G.; Zhou, G. Metabolomics integrated with transcriptomics reveals redirection of the phenylpropanoids metabolic flux in Ginkgo biloba. J. Agric. Food Chem. 2019, 67, 3284–3291. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chen, L.; Chen, H.; Chen, S.; Liu, Y. Analysis of flavonoid metabolites in citrus peels (Citrus reticulata “Dahongpao”) using UPLC-ESI-MS/MS. Molecules 2019, 24, 2680. [Google Scholar] [CrossRef]
- Wang, Y.; Liang, X.; Li, Y.; Fan, Y.; Li, Y.; Cao, Y.; An, W.; Shi, Z.; Zhao, J.; Guo, S. Changes in metabolome and nutritional quality of Lycium barbarum fruits from three typical growing areas of China as revealed by widely targeted metabolomics. Metabolites 2020, 10, 46. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [PubMed]
- Woldu, Y.; Abegaz, B. Isoflavonoids from Salsola somalensis. Phytochemistry 1990, 29, 2013–2015. [Google Scholar] [CrossRef]
- Xu, S.; Ge, X.; Li, S.; Guo, X.; Dai, D.; Yang, T. Discrimination of Different Parts of Saffron by Metabolomic-Based Ultra-Performance Liquid Chromatography Coupled with High-Definition Mass Spectrometry. Chem. Biodivers. 2019, 16, e1900363. [Google Scholar]
- Sun, X.; Li, L.; Pei, J.; Liu, C.; Huang, L.F. Metabolome and transcriptome profiling reveals quality variation and underlying regulation of three ecotypes for Cistanche deserticola. Plant Mol. Biol. 2020, 102, 253–269. [Google Scholar] [CrossRef] [PubMed]

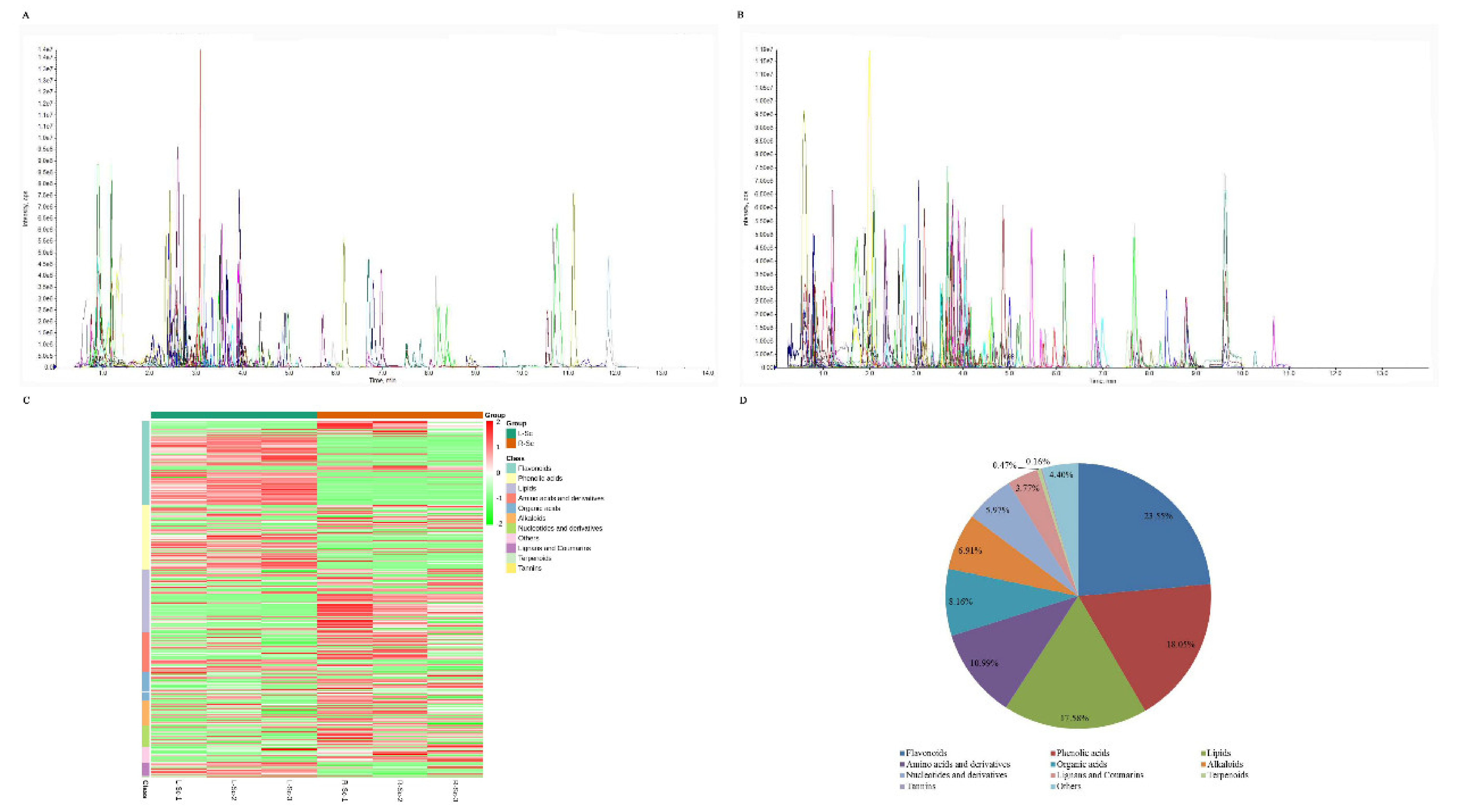
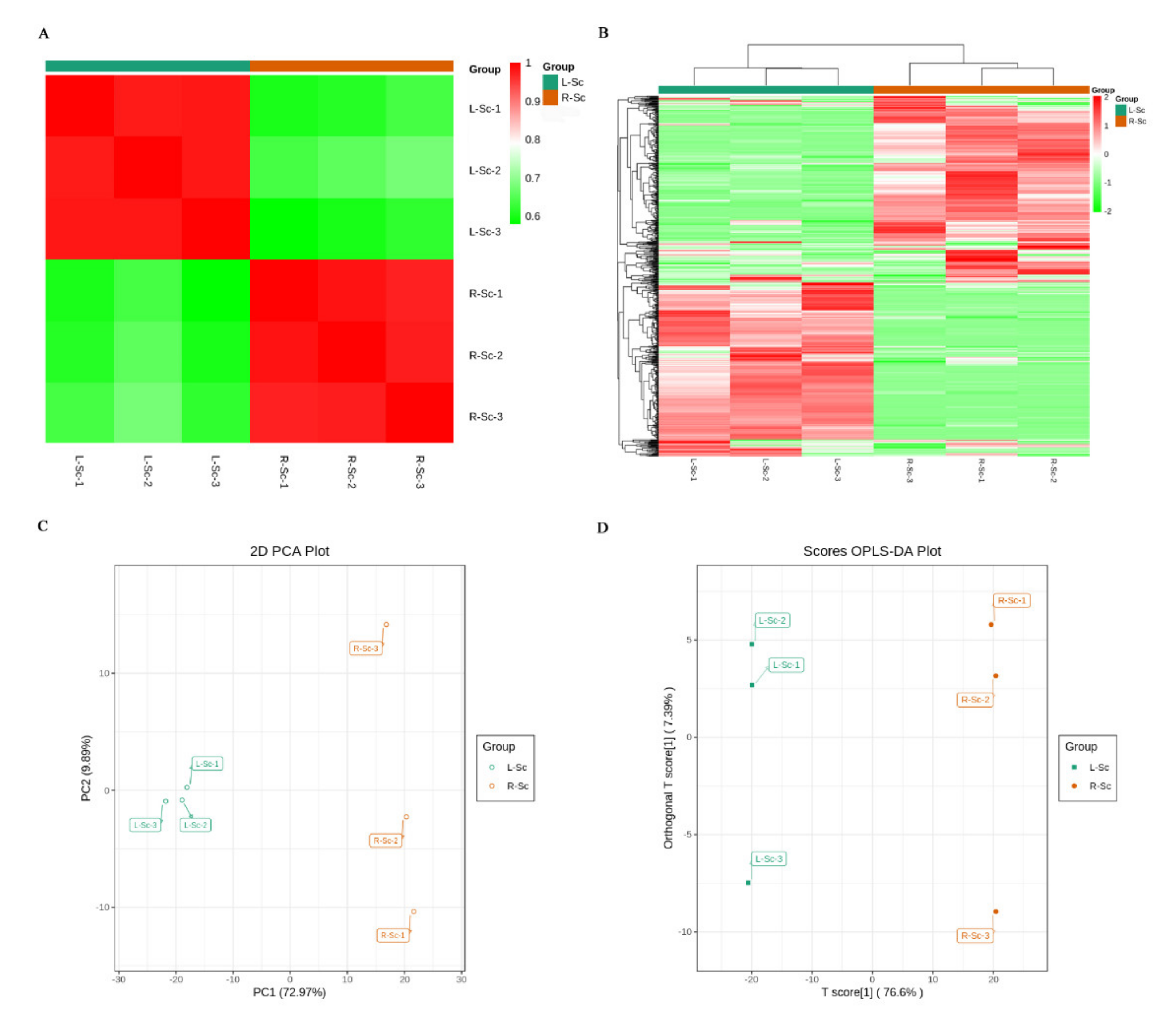
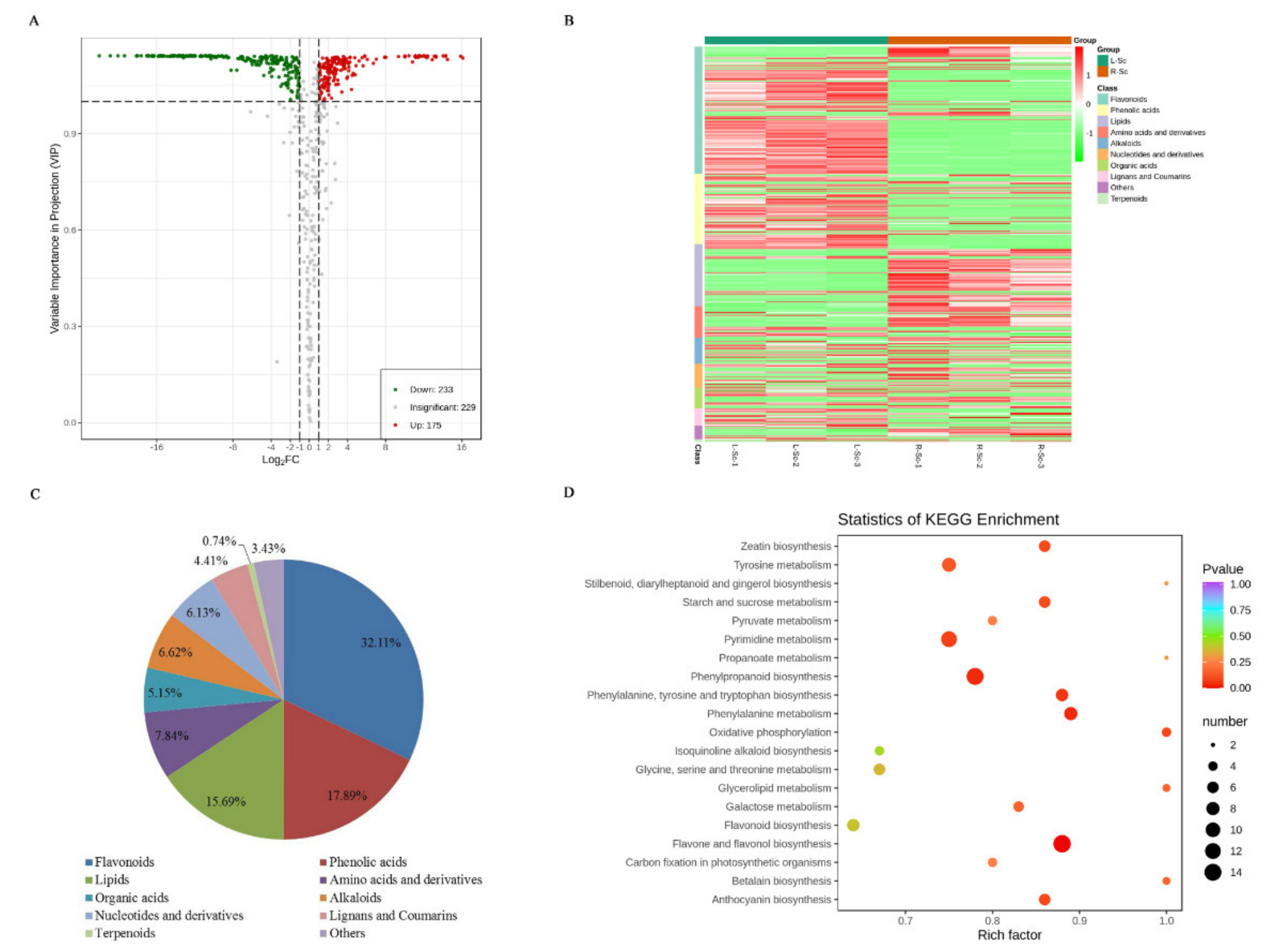
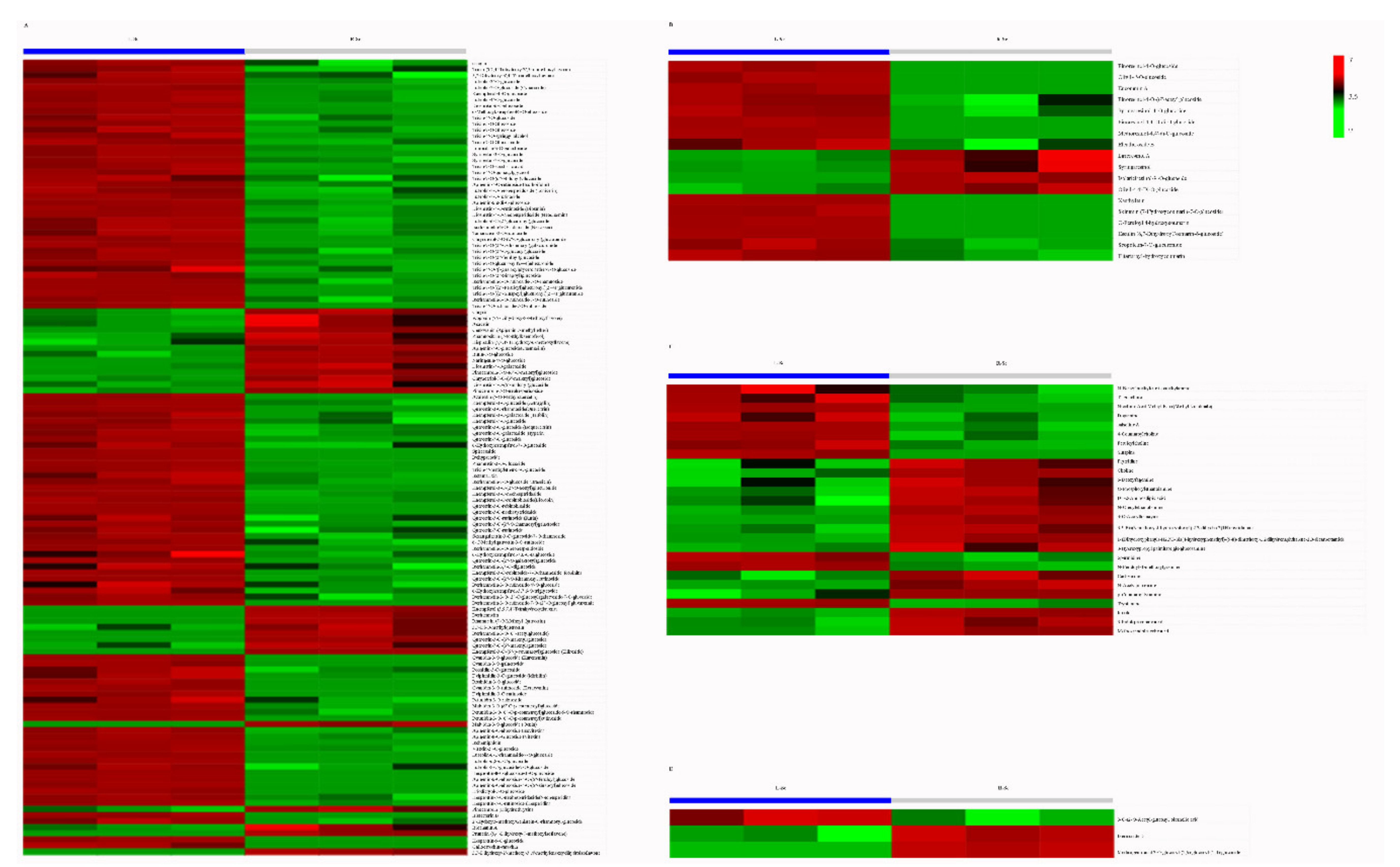
| Primary Classification | Secondary Classification | Number of Metabolites |
|---|---|---|
| Flavonoids | Flavones | 63 |
| Flavonols | 48 | |
| Anthocyanins | 13 | |
| C-flavonoids | 11 | |
| Dihydroflavones | 7 | |
| Isoflavones | 4 | |
| Dihydroflavonols | 2 | |
| Dihydroisoflavone | 1 | |
| Flavanol | 1 | |
| Phenolic acids | Phenolic acids | 115 |
| Lipids | Free fatty acids | 39 |
| Lysophosphatidycholines | 30 | |
| Lysophosphatidyl ethanolamines | 26 | |
| Glycerol esters | 17 | |
| Amino acids and derivatives | Amino acids and derivatives | 70 |
| Organic acids | Organic acids | 52 |
| Alkaloids | Alkaloids | 26 |
| Phenolamines | 11 | |
| Plumeranes | 7 | |
| Nucleotides and derivatives | Nucleotides and derivatives | 38 |
| Lignans and Coumarins | Lignans | 17 |
| Coumarins | 7 | |
| Terpenoids | Triterpene | 1 |
| Triterpene Saponin | 1 | |
| Sesquiterpenoid | 1 | |
| Tannins | Tannin | 1 |
| Others | Saccharides and Alcohols | 17 |
| Vitamins | 4 | |
| Others | 7 |
| Primary Classification | Secondary Classification | L-Sc vs. R-Sc | |
|---|---|---|---|
| Down | Up | ||
| Flavonoids | Flavones | 41 | 14 |
| Flavonols | 35 | 8 | |
| Anthocyanins | 11 | 1 | |
| C-flavonoids | 10 | 0 | |
| Dihydroflavones | 3 | 1 | |
| Isoflavones | 2 | 2 | |
| Dihydroflavonol | 1 | 0 | |
| Flavanol | 1 | 0 | |
| Dihydroisoflavone | 0 | 1 | |
| Lignans and Coumarins | Lignans | 8 | 4 |
| Coumarins | 6 | 0 | |
| Alkaloids | Alkaloids | 8 | 10 |
| Phenolamines | 2 | 3 | |
| Plumeranes | 1 | 3 | |
| Terpenoids | Triterpene | 1 | 0 |
| Triterpene Saponin | 0 | 1 | |
| Sesquiterpenoid | 0 | 1 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Chen, Y.; Duan, Y.; Zhao, Y.; Zhang, D.; Zang, L.; Ya, H. Widely Targeted Metabolomics Analysis of Different Parts of Salsola collina Pall. Molecules 2021, 26, 1126. https://doi.org/10.3390/molecules26041126
Li S, Chen Y, Duan Y, Zhao Y, Zhang D, Zang L, Ya H. Widely Targeted Metabolomics Analysis of Different Parts of Salsola collina Pall. Molecules. 2021; 26(4):1126. https://doi.org/10.3390/molecules26041126
Chicago/Turabian StyleLi, Shipeng, Ye Chen, Ying Duan, Yinhui Zhao, Di Zhang, Liyan Zang, and Huiyuan Ya. 2021. "Widely Targeted Metabolomics Analysis of Different Parts of Salsola collina Pall" Molecules 26, no. 4: 1126. https://doi.org/10.3390/molecules26041126
APA StyleLi, S., Chen, Y., Duan, Y., Zhao, Y., Zhang, D., Zang, L., & Ya, H. (2021). Widely Targeted Metabolomics Analysis of Different Parts of Salsola collina Pall. Molecules, 26(4), 1126. https://doi.org/10.3390/molecules26041126






