Aggregate Sampling to Detect Pathogens and Antimicrobial Resistance Genes Associated with Bovine Respiratory Disease in US Feedlots: A Pilot Study
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Sample Collection and Processing
2.3. Statistical Analysis
3. Results
4. Discussion
Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smith, R.A.; Step, D.L.; Woolums, A.R. Bovine Respiratory Disease: Looking Back and Looking Forward, What Do We See? Vet. Clin. N. Am. Food Anim. Pract. 2020, 36, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Chai, J.; Capik, S.F.; Kegley, B.; Richeson, J.T.; Powell, J.G.; Zhao, J. Bovine respiratory microbiota of feedlot cattle and its association with disease. Vet. Res. 2022, 53, 4. [Google Scholar] [CrossRef]
- Grissett, G.P.; White, B.J.; Larson, R.L. Structured literature review of responses of cattle to viral and bacterial pathogens causing bovine respiratory disease complex. J. Vet. Intern. Med. 2015, 29, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J. What is the evidence that bovine coronavirus is a biologically significant respiratory pathogen in cattle? Can. Vet. J. 2019, 60, 147. [Google Scholar]
- Rahe, M.C.; Magstadt, D.R.; Groeltz-Thrush, J.; Gauger, P.C.; Zhang, J.; Schwartz, K.J.; Siepker, C.L. Bovine coronavirus in the lower respiratory tract of cattle with respiratory disease. J. VET Diagn. Investig. 2022, 34, 482–488. [Google Scholar] [CrossRef] [PubMed]
- Prickett, J.R.; Zimmerman, J.J. The development of oral fluid-based diagnostics and applications in veterinary medicine. Ani Health Res. Rev. 2010, 11, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Prickett, J.; Simer, R.; Christopher-Hennings, J.; Yoon, K.J.; Evans, R.B.; Zimmerman, J.J. Detection of Porcine reproductive and respiratory syndrome virus infection in porcine oral fluid samples: A longitudinal study under experimental conditions. J. Vet. Diagn. Investig. 2008, 20, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Rotolo, M.L.; Main, R.G.; Zimmerman, J.J. Herd-level infectious disease surveillance of livestock populations using aggregate samples. Ani Health Res. Rev. 2018, 19, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Hayes, M.C.; Ralyea, R.D.; Murphy, S.C.; Carey, N.R.; Scarlett, J.M.; Boor, K.J. Identification and Characterization of Elevated Microbial Counts in Bulk Tank Raw Milk. J. Dairy Sci. 2001, 84, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Drew, T.W.; Yapp, F.; Paton, D.J. The detection of bovine viral diarrhea virus in bulk milk samples by the use of a single-tube RT-PCR. Vet. Microbiol. 1999, 64, 145–154. [Google Scholar] [CrossRef]
- Deng, Y.; Xu, X.; Zheng, X.; Ding, J.; Li, S.; Chui, H.-K.; Wong, T.-K.; Poon, L.L.; Zhang, T. Use of sewage surveillance for COVID-19 to guide public health response: A case study in Hong Kong. Sci. Total Environ. 2022, 821, 153250. [Google Scholar] [CrossRef]
- Wurtzer, S.; Waldman, P.; Levert, M.; Cluzel, N.; Almayrac, J.L.; Charpentier, C.; Masnada, S.; Gillon-Ritz, M.; Mouchel, J.M.; Maday, Y.; et al. SARS-CoV-2 genome quantification in wastewaters at regional and city scale allows precise monitoring of the whole outbreaks dynamics and variants spreading in the population. Sci. Total Environ. 2022, 810, 152213. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.D.; Yondon, M.; Bailey, E.S.; Duman, E.K.; Simmons, R.A.; Greer, A.G.; Gray, G.C. Environmental bioaerosol surveillance as an early warning system for pathogen detection in North Carolina swine farms: A pilot study. Transbound. Emerg. Dis. 2021, 68, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Sharon, K. Hietala SK, Hullinger PJ, Crossley BM, Kinde H, Ardans AA. Environmental Air Sampling to Detect Exotic Newcastle Disease Virus in Two California Commercial Poultry Flocks. J. Vet. Diag Investig. 2005, 17, 198–200. [Google Scholar]
- Echavarria, M.; Kolavic, S.A.; Cersovsky, S.; Mitchell, F.; Sanchez, J.L.; Polyak, C.; Innis, B.L.; Binn, L.N. Detection of Adenoviruses (AdV) in Culture-Negative Environmental Samples by PCR during an AdV-Associated Respiratory Disease Outbreak. J. Clin. Microbiol. 2000, 38, 2982–2984. [Google Scholar] [CrossRef] [PubMed]
- Renter, D.G.; Smith, D.R.; King, R.; Stilborn, R.; Berg, J.; Berezowski, J.; McFall, M. Detection and determinants of Escherichia coli O157:H7 in Alberta feedlot pens immediately prior to slaughter. Can. J. Vet. Res. 2008, 72, 217–227. [Google Scholar]
- Smith, D.R.; Moxley, R.A.; Clowser, S.L.; Folmer, J.D.; Hinkley, S.; Erickson, G.E.; Klopfenstein, T.J. Use of Rope Devices to Describe and Explain the Feedlot Ecology of Salmonella by Time and Place. Foodborne Pathog. Dis. 2005, 2, 61–69. [Google Scholar] [CrossRef]
- Abi Younes, J.N.; Campbell, J.R.; Otto, S.J.; Gow, S.P.; Woolums, A.R.; Jelinski, M.; Lacoste, S.; Waldner, C.L. Variation in Pen-Level Prevalence of BRD Bacterial Pathogens and Antimicrobial Resistance Following Feedlot Arrival in Beef Calves. Antibiotics 2024, 13, 322. [Google Scholar] [CrossRef]
- Carlson, J.M. Environmental Sampling Techniques for Herd-Level Surveillance of Bovine Viral Diarrhea Virus. Master’s Thesis, Department of Veterinary and Biomedical Sciences, University of Nebraska-Lincoln, Lincoln, NE, USA, 2019. [Google Scholar]
- Plauzolles, A.; Toumi, E.; Bonnet, M.; Pénaranda, G.; Bidaut, G.; Chiche, L.; Allardet-Servent, J.; Retornaz, F.; Goutorbe, B.; Halfon, P. Human Stool Preservation Impacts Taxonomic Profiles in 16S Metagenomics Studies. Front. Cell Infect. Microbiol. 2022, 12, 722886. [Google Scholar] [CrossRef]
- USDA National Animal Health Emergency Management System: Guidelines for Cleaning and Disinfection. Available online: https://www.aphis.usda.gov/sites/default/files/cleaning_disfection.pdf (accessed on 10 June 2024).
- Loy, J.D.; Leger, L.; Workman, A.M.; Clawson, M.L.; Bulut, E.; Wang, B. Development of a multiplex real-time PCR assay using two thermocycling platforms for detection of major bacterial pathogens associated with bovine respiratory disease complex from clinical samples. J. Vet. Diagn. Investig. 2018, 30, 837–847. [Google Scholar] [CrossRef]
- Dutta, E.; Loy, J.D.; Deal, C.A.; Wynn, E.L.; Clawson, M.L.; Clarke, J.; Wang, B. Development of a Multiplex Real-Time PCR Assay for Predicting Macrolide and Tetracycline Resistance Associated with Bacterial Pathogens of Bovine Respiratory Disease. Pathogens 2021, 10, 64. [Google Scholar] [CrossRef]
- Dohoo, I.R.; Martin, S.W.; Stryhn, H. Veterinary Epidemiologic Research, 1st ed.; AVC Inc.: Fremont, CA, USA, 2003; pp. 90–117. [Google Scholar]
- Stan Development Team. RStan: The R interface to Stan. R Package Version 4.2.1. Available online: https://mc-stan.org (accessed on 1 May 2024).
- USDA National Animal Health Monitoring System. Feedlot 2011 Part IV: Health and Health Management on US feedlots with a Capacity of 1000 or More Head. Available online: https://www.aphis.usda.gov/sites/default/files/feed11_dr_partiv.pdf (accessed on 10 May 2024).
- Walz, P.H.; Newcomer, B.W.; Riddell, K.P.; Scruggs, D.W.; Cortese, V.S. Virus detection by PCR following vaccination of naive calves with intranasal or injectable multivalent modified-live viral vaccines. J. VET Diagn. Investig. 2017, 29, 628–635. [Google Scholar] [CrossRef] [PubMed]
- Kos, D.; Schreiner, B.; Thiessen, S.; McAllister, T.; Jelinski, M.; Ruzzini, A. Insight into antimicrobial resistance at a new beef cattle feedlot in western Canada. Vet. Microbiol. 2023, 8, 00317–00323. [Google Scholar] [CrossRef] [PubMed]
- Crosby, W.B.; Pinnell, L.J.; Richeson, J.T.; Wolfe, C.; Castle, J.; Loy, J.D.; Gow, S.P.; Seo, K.S.; Capik, S.F.; Woolums, A.R.; et al. Does swab type matter? Comparing methods for Mannheimia haemolytica recovery and upper respiratory microbiome characterization in feedlot cattle. Anim. Microbiome 2022, 4, 49. [Google Scholar] [CrossRef]
- White, B.J.; Renter, D.G. Bayesian Estimation of the Performance of Using Clinical Observations and Harvest Lung Lesions for Diagnosing Bovine Respiratory Disease in Post-weaned Beef Calves. J. Vet. Diagn. Investig. 2009, 4, 446–453. [Google Scholar] [CrossRef] [PubMed]
- White, B.J.; Goehl, D.R.; Amrine, D.E.; Booker, C.; Wildman, B.; Perrett, T. Bayesian evaluation of clinical diagnostic test characteristics of visual observations and remote monitoring to diagnose bovine respiratory disease in beef calves. Prev. Vet. Med. 2016, 126, 74–80. [Google Scholar] [CrossRef]
- Buczinski, S.; Ollivett, T.L.; Dendukuri, N. Bayesian estimation of the accuracy of the calf respiratory scoring chart and ultrasonography for the diagnosis of bovine respiratory disease in pre-weaned dairy calves. Prev. Vet. Med. 2015, 119, 227–231. [Google Scholar] [CrossRef]
- Timsit, E.; Dendukuri, N.; Schiller, I.; Buczinski, S. Diagnostic accuracy of clinical illness for bovine respiratory disease (BRD) diagnosis in beef cattle placed in feedlots: A systematic literature review and hierarchical Bayesian latent-class meta-analysis. Prev. Vet. Med. 2016, 135, 67–73. [Google Scholar] [CrossRef]
- Branscum, A.J.; Gardner, I.A.; Johnson, W.O. Estimation of diagnostic-test sensitivity and specificity through Bayesian modeling. Prev. Vet. Med. 2005, 68, 145–163. [Google Scholar] [CrossRef] [PubMed]
- Hanson, T.; Johnson, W.O.; Gardner, I.A. Hierarchical models for estimating herd prevalence and test accuracy in the absence of a gold standard. JABES 2003, 8, 223–239. [Google Scholar] [CrossRef]
- FASS. Guide for the Care and Use of Agricultural Animals in Research and Teaching, 3rd ed.; Federation of Animal Science Societies: Champaign, IL, USA, 2010. [Google Scholar]
| Category | Placement Weight (lbs.) | Morbidity (%) | Mortality (%) |
|---|---|---|---|
| Low (n = 2) | 849.5 (227.0) | 2.2 (2.1) | 1.1 (0.5) |
| Moderate (n = 5) | 621.7 (24.4) | 24.4 (5.5) | 3.6 (3.9) |
| High (n = 3) | 647.5 (90.5) | 40.5 (9.8) | 4.7 (3.9) |
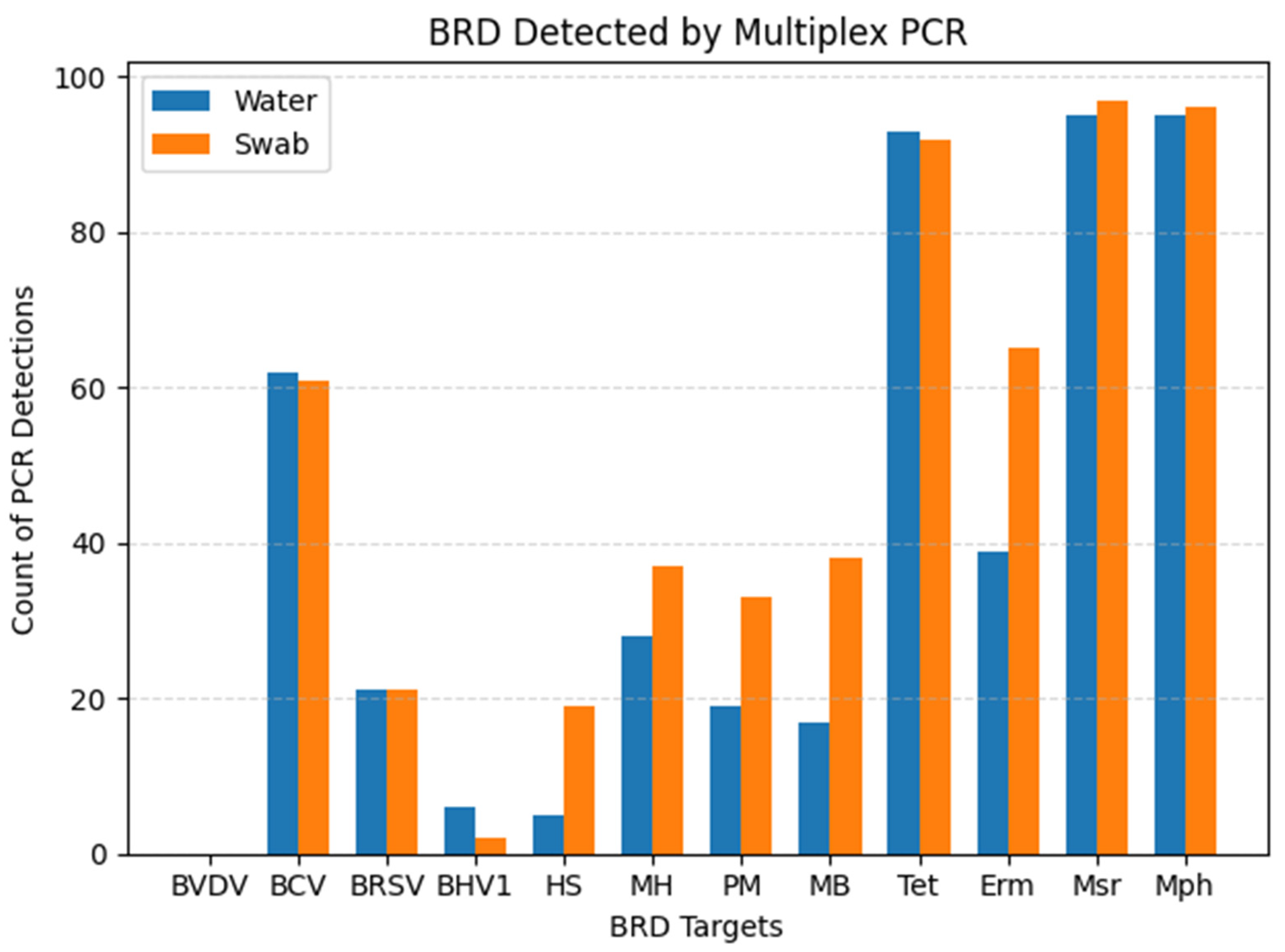
| BRD | Swab +/ Water + | Swab −/ Water − | Swab +/ Water − | Swab −/ Water + | PPA 1 (%) | Kappa 2 (95% CI) |
|---|---|---|---|---|---|---|
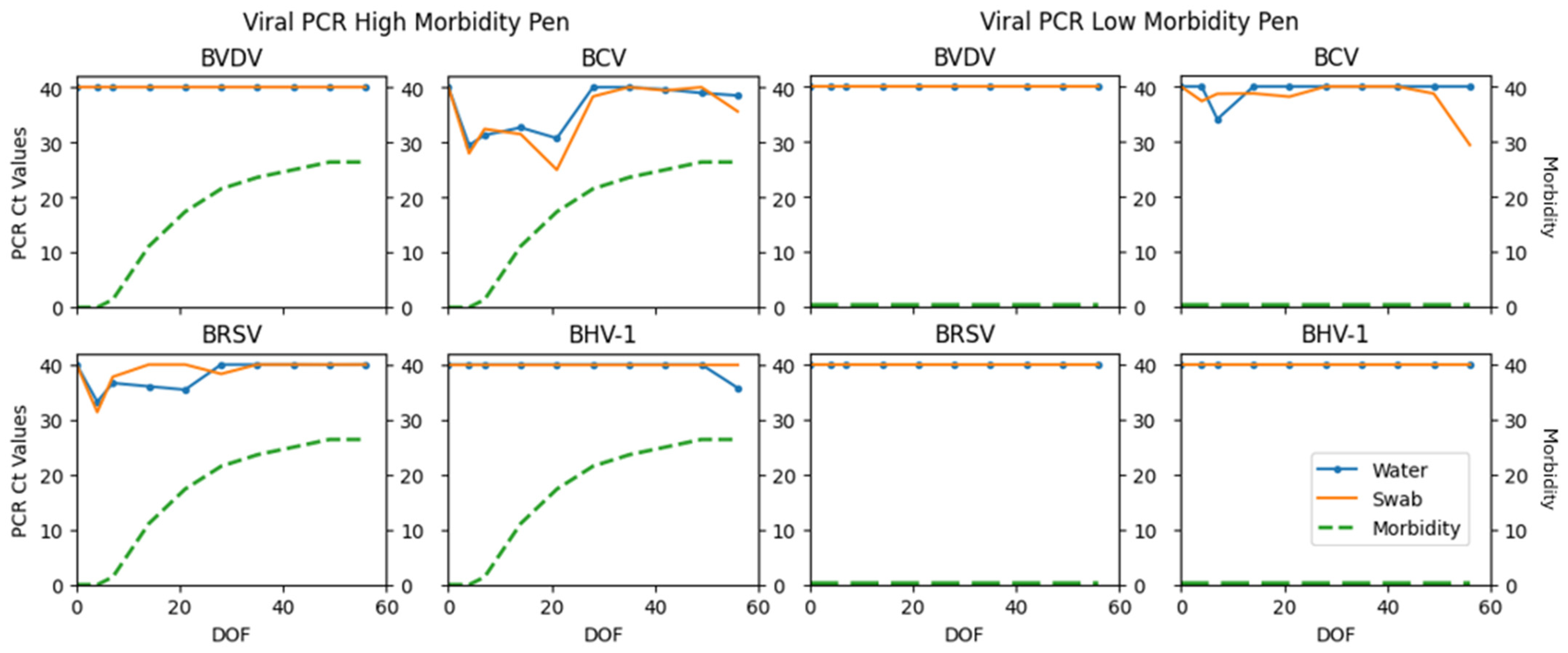
| BVDV | 0 | 100 | 0 | 0 | 0 | −1 |
| BCV | 45 | 22 | 16 | 17 | 73.17 | 0.30 (0.11–0.49) |
| BRSV | 15 | 73 | 6 | 6 | 71.43 | 0.64 (0.45–0.83) |
| BHV-1 | 2 | 94 | 0 | 4 | 50.00 | 0.49 (0.06–0.91) |
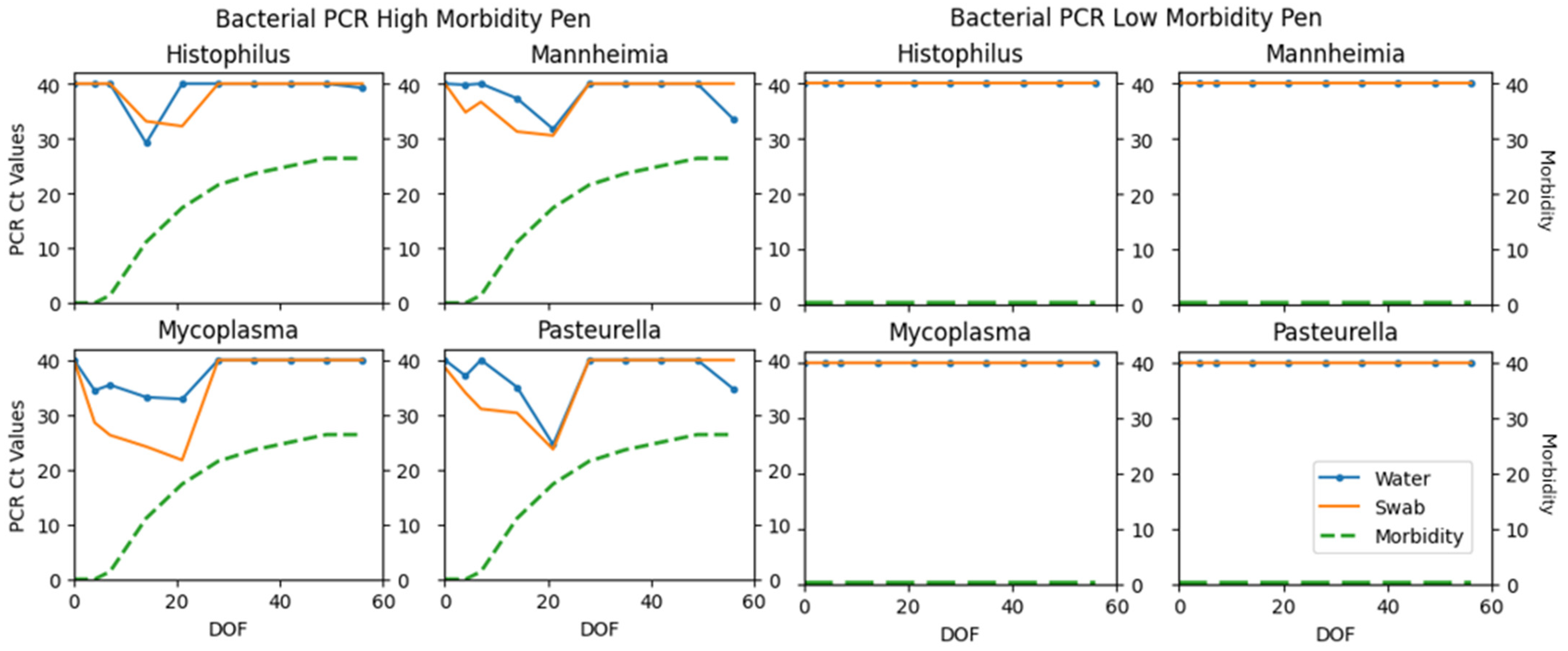
| H. somni | 2 | 79 | 16 | 3 | 17.39 | 0.10 (0.02–0.31) |
| M. haem. | 24 | 59 | 13 | 4 | 73.85 | 0.62 (0.46–0.78) |
| P. mult. | 16 | 66 | 15 | 3 | 64.00 | 0.53 (0.35–0.71) |
| M. bovis | 16 | 61 | 22 | 1 | 58.18 | 0.45 (0.29–0.62) |
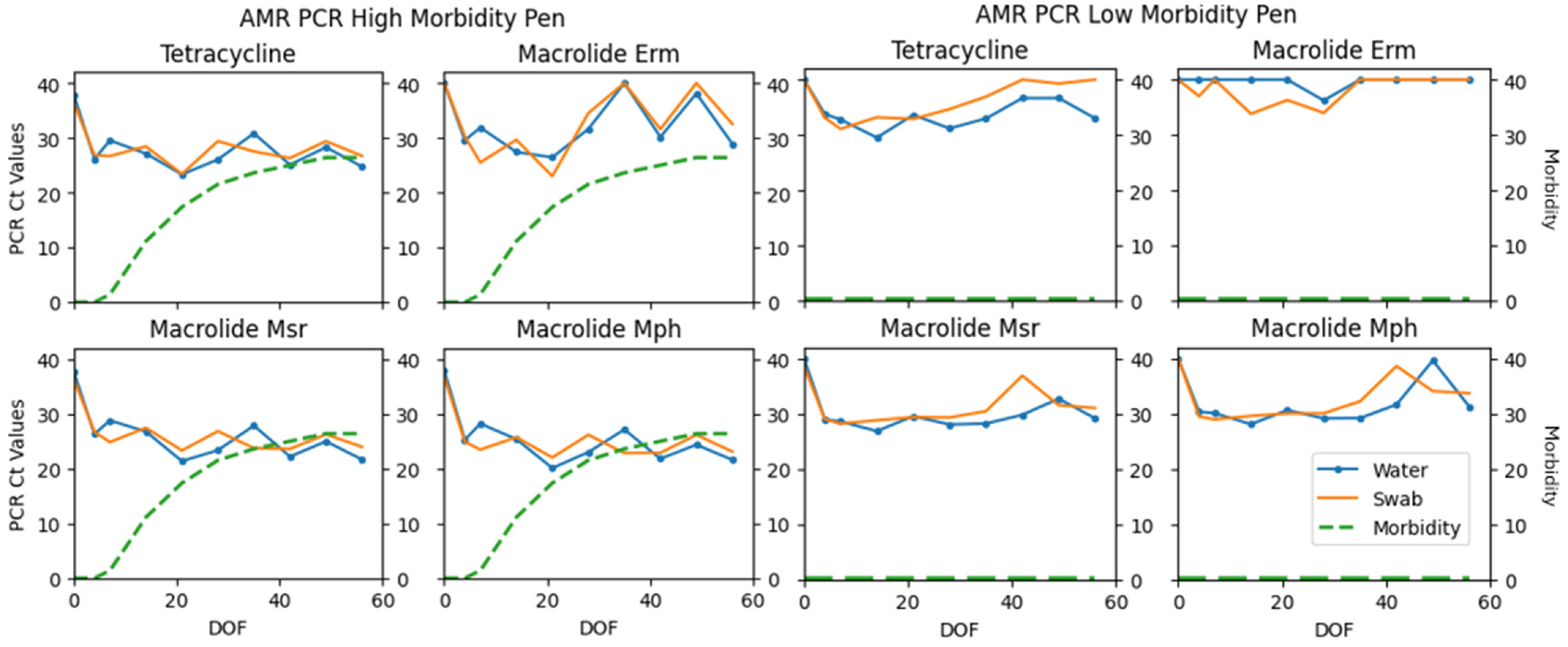
| Tet | 91 | 6 | 1 | 2 | 98.38 | 0.78 (0.55–0.99) |
| Erm | 36 | 32 | 29 | 3 | 69.23 | 0.40 (0.25–0.55) |
| Msr | 34 | 2 | 3 | 1 | 97.92 | 0.48 (0.05–0.91) |
| Mph | 95 | 4 | 1 | 0 | 99.48 | 0.88 (0.66–0.99) |
| BRD | Sensitivity (%) | Specificity (%) | ||
|---|---|---|---|---|
| Water | Swab | Water | Swab | |
| BVDV | - | - | - | - |
| BCV | 57 (4–98) | 56 (4–97) | 45 (3–96) | 46 (3–96) |
| BRSV | 61 (1–99) | 62 (1–99) | 76 (4–100) | 76 (4–100) |
| BHV-1 | 38 (1–98) | 27 (1–93) | 64 (2–97) | 75 (7–99) |
| H. somni | 18 (1–86) | 31 (3–92) | 82 (15–97) | 69 (7–98) |
| M. haem. | 23 (1–95) | 31 (1–98) | 40 (2–99) | 30 (1–97) |
| P. mult. | 53 (1–97) | 68 (3–99) | 80 (8–98) | 69 (3–99) |
| M. bovis | 48 (1–97) | 73 (4–96) | 82 (10–99) | 64 (1–98) |
| Tet | 98 (95–99) | 98 (97–99) | 77 (41–90) | 81 (47–92) |
| Erm | 86 (18–96) | 87 (21–97) | 65 (3–98) | 63 (3–97) |
| Msr | 94 (92–97) | 96 (94–98) | 74 (40–91) | 78 (39–92) |
| Mph | 97 (95–99) | 98 (96–99) | 76 (49–92) | 80 (50–93) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jobman, E.; Vander Ley, B.; Loy, J.D.; Loy, D.S.; Meyer, N.; Thomson, D.; Lowe, J.; Terrell, S. Aggregate Sampling to Detect Pathogens and Antimicrobial Resistance Genes Associated with Bovine Respiratory Disease in US Feedlots: A Pilot Study. Vet. Sci. 2025, 12, 244. https://doi.org/10.3390/vetsci12030244
Jobman E, Vander Ley B, Loy JD, Loy DS, Meyer N, Thomson D, Lowe J, Terrell S. Aggregate Sampling to Detect Pathogens and Antimicrobial Resistance Genes Associated with Bovine Respiratory Disease in US Feedlots: A Pilot Study. Veterinary Sciences. 2025; 12(3):244. https://doi.org/10.3390/vetsci12030244
Chicago/Turabian StyleJobman, Erin, Brian Vander Ley, John Dustin Loy, Duan Sriyotee Loy, Nathan Meyer, Dan Thomson, James Lowe, and Shane Terrell. 2025. "Aggregate Sampling to Detect Pathogens and Antimicrobial Resistance Genes Associated with Bovine Respiratory Disease in US Feedlots: A Pilot Study" Veterinary Sciences 12, no. 3: 244. https://doi.org/10.3390/vetsci12030244
APA StyleJobman, E., Vander Ley, B., Loy, J. D., Loy, D. S., Meyer, N., Thomson, D., Lowe, J., & Terrell, S. (2025). Aggregate Sampling to Detect Pathogens and Antimicrobial Resistance Genes Associated with Bovine Respiratory Disease in US Feedlots: A Pilot Study. Veterinary Sciences, 12(3), 244. https://doi.org/10.3390/vetsci12030244






