New Structure Sheds Light on Selective HIV-1 Genomic RNA Packaging
Abstract
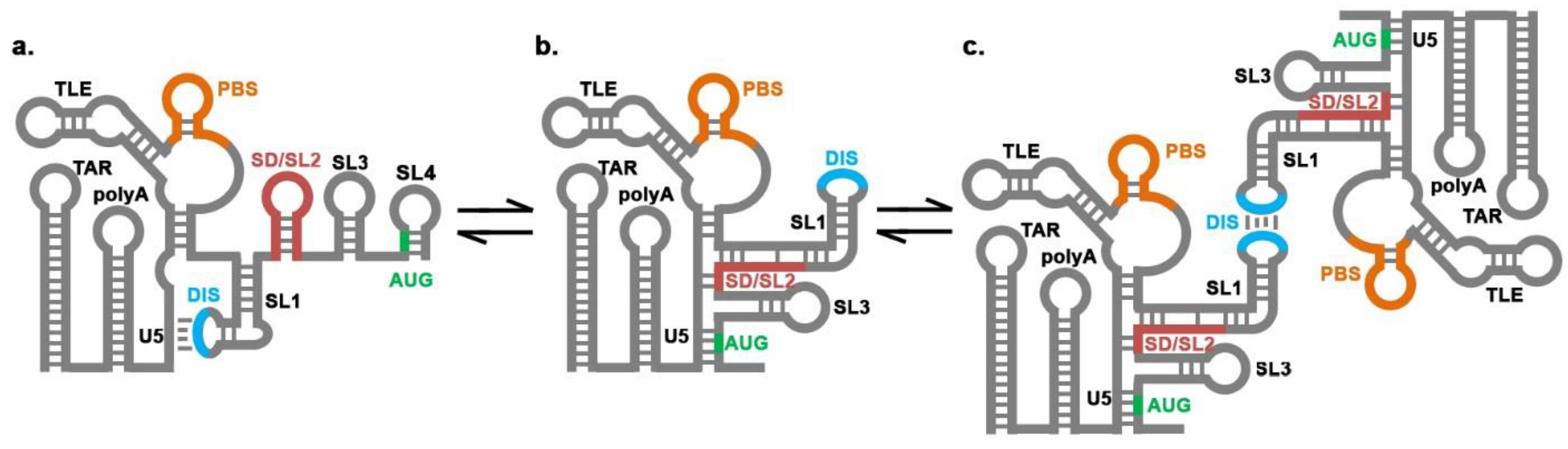
:
Acknowledgments
References
- Keane, S.C.; Heng, X.; Lu, K.; Kharytonchyk, S.; Ramakrishnan, V.; Carter, G.; Barton, S.; Hosic, A.; Florwick, A.; Santos, J.; et al. RNA structure. Structure of the HIV-1 RNA packaging signal. Science 2015, 348, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Bell, N.M.; Lever, A.M. HIV Gag polyprotein: Processing and early viral particle assembly. Trends Microbiol. 2013, 21, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Sundquist, W.I.; Krausslich, H.G. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2012, 2, a006924. [Google Scholar] [CrossRef] [PubMed]
- Kutluay, S.B.; Zang, T.; Blanco-Melo, D.; Powell, C.; Jannain, D.; Errando, M.; Bieniasz, P.D. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell 2014, 159, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Luban, J.; Goff, S.P. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J. Virol. 1994, 68, 3784–3793. [Google Scholar] [PubMed]
- Moore, M.D.; Nikolaitchik, O.A.; Chen, J.; Hammarskjold, M.L.; Rekosh, D.; Hu, W.S. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS Pathog. 2009, 5, e1000627. [Google Scholar] [CrossRef] [PubMed]
- Nikolaitchik, O.A.; Dilley, K.A.; Fu, W.; Gorelick, R.J.; Tai, S.H.; Soheilian, F.; Ptak, R.G.; Nagashima, K.; Pathak, V.K.; Hu, W.S. Dimeric RNA recognition regulates HIV-1 genome packaging. PLoS Pathog. 2013, 9, e1003249. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Grunwald, D.; Sardo, L.; Galli, A.; Plisov, S.; Nikolaitchik, O.A.; Chen, D.; Lockett, S.; Larson, D.R.; Pathak, V.K.; et al. Cytoplasmic HIV-1 RNA is mainly transported by diffusion in the presence or absence of Gag protein. Proc. Natl. Acad. Sci. USA 2014, 111, E5205–E5213. [Google Scholar] [CrossRef] [PubMed]
- Chamanian, M.; Purzycka, K.J.; Wille, P.T.; Ha, J.S.; McDonald, D.; Gao, Y.; le Grice, S.F.; Arts, E.J. A cis-acting element in retroviral genomic RNA links Gag-Pol ribosomal frameshifting to selective viral RNA encapsidation. Cell Host Microbe 2013, 13, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Jouvenet, N.; Simon, S.M.; Bieniasz, P.D. Visualizing HIV-1 assembly. J. Mol. Biol. 2011, 410, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Jouvenet, N.; Bieniasz, P.D.; Simon, S.M. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature 2008, 454, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Jouvenet, N.; Simon, S.M.; Bieniasz, P.D. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc. Natl. Acad. Sci. USA 2009, 106, 19114–19119. [Google Scholar] [CrossRef] [PubMed]
- Kuzembayeva, M.; Dilley, K.; Sardo, L.; Hu, W.S. Life of psi: How full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology 2014, 454–455, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Heng, X.; Summers, M.F. Structural determinants and mechanism of HIV-1 genome packaging. J. Mol. Biol. 2011, 410, 609–633. [Google Scholar] [CrossRef] [PubMed]
- Levin, J.G.; Guo, J.; Rouzina, I.; Musier-Forsyth, K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: Critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005, 80, 217–286. [Google Scholar] [PubMed]
- McBride, M.S.; Panganiban, A.T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J. Virol. 1996, 70, 2963–2973. [Google Scholar] [PubMed]
- McBride, M.S.; Panganiban, A.T. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J. Virol. 1997, 71, 2050–2058. [Google Scholar] [PubMed]
- Mann, R.; Baltimore, D. Varying the position of a retrovirus packaging sequence results in the encapsidation of both unspliced and spliced RNAs. J. Virol. 1985, 54, 401–407. [Google Scholar] [PubMed]
- Adam, M.A.; Miller, A.D. Identification of a signal in a murine retrovirus that is sufficient for packaging of nonretroviral RNA into virions. J. Virol. 1988, 62, 3802–3806. [Google Scholar] [PubMed]
- Mougel, M.; Barklis, E. A role for two hairpin structures as a core RNA encapsidation signal in murine leukemia virus virions. J. Virol. 1997, 71, 8061–8065. [Google Scholar] [PubMed]
- Mann, R.; Mulligan, R.C.; Baltimore, D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell 1983, 33, 153–159. [Google Scholar] [CrossRef]
- Banks, J.D.; Linial, M.L. Secondary structure analysis of a minimal avian leukosis-sarcoma virus packaging signal. J. Virol. 2000, 74, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Doria-Rose, N.A.; Vogt, V.M. In vivo selection of Rous sarcoma virus mutants with randomized sequences in the packaging signal. J. Virol. 1998, 72, 8073–8082. [Google Scholar] [PubMed]
- Banks, J.D.; Yeo, A.; Green, K.; Cepeda, F.; Linial, M.L. A minimal avian retroviral packaging sequence has a complex structure. J. Virol. 1998, 72, 6190–6094. [Google Scholar] [PubMed]
- Banks, J.D.; Kealoha, B.O.; Linial, M.L. An Mpsi-containing heterologous RNA, but not env mRNA, is efficiently packaged into avian retroviral particles. J. Virol. 1999, 73, 8926–8933. [Google Scholar] [PubMed]
- Abd El-Wahab, E.W.; Smyth, R.P.; Mailler, E.; Bernacchi, S.; Vivet-Boudou, V.; Hijnen, M.; Jossinet, F.; Mak, J.; Paillart, J.C.; Marquet, R. Specific recognition of the HIV-1 genomic RNA by the Gag precursor. Nat. Commun. 2014, 5, 4304. [Google Scholar] [CrossRef] [PubMed]
- Cockrell, A.S.; van Praag, H.; Santistevan, N.; Ma, H.; Kafri, T. The HIV-1 Rev/RRE system is required for HIV-1 5′ UTR cis elements to augment encapsidation of heterologous RNA into HIV-1 viral particles. Retrovirology 2011, 8, 51. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Heng, X.; Garyu, L.; Monti, S.; Garcia, E.L.; Kharytonchyk, S.; Dorjsuren, B.; Kulandaivel, G.; Jones, S.; Hiremath, A.; et al. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science 2011, 334, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Heng, X.; Kharytonchyk, S.; Garcia, E.L.; Lu, K.; Divakaruni, S.S.; LaCotti, C.; Edme, K.; Telesnitsky, A.; Summers, M.F. Identification of a minimal region of the HIV-1 5′-leader required for RNA dimerization, NC binding, and packaging. J. Mol. Biol. 2012, 417, 224–239. [Google Scholar] [CrossRef] [PubMed]
- Huthoff, H.; Berkhout, B. Two alternating structures of the HIV-1 leader RNA. RNA 2001, 7, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Seif, E.; Niu, M.; Kleiman, L. Annealing to sequences within the primer binding site loop promotes an HIV-1 RNA conformation favoring RNA dimerization and packaging. RNA 2013, 19, 1384–1393. [Google Scholar] [CrossRef] [PubMed]
- Pappalardo, L.; Kerwood, D.J.; Pelczer, I.; Borer, P.N. Three-dimensional folding of an RNA hairpin required for packaging HIV-1. J. Mol. Biol. 1998, 282, 801–818. [Google Scholar] [CrossRef] [PubMed]
- Amarasinghe, G.K.; de Guzman, R.N.; Turner, R.B.; Summers, M.F. NMR structure of stem-loop SL2 of the HIV-1 psi RNA packaging signal reveals a novel A-U-A base-triple platform. J. Mol. Biol. 2000, 299, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Amarasinghe, G.K.; Zhou, J.; Miskimon, M.; Chancellor, K.J.; McDonald, J.A.; Matthews, A.G.; Miller, R.R.; Rouse, M.D.; Summers, M.F. Stem-loop SL4 of the HIV-1 psi RNA packaging signal exhibits weak affinity for the nucleocapsid protein. structural studies and implications for genome recognition. J. Mol. Biol. 2001, 314, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, D.C.; Stover, C.C.; Noznitsky, J.; Wu, Z.; Summers, M.F. Structure of the intact stem and bulge of HIV-1 Psi-RNA stem-loop SL1. J. Mol. Biol. 2003, 326, 529–542. [Google Scholar] [CrossRef]
- De Guzman, R.N.; Wu, Z.R.; Stalling, C.C.; Pappalardo, L.; Borer, P.N.; Summers, M.F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 1998, 279, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Amarasinghe, G.K.; de Guzman, R.N.; Turner, R.B.; Chancellor, K.J.; Wu, Z.R.; Summers, M.F. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2000, 301, 491–511. [Google Scholar] [CrossRef] [PubMed]
- Mujeeb, A.; Clever, J.L.; Billeci, T.M.; James, T.L.; Parslow, T.G. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat. Struct. Biol. 1998, 5, 432–436. [Google Scholar] [CrossRef] [PubMed]
- Kieken, F.; Paquet, F.; Brule, F.; Paoletti, J.; Lancelot, G. A new NMR solution structure of the SL1 HIV-1Lai loop-loop dimer. Nucleic Acids Res. 2006, 34, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Mihailescu, M.R.; Marino, J.P. A proton-coupled dynamic conformational switch in the HIV-1 dimerization initiation site kissing complex. Proc. Natl. Acad. Sci. USA 2004, 101, 1189–1194. [Google Scholar] [CrossRef] [PubMed]
- Ennifar, E.; Yusupov, M.; Walter, P.; Marquet, R.; Ehresmann, B.; Ehresmann, C.; Dumas, P. The crystal structure of the dimerization initiation site of genomic HIV-1 RNA reveals an extended duplex with two adenine bulges. Structure 1999, 7, 1439–1449. [Google Scholar] [CrossRef]
- Yuan, Y.; Kerwood, D.J.; Paoletti, A.C.; Shubsda, M.F.; Borer, P.N. Stem of SL1 RNA in HIV-1: Structure and nucleocapsid protein binding for a 1 × 3 internal loop. Biochemistry 2003, 42, 5259–5269. [Google Scholar] [CrossRef] [PubMed]
- Ulyanov, N.B.; Mujeeb, A.; Du, Z.; Tonelli, M.; Parslow, T.G.; James, T.L. NMR structure of the full-length linear dimer of stem-loop-1 RNA in the HIV-1 dimer initiation site. J. Biol. Chem. 2006, 281, 16168–16177. [Google Scholar] [CrossRef] [PubMed]
- Mujeeb, A.; Parslow, T.G.; Zarrinpar, A.; Das, C.; James, T.L. NMR structure of the mature dimer initiation complex of HIV-1 genomic RNA. FEBS Lett. 1999, 458, 387–392. [Google Scholar] [CrossRef]
- Yu, E.T.; Hawkins, A.; Eaton, J.; Fabris, D. MS3D structural elucidation of the HIV-1 packaging signal. Proc. Natl. Acad. Sci. USA 2008, 105, 12248–12253. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, J.D.; Li, H.; Kenyon, J.C.; Symmons, M.; Klenerman, D.; Lever, A.M. Three-dimensional RNA structure of the major HIV-1 packaging signal region. Structure 2013, 21, 951–962. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.P.; Cantara, W.A.; Olson, E.D.; Musier-Forsyth, K. Small-angle X-ray scattering-derived structure of the HIV-1 5′ UTR reveals 3D tRNA mimicry. Proc. Natl. Acad. Sci. USA 2014, 111, 3395–3400. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, V.; Dey, A.; Habib, D.; Summers, M.F. NMR structure of the 101-nucleotide core encapsidation signal of the Moloney murine leukemia virus. J. Mol. Biol. 2004, 337, 427–442. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, V.; Summers, M.F. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature 2004, 431, 586–590. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, J.C.; Prestwood, L.J.; le Grice, S.F.; Lever, A.M. In-gel probing of individual RNA conformers within a mixed population reveals a dimerization structural switch in the HIV-1 leader. Nucleic Acids Res. 2013, 41, e174. [Google Scholar] [CrossRef] [PubMed]
- Retroviruses; Cold Spring Laboratory Press: Plainview, New York, NY, USA, 1997.
- D’Souza, V.; Summers, M.F. How retroviruses select their genomes. Nat. Rev. Microbiol. 2005, 3, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.P.; Datta, S.A.; Rein, A.; Rouzina, I.; Musier-Forsyth, K. Matrix domain modulates HIV-1 Gag’s nucleic acid chaperone activity via inositol phosphate binding. J. Virol. 2011, 85, 1594–1603. [Google Scholar] [CrossRef] [PubMed]
- Rein, A.; Datta, S.A.; Jones, C.P.; Musier-Forsyth, K. Diverse interactions of retroviral Gag proteins with RNAs. Trends Biochem. Sci. 2011, 36, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Datta, S.A.; Mitra, M.; Gorelick, R.J.; Rein, A.; Levin, J.G. Fundamental differences between the nucleic acid chaperone activities of HIV-1 nucleocapsid protein and Gag or Gag-derived proteins: Biological implications. Virology 2010, 405, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Roldan, A.; Warren, O.U.; Russell, R.S.; Liang, C.; Wainberg, M.A. A HIV-1 minimal gag protein is superior to nucleocapsid at in vitro annealing and exhibits multimerization-induced inhibition of reverse transcription. J. Biol. Chem. 2005, 280, 17488–17496. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olson, E.D.; Cantara, W.A.; Musier-Forsyth, K. New Structure Sheds Light on Selective HIV-1 Genomic RNA Packaging. Viruses 2015, 7, 4826-4835. https://doi.org/10.3390/v7082846
Olson ED, Cantara WA, Musier-Forsyth K. New Structure Sheds Light on Selective HIV-1 Genomic RNA Packaging. Viruses. 2015; 7(8):4826-4835. https://doi.org/10.3390/v7082846
Chicago/Turabian StyleOlson, Erik D., William A. Cantara, and Karin Musier-Forsyth. 2015. "New Structure Sheds Light on Selective HIV-1 Genomic RNA Packaging" Viruses 7, no. 8: 4826-4835. https://doi.org/10.3390/v7082846
APA StyleOlson, E. D., Cantara, W. A., & Musier-Forsyth, K. (2015). New Structure Sheds Light on Selective HIV-1 Genomic RNA Packaging. Viruses, 7(8), 4826-4835. https://doi.org/10.3390/v7082846





