Abstract
Leptospirosis is a zoonotic disease caused by pathogenic Leptospira, prevalent in tropical/sub-tropical regions. This study aimed to clarify the prevailing leptospiral species, clinical features, and risk factors of leptospirosis in north-central Bangladesh in 2024. Venous blood and urine samples were collected from 117 patients with clinically suspected leptospirosis. Among these cases, 75 (64%) tested positive for Leptospira infection by IgM ELISA test and/or PCR. By phylogenetic analysis of the 16S rRNA gene, all the samples tested were classified into L. wolffii (pathogenic group P2), showing high sequence identity to those of the type strain Khorat-H2 (97–99%) and L. wolffii reported in Bangladesh previously. Confirmed leptospirosis patients were mostly male (93%), aged 15–60 years (93%), living in rural areas in low socioeconomic conditions. Variable symptoms were presented by patients, with jaundice (84%), nausea/vomiting (84%), and myalgia (67%) being common. Some patients showed severe symptoms involving the nervous system (disorientation and neck stiffness) and the respiratory tract (cough, shortness of breath, and hemoptysis). Major risk factors for leptospirosis were exposures to mud/wet soil, sanding water, heavy rain, working in a paddy field, and cattle. In conclusion, L. wolffii was revealed to be circulating endemically in north-central Bangladesh, since its first detection in 2018, associated with variable and severe clinical symptoms in humans.
1. Introduction
Leptospirosis is a reemerging, neglected zoonotic infectious disease caused by spirochetes of the genus Leptospira. This disease is widespread globally, except in Antarctica, with frequent outbreaks in regions such as the Indian subcontinent, Oceania, and Latin America [1], and is of particular concern in tropical and subtropical regions, including Bangladesh, posing a significant public health threat [2]. It was estimated that there are approximately 1.03 million cases of leptospirosis globally every year, resulting in 58,900 deaths [3]. Median series mortality is shown as 2.2% (0–40%) among untreated cases of leptospirosis, while higher mortality is noted for patients with jaundice, renal failure, and those aged over 60 years [4]. Environmental factors, urbanization, inadequate sanitation, and climate change-induced flooding are expected to exacerbate the disease burden [5]. Despite its high prevalence, leptospirosis remains underdiagnosed and underreported due to its protean clinical manifestations, lack of awareness among healthcare providers, limited diagnostic facilities in poor-resource settings, and the absence of sensitive tests for early-stage detection [5,6].
The transmission cycle of Leptospira involves reservoir hosts (livestock, pets, rodents, and wildlife), environmental factors, and humans. Leptospira is widely distributed to numerous mammalian species including cattle, horse, pig, dog, camels, and rodents [7,8], as well as amphibia and reptilia [9], indicating a global risk of leptospirosis in humans. In these animals, leptospires circulate in the bloodstream and colonize the brush border of the proximal renal tubular epithelium via glomerular or peritubular capillaries. Pathogenic Leptospira are excreted into the environment via urine from reservoir hosts which are asymptomatic carriers. Humans are infected with leptospires through contact with the contaminated environment [5].
The genus Leptospira comprises 68 species, classified into two primary clades: S (saprophytes) and P (pathogens). These clades are further subdivided into four subclades: S1 and S2 (saprophyte subclades), P1 (formerly categorized as the pathogen group), and P2 (previously referred to as the “intermediate” group) [10]. Saprophytic Leptospira species, belonging to the S clade, are free-living spirochetes commonly found in soil and surface water, and they typically do not cause disease in mammals. In contrast, the P1 subclade includes species with high pathogenicity, which are capable of infecting a wide range of animal species and are transmissible to humans. The P2 subclade contains species exhibiting moderate pathogenicity, affecting both humans and animals [11]. These species are further designated into hundreds of serovars, which are grouped into serogroups based on antigenic relatedness [12].
In Bangladesh, leptospirosis was shown to be prevalent, especially in flood-prone rural areas, due to risk factors such as a tropical climate, inadequate sanitation, frequent flooding, many animal reservoirs, and an agriculture-based economy which significantly increases the risk of leptospirosis [2,13]. Serological testing indicated that leptospirosis accounts for 8.6% of outpatient fever cases [13], 10.6% of healthy individuals in rural areas, and 12.0% in the urban febrile population [14]. Since 2019, the Institute of Epidemiology, Disease Control and Research (IEDCR), Bangladesh, has been conducting a nationwide surveillance of leptospirosis across eight sentinel sites by detection of IgM antibodies and confirmatory PCR testing (https://iedcr.portal.gov.bd/site/page/4dd56857-84df-4269-b341-04e9e8a55725, accessed on 14 March 2025). Between 2019 and 2023, 4.4% to 8.3% of clinically suspected cases (a total of 5325 samples) were reported to be positive for Leptospira (IEDCR, 2024). Compared with these findings, relatively higher detection rates of Leptospira by the genetic method were revealed in Mymensingh, the north-central region of Bangladesh, in 2018 (17.6%) and 2019 (35.7%), showing the dominant species as L. interrogans and L. wolffii, respectively [15,16]. Thereafter, between 2021 and 2022, leptospirosis accounted for 47% among suspected cases (n = 186), with the detection of only L. wolffii [17]. The predominance of L. wolffii was notable because it belongs to the P2 subclade and its pathogenicity and epidemiological features have been rarely described in this region.
The present study aimed to clarify the prevalence of leptospirosis and prevailing leptospiral species in north-central Bangladesh, to monitor the persistence or change of dominant species since the initial study in 2018. In addition, clinical features and risk factors of leptospirosis that might have been associated with the dominant species were investigated to explore further preventive measures.
2. Materials and Methods
2.1. Collection of Specimens
This study was conducted as a cross-sectional, observational study. Venous blood and urine samples were collected from patients with clinically suspected leptospirosis who visited Mymensingh Medical College Hospital in Mymensingh city and agreed to participate in this study, for a period from March 2024 to February 2025. Mymensingh is a central city in Mymensingh Division, located about 100 km north of the capital city Dhaka (Figure 1). Inclusion criteria for cases were as follows: (1) fever for more than five days with or without malaise, headache, myalgia, anorexia, abdominal pain, or conjunctival suffusion, (2) febrile patients with any of the symptoms of hepatic involvement such as jaundice, renal involvement such as oliguria and hematuria, and pulmonary symptoms such as cough and hemoptysis. Exclusion criteria were (1) patients suffering from a febrile illness where any underlying etiology other than leptospiral illness had already been established, and (2) patients who did not give written informed consent. The blood sample (5 mL) was collected aseptically, placed in a sterile test tube, and allowed to stand for 30 min to facilitate clot formation, followed by centrifuging at 3000 rpm for 5 min. The serum was separated and transferred into a sterile tube for ELISA and polymerase chain reaction (PCR) analysis.
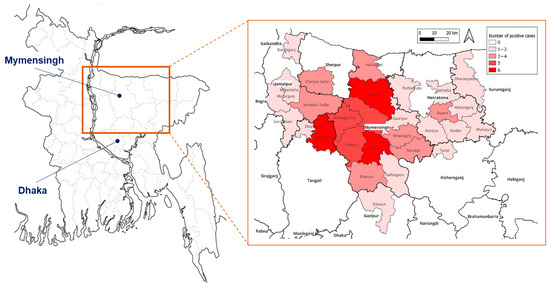
Figure 1.
Location of Mymensingh in Bangladesh (left) and incidence of leptospirosis patients in each place of residence (extracted map in a square).
2.2. Detection of Leptospira-Specific IgM by ELISA
Leptospira-specific IgM in serum is detectable from day 5 onwards after symptom onset and usually persists for several months. Detection of the IgM antibodies was performed using the Panbio™ Leptospira IgM ELISA kit (Abbott, Chicago, IL, USA) [18] following the manufacturer’s protocol.
2.3. Urine Culture for Leptospira
In total, 500 µL of the urine sample was inoculated into semi-solid Ellinghausen and McCullough modified by Johnson and Harris (EMJH) media supplemented with 2% bovine serum albumin and incubated at 30 °C [19]. The cultures were maintained for up to 2 months with regular examinations for growth. For any presumptive positive culture, two serial subcultures were performed every 7–14 days to ensure bacterial growth. The growth of Leptospira in the cultures was examined weekly using dark-field microscopy.
2.4. Detection of Leptospira by PCR
From the serum sample, genomic DNA was extracted according to a method described previously [20]. Genomic DNA in the serum sample was extracted using the phenol–chloroform method. From urine sample, the direct lysis method described previously [21] was employed for DNA extraction. For the culture sample, the pellet of culture fluid after centrifugation was suspended in 20 µL of 10 mM Tris-1 mM EDTA (TE; pH 8.0). After boiling, supernatant followed by centrifugation was used as the DNA sample. Nested PCR, targeting 16S rRNA of Leptospira including the pathogenic and saprophytic clades, was performed by using the protocol and primers described previously [20] for serum and urine samples, and cultured samples. As a thermostable DNA polymerase for PCR, TaKaRa Ex Taq® (Takara, BIO INC, Kusatsu, Japan) was used. The second-round PCR product was electrophoresed on agarose gel and visualized as a band using a gel documentation system. In addition, conventional PCR to detect the pathogenic species of Leptospira targeting lig gene was performed as described previously [22] for genus-specific PCR-positive samples. Double-distilled water was used as a negative control. Though positive control was not available, the PCR products in this study were sequenced to confirm its derivation from the target gene, as shown below.
2.5. Sequencing and Phylogenetic Analysis
Species of Leptospira was identified by determination of the nucleotide sequence of the partial 16S rRNA by Sanger sequencing with the PCR product (289 bp) using a BigDye Terminator v3.1 Cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) on an automated sequencer (Applied Biosystems® PRISM3130 Genetic Analyzer, Thermo Fisher Scientific, Foster City, CA, USA). The sequence data obtained were analyzed using BLAST on the NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 15 March 2025), to search for the most similar sequence in the GenBank database. The Clustal Omega program (https://www.ebi.ac.uk/jdispatcher/msa/clustalo, accessed on 15 March 2025) was used to perform the multiple nucleotide alignment and obtain sequence identity. A phylogenetic dendrogram of the 16S rRNA was constructed with the maximum likelihood method using the MEGA ver.6 software package, together with sequences of representative Leptospira strains in GenBank database. The representative sequence data of Leptospira samples in this study were deposited in the GenBank under the accession numbers PV367219-PV367223, PV367229, and PV367230.
2.6. Patients’ Information, Risk Factors, and Statistical Analysis
Clinical symptoms of the individual patients, their sociodemographic factors, and risk factors were recorded on data sheets, together with the laboratory test results. The difference in the variables between different patient groups was statistically analyzed with Fisher’s exact test using the js-STAR XR ver.1.1.9 software (https://www.kisnet.or.jp/nappa/software/star/index.htm, accessed on 10 April 2025). The difference in the Leptospira-positive rates by the different methods was analyzed with McNemar’s test. A p-value < 0.05 was considered to be statistically significant.
3. Results
3.1. Detection of Leptospira by ELISA, PCR, and Culture
During the study period, 117 clinically suspected cases of leptospirosis were selected by the criteria. Of these patients, 88 were tested by both IgM ELISA and nested PCR (urine and serum), and the remaining 29 were examined only by nested PCR for serum samples. Positive rate of Leptospira-specific IgM by ELISA was 69.3% (61/88), which was higher than those by nested PCR for serum samples (29.1%, 34/117) and urine samples (23.9%, 21/88). Among a total of 117 cases, 75 (64%) were tested positive for Leptospira infection by any of the diagnostic methods. All the PCR-positive urine samples and most of the serum samples (20 among 26) were IgM-positive (Figure 2). However, 39% (34/88) and 7% (6/88) of samples were positive in only ELISA and PCR for serum, respectively.
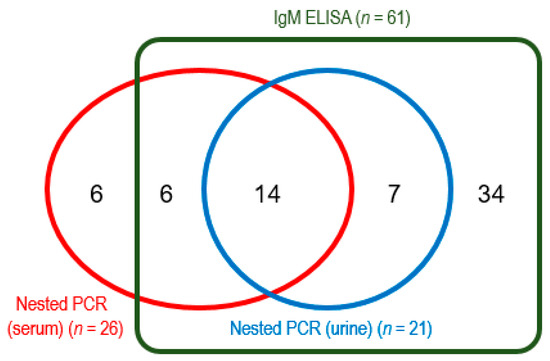
Figure 2.
Number of positive samples by IgM ELISA and nested PCR for urine and serum, among 88 samples. The remaining 29 samples were tested only by PCR (8 samples were positive).
The nested PCR for serum and urine showed a higher detection rate among cases with a shorter period of fever (5–7 days) than those with a longer period (8 or more days) (Table 1). In contrast, in cases with the longer fever periods, IgM-ELISA-positive rates were significantly higher than those by nested PCR for serum. Culture of Leptospira was attempted for 20 PCR-positive urine samples. The growth of Leptospira was confirmed in four samples, among which three samples were derived from cases with 8–14-day duration of fever (Table 1).

Table 1.
Diagnosis of leptospirosis by different detection methods and duration of fever.
3.2. Genetic Analysis of 16S rRNA and lig Gene
Partial 16S rRNA sequences were determined for the selected 17 samples (13 serum and 4 urine culture samples) obtained from July to October 2024 (Table S1). Among these samples, sequence identity was 98.6–100% (Table S2). Phylogenetic analysis revealed that all the samples belonged to L. wolffii in the P2 subclade, clustering with L. wolffii type strain Khorat-H2 [23] and those reported in Bangladesh previously [15,16,17], showing sequence identity of 95–100% (97–99% with Khorat-H2 strain) (Figure 3 and Table S3). A specific amplicon was not obtained from any 16S rRNA-positive samples in the PCR targeting lig gene.
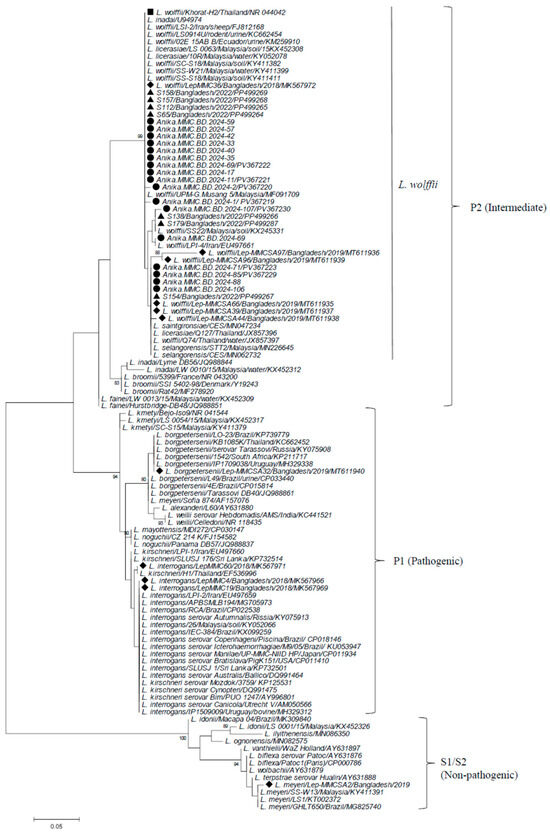
Figure 3.
Phylogenetic dendrogram based on partial 16S rRNA gene sequences of Leptospira constructed by the maximum likelihood method using the MEGA6 program, following alignment with the ClustalW algorithm. The trees were statistically supported by bootstrapping with 1000 replicates, and phylogenetic distances were measured by the Kimura 2-parameter model with uniform rates among sites. Samples analyzed in the present study are marked with closed circles, while those in the previous studies in Mymensingh, Bangladesh, are shown with triangles (2022) and diamonds (2018–2016), respectively. L. wolffii prototype strain Khorat-H2 is indicated by a closed square. Bootstrap values more than 80% are shown. The scale bar represents the genetic distance. Subclusters are shown on the right using the designations by Guglielmini et al. (P1, P2, and S1/S2) [24] with former designations (pathogenic, intermediate, and non-pathogenic). A cluster of L. wolffii and closely related species to L. wolffii in the P2 subclade are shown by a vertical line.
3.3. Sociodemographics, Clinical Findings, and Risk Factors of Leptospirosis
Confirmed leptospirosis patients were mostly male (93%), aged 15–60 years (93%), and lived in rural area (92%), in low socioeconomic conditions (poor) (91%), with half of the patients being farmers (Table 2). The peak of leptospirosis cases was found in September, the end of the rainy (monsoon) season, though cases occurred throughout the year. Geographically, patients’ residences were distributed across Mymensingh district and neighboring regions, with higher case concentrations (5–6 patients) observed particularly in Trishal and Fulbaria upazilas of Mymensingh district, and Madhupur upazila of the adjoining Tangail district (Figure 1).

Table 2.
Sociodemographic characteristics of leptospirosis patients (n = 75) and non-patients (n = 42).
Clinical symptoms of leptospirosis patients were variable, as shown in Table 3, though their incidence rates were not statistically compared with non-leptospirosis cases. Malaise (96%) and jaundice (84%) were most prominent, with also frequent incidence of myalgia (66.7%), and manifestations of gastrointestinal tract (e.g., nausea/vomiting) and musculoskeletal system (e.g., calf muscle pain). Some patients presented severe symptoms involving nervous systems (e.g., disorientation and neck stiffness) and respiratory tract (e.g., shortness of breath and hemoptysis). Leukocytosis associated with thrombocytopenia, low hemoglobin, and raised bilirubin/creatinine/ALT were commonly seen in patients (>60%), with high incidence of pus cells in urine (82.9%) (Table 3). In agreement with the laboratory findings, nephritis and hepatitis were indicated by ultrasonography for suspected cases (63% and 29%, respectively). Additionally, evidence of plural effusion was found in the four leptospirosis patients by chest X-ray and High-Resolution Computed Tomography (HRCT), indicating alveolar hemorrhage in one case.

Table 3.
Clinical features and laboratory findings in leptospirosis patients.
Among the leptospirosis cases, rural patients had high frequencies of exposure to mud or wet soil (78.3%) and standing water (75.4%), often due to activities like working in paddy fields (56.5%), fishing (39.1%), and swimming (33.3%). They also had significant animal exposure, particularly to cattle (75.4%), dogs (73.9%), and rodents (65.2%). Both rural and urban patients were mostly affected by heavy rain, while involvement of wound and travel history was less prominent (Table 4).

Table 4.
Risk factors of leptospirosis patients living in rural and urban areas (n = 75).
4. Discussion
Leptospirosis is one of the major globally widespread zoonotic diseases, causing a presumptive high disease burden in developing countries. In Bangladesh, a high seroprevalence of Leptospira (38%) evaluated by the microscopic agglutination test (MAT) was reported in a rural flood-prone area in the early 1990s [2]. Thereafter, in Dhaka (2001), the prevalence of leptospirosis diagnosed among febrile patients was reported as 18% during a dengue outbreak [25], and 8.4% among febrile patients in a low-income area [13]. In Mymensingh, a city of north-central Bangladesh, leptospirosis was diagnosed by serological tests and PCR in 17.6–47% of suspected febrile patients from 2018 to 2022 [15,16,17]. In the present study conducted in the same study site/hospital (March 2024–February 2025), 64% of suspected cases examined were diagnosed as leptospirosis by PCR or IgM ELISA (PCR-positive rate, 29.1%). Though detection rates might be affected by the number of cases, differences in detection methods, and selection of cases via differential diagnosis, a series of studies in Mymensingh indicated that leptospirosis has been endemically persisting at high prevalence as a febrile disease in this region.
In South Asia and Southeast Asia, L. interrogans has been described as the most common species, as summarized in Table 5. Other species often identified are L. borgpetersenii, L. kirschneri, and L. weilii, though L. wolffii has been rarely reported. However, in our present study, only L. wolffii was identified from samples selected throughout the study period. In Mymensingh, this species was first reported in only one sample with other common species (L. interrogans) in 2018 [15]. Thereafter, L wolffii was predominantly detected in 2019 [16], with the sole detection of L. wolffii from 2021 until 2024 [17], including our present study. This suggests that L. wolffii has been endemic to north-central Bangladesh, at least since 2019. L. wolffii was assigned as a new species for the type strain Khorat-H2 from a patient in Thailand [23]. L. wolffii was originally classified into a group with intermediate pathogenicity [26] and genetically assigned to pathogenic subclade P2 [10], which was further grouped into P2-2 subgroup as a sole member, distinct from the other two subgroups P2-1 (e.g., L. fainei) and P2-3 (e.g., L. licarasiae) [24]. Detection of L. wolffii from humans was reported in Iran, including a study with its dominance [27,28], India [29], Malaysia [30], Ecuador [31], and Argentina [32]. Recently, in northeastern India (Sikkim and Assam) located near Bangladesh, a fatal case of L. wolffii infection [33] and an outbreak of leptospirosis due to L. wolffii were reported [34]. This species has also been detected in animals (cow, goat, buffalo, dog, raccoon dog, and Eurasian otter) in Iran [28], India [29], and South Korea [35]. Furthermore, L. wolffii DNA was detected in environmental water and soil in Malaysia [36,37,38], Thailand [39,40], and a southern island in Japan [41]. These findings suggest that L. wolffii may be distributed to humans and animals primarily in a wide region from eastern to western Asia. Although its incidence in humans with leptospirosis appears to be low, except in Bangladesh (Mymensingh) and Iran, monitoring of L. wolffii may be necessary in Asia due to its potential for widespread distribution in the environment.

Table 5.
Major species of Leptospira in humans in South and Southeast Asia reported in representative studies.
Little is known about the clinical features of leptospirosis due to L. wolffii, as well as other Leptospira previously referred to as “intermediate pathogenic group”, as it has been just described as being associated with mild disease [52] or minimal/no disease [26]. In the present study in 2024 and the previous study in 2021–2022 [17] in the same study site in Bangladesh, only L wolffii was identified, though not all the samples could be analyzed. Accordingly, clinical and demographic findings on cases obtained from these studies are deemed to generally represent those due to L. wolffii infection. In these studies, jaundice, nausea/vomiting, and myalgia were found in 66–84% of cases, with 15% having conjunctival suffusion. In contrast, among leptospirosis outbreaks worldwide, jaundice and myalgia were found in about 50% and 4%, respectively [53]. In the early studies in Bangladesh (2001), jaundice and conjunctival suffusion were rare (5% and 2%, respectively), while myalgia was common (67–85%) [13,25]. In a study in Malaysia where L. interrogans and L kirschneri (P1) were dominant, gastrointestinal signs (59%) and respiratory signs (46%) were common, with myalgia being less frequent (30%) [30]. In our present study, gastrointestinal symptoms (nausea/vomiting, diarrhea, and abdominal pain) were also frequently reported, along with respiratory symptoms such as cough and shortness of breath. Relevant laboratory tests revealed the high incidence of elevated serum bilirubin, creatinine, thrombocytopenia, and leukocytosis, with the ultrasonographic findings of nephritis, hepatitis, and ascites indicating a severe systemic infection with multi-organ involvement. Hemoptysis was present in three patients; among them, one patient showed evidence of pulmonary hemorrhage in HRCT, suggesting severe pulmonary hemorrhagic syndrome (SPHS), a serious complication of leptospirosis. These findings suggest that L. wolffii may be responsible for severe diseases comparable to leptospirosis caused by pathogenic Leptospira. For other intermediate pathogenic species, available information is limited, though mild to severe cases were reported for L. fainei, L. liceraciae, and L. venezuelensis [54,55,56]. Phylogenetically, L. wolffii is somewhat distinct from other species among the P2 subclade [24], with an almost similar profile of virulence factors [57]. Therefore, among the P2 subclade, L. wolffii is suggested to have high pathogenicity to humans, similar to the leptospiral species of the P1 subclade. We observed higher rates of genetic and serological detection of Leptospira in the early and late stages of disease, respectively. This was also described previously [17,58], being in line with the kinetics of leptospiral cells and antibody in the blood of patients [59], suggesting that the immune response to Leptospira is common irrespective of its species.
In the present study, the peak incidence of leptospirosis was observed in the end of rainy season (September), which had been similarly reported in Thailand [60] and India [61]. Such seasonality of leptospirosis may be related to the risk factors of the disease found in the present study, exposure to standing water, mud/wet soil, and contact with rodents and cattle, which have also been described as environmental factors of this disease [62]. L. wolffii, which was dominant in human cases in this study, has been commonly detected in various animal species, environmental water, and soil, as described above. Therefore, for prevention of leptospirosis in north-central Bangladesh, reducing the opportunity to come into contact with the risk factors may be primarily important, particularly in rural areas during the rainy season.
The present study has some limitations. Due to a limited study period, the number of suspected cases was relatively smaller for more accurate evaluation of the prevalence, clinical features, and risk factors of leptospirosis in the study site. Because sufficient amount of blood samples was not obtained from some patients, serum was used for nested PCR as well as IgM ELISA for all the subjects, and positive controls of target genes were not available, which might lead to a potential underestimation of positive cases. However, our present and previous studies [16,17] strongly suggested the predominance of L. wolffii belonging to the P2 subclade and its involvement in severe symptoms in humans in north-central Bangladesh. Accordingly, further studies may be required to determine the prevalence of L. wolffii in other divisions/regions in Bangladesh as well as South Asian countries, and its relevance to clinical symptoms in human cases. In addition, studying the possible distribution of L. wolffii in animal species and the environment could be useful to devise preventive measures for leptospirosis in the endemic areas.
5. Conclusions
In conclusion, the present study revealed the high prevalence of leptospirosis in north-central Bangladesh due to L. wolffii (P2 subclade), which may be persisting endemically as a dominant species. Furthermore, the probable association of L. wolffii with severe symptoms was suggested. These observations underscore the importance of P2 subclade Leptospira as a significant and probably emerging cause of human leptospirosis. The risk factors of L. wolffii-associated leptospirosis were generally related to a contaminated environment, as observed for other virulent Leptospira. Therefore, for the control of leptospirosis in Bangladesh, further surveillance and clinical studies are necessary, as well as improvement of the hygienic conditions of the population.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/tropicalmed10100290/s1, Table S1: Selected Leptospira samples sequenced for 16S rRNA gene; Table S2: Nucleotide sequence identities (%) of partial 16S rRNA among Leptospira samples; Table S3: Nucleotide sequence identities (%) of partial 16S rRNA among Leptospira samples in the present study and previously identified strains in Bangladesh, the prototype strain (Khorat2), and worldwide circulating strains (Malaysia, Ecuador, and Iran).
Author Contributions
Conceptualization, S.A.T. and S.K.P.; supervision, N.H.; methodology, S.A.T., S.K.P. and M.S.A.; investigation, S.A.T., M.R.N., S.J.T., P.I.D., A.A.M. and M.S.A.; data curation, S.A.T., M.S.A. and N.K.; resources, S.A.T., S.K.P., M.R.H., S.N.N., A.I., S.A.N., M.S.I. and S.R.S.; writing—original draft preparation, S.A.T.; writing—review and editing, M.S.A. and N.K.; funding, N.K.; supervision and project administration, N.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by JSPS (Japan Society for the Promotion of Science) KAKENHI, Grant Number JP23KK0171.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Mymensingh Medical College (IRB approval no. MMC/IRB/2024/667, approval date: 24 June 2024).
Informed Consent Statement
Written informed consent was obtained from all the patients who participated in this study.
Data Availability Statement
The data presented in this study are available from the corresponding author upon reasonable request, due to ethical restrictions on data sharing.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Pappas, G.; Papadimitriou, P.; Siozopoulou, V.; Christou, L.; Akritidis, N. The globalization of leptospirosis: Worldwide incidence trends. Int. J. Infect. Dis. 2008, 12, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Morshed, M.G.; Konishi, H.; Terada, Y.; Arimitsu, Y.; Nakazawa, T. Seroprevalence of leptospirosis in a rural flood prone district of Bangladesh. Epidemioly Infect. 1994, 112, 527–531. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Costa, F.; Hagan, J.E.; Calcagno, J.; Kane, M.; Torgerson, P.; Martinez-Silveira, M.S.; Stein, C.; Abela-Ridder, B.; Ko, A.I. Global Morbidity and Mortality of Leptospirosis: A Systematic Review. PLoS Negl. Trop. Dis. 2015, 9, e0003898. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.J.; Paris, D.H.; Newton, P.N. A Systematic Review of the Mortality from Untreated Leptospirosis. PLoS Negl. Trop. Dis. 2015, 9, e0003866. [Google Scholar] [CrossRef]
- Lau, C.L.; Smythe, L.D.; Craig, S.B.; Weinstein, P. Climate change, flooding, urbanisation and leptospirosis: Fuelling the fire? Trans. R. Soc. Trop. Med. Hyg. 2010, 104, 631–638. [Google Scholar] [CrossRef]
- Karpagam, K.B.; Ganesh, B. Leptospirosis: A neglected tropical zoonotic infection of public health importance-an updated review. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 835–846. [Google Scholar] [CrossRef]
- Putz, E.J.; Nally, J.E. Investigating the Immunological and Biological Equilibrium of Reservoir Hosts and Pathogenic Leptospira: Balancing the Solution to an Acute Problem? Front. Microbiol. 2020, 11, 2005. [Google Scholar] [CrossRef]
- Sarabandi, R.; Sarani, A.; Rasekh, M.; Sadr, S.; Abdollahpour, G.; Nazemian, S.; Khajehmohammadi, M.; Borji, H. Serovar typing and risk factors of Leptospira infection in dromedary camels (Camelus dromedarius) of Sistan and Baluchestan, Iran: An exploratory study, with a worldwide update of Leptospira infections in camels. IJID One Health 2025, 7, 100065. [Google Scholar] [CrossRef]
- Hagedoorn, N.N.; Maze, M.J.; Carugati, M.; Cash-Goldwasser, S.; Allan, K.J.; Chen, K.; Cossic, B.; Demeter, E.; Gallagher, S.; German, R.; et al. Global distribution of Leptospira serovar isolations and detections from animal host species: A systematic review and online database. Trop. Med. Int. Health 2024, 29, 161–172. [Google Scholar] [CrossRef]
- Vincent, A.T.; Schiettekatte, O.; Goarant, C.; Neela, V.K.; Bernet, E.; Thibeaux, R.; Ismail, N.; Mohd Khalid, M.K.N.; Amran, F.; Masuzawa, T.; et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 2019, 13, e0007270. [Google Scholar] [CrossRef]
- Arent, Z.; Pardyak, L.; Dubniewicz, K.; Plachno, B.; Kotula-Balak, M. Leptospira taxonomy: Then and now. Med. Water 2022, 78, 489–496. [Google Scholar] [CrossRef]
- Caimi, K.; Ruybal, P. Leptospira spp., a genus in the stage of diversity and genomic data expansion. Infect. Genet. Evol. 2020, 81, 104241. [Google Scholar] [CrossRef] [PubMed]
- Kendall, E.A.; LaRocque, R.C.; Bui, D.M.; Galloway, R.; Ari, M.D.; Goswami, D.; Breiman, R.F.; Luby, S.; Abdullah Brooks, W. Leptospirosis as a cause of fever in urban Bangladesh. Am. J. Trop. Med. Hyg. 2010, 82, 1127–1130. [Google Scholar] [CrossRef] [PubMed]
- Tamanna, S.; Rahman, F.; Tang, T.H.; Ahmed, S.A.; Ang, K.C.; Zannat, K.; Jilani, M.S.A.; Mohiuddin, M.; Haq, J.A. Seroprevalence of Leptospira infection in selected rural and urban areas of Bangladesh by rLipL32 based ELISA. IMC J. Med. Sci. 2017, 11, 50–55. Available online: https://doaj.org/article/89c72e3fa7824f7aac1b20accdd33a64 (accessed on 20 April 2025). [CrossRef][Green Version]
- Aziz, M.A.; Aung, M.S.; Paul, S.K.; Ahmed, S.; Haque, N.; Roy, S.; Al Amin, M.; Paul, A.; Miah, M.A.H.; Alam, M.K.; et al. First molecular identification of two Leptospira species (Leptospira interrogans and Leptospira wolffii) in Bangladesh. New Microb. New Infect. 2019, 31, 100570. [Google Scholar] [CrossRef]
- Rahman, S.; Paul, S.K.; Aung, M.S.; Ahmed, S.; Haque, N.; Raisul, M.N.I.; Choity, J.K.; Nila, S.S.; Ara, H.; Roy, S.; et al. Predominance of Leptospira wolffii in north-central Bangladesh, 2019. New Microb. New Infect. 2020, 38, 100765. [Google Scholar] [CrossRef]
- Sultana, M.; Paul, S.K.; Nasreen, S.A.; Haque, N.; Hasan, M.K.; Islam, A.; Nila, S.S.; Jahan, A.; Sathi, F.A.; Hossain, T.; et al. Epidemiological Features of Leptospirosis and Identification of Leptospira wolffii as a Persistently Prevailing Species in North-Central Bangladesh. Infect. Dis. Rep. 2024, 16, 638–649. [Google Scholar] [CrossRef]
- Dhawan, S.; Althaus, T.; Lubell, Y.; Suwancharoen, D.; Blacksell, S.D. Evaluation of the Panbio Leptospira IgM ELISA among Outpatients Attending Primary Care in Southeast Asia. Am. J. Trop. Med. Hyg. 2021, 104, 1777–1781. [Google Scholar] [CrossRef]
- Goarant, C.; Girault, D.; Thibeaux, R.; Soupé-Gilbert, M.E. Isolation and Culture of Leptospira from Clinical and Environmental Samples. In Leptospira spp. Methods in Molecular Biology; Koizumi, N., Picardeau, M., Eds.; Humana: New York, NY, USA, 2020; Volume 2134, pp. 1–9. [Google Scholar] [CrossRef]
- Djadid, N.D.; Ganji, Z.F.; Gouya, M.M.; Rezvani, M.; Zakeri, S. A simple and rapid nested polymerase chain reaction-restriction fragment length polymorphism technique for differentiation of pathogenic and nonpathogenic Leptospira spp. Diagn. Microbiol. Infect. Dis. 2009, 63, 251–256. [Google Scholar] [CrossRef]
- Koizumi, N.; Nakajima, C.; Harunari, T.; Tanikawa, T.; Tokiwa, T.; Uchimura, E.; Furuya, T.; Mingala, C.N.; Villanueva, M.A.; Ohnishi, M.; et al. A new loop-mediated isothermal amplification method for rapid, simple, and sensitive detection of Leptospira spp. in urine. J. Clin. Microbiol. 2012, 50, 2072–2074. [Google Scholar] [CrossRef]
- Palaniappan, R.U.; Chang, Y.F.; Chang, C.F.; Pan, M.J.; Yang, C.W.; Harpending, P.; McDonough, S.P.; Dubovi, E.; Divers, T.; Qu, J.; et al. Evaluation of lig-based conventional and real time PCR for the detection of pathogenic leptospires. Mol. Cell. Probes 2005, 19, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Slack, A.T.; Kalambaheti, T.; Symonds, M.L.; Dohnt, M.F.; Galloway, R.L.; Steigerwalt, A.G.; Chaicumpa, W.; Bunyaraksyotin, G.; Craig, S.; Harrower, B.J.; et al. Leptospira wolffii sp. nov., isolated from a human with suspected leptospirosis in Thailand. Int. J. Syst. Evol. Microbiol. 2008, 58, 2305–2308. [Google Scholar] [CrossRef] [PubMed]
- Guglielmini, J.; Bourhy, P.; Schiettekatte, O.; Zinini, F.; Brisse, S.; Picardeau, M. Genus-wide Leptospira core genome multilocus sequence typing for strain taxonomy and global surveillance. PLoS Negl. Trop. Dis. 2019, 13, e0007374. [Google Scholar] [CrossRef] [PubMed]
- LaRocque, R.C.; Breiman, R.F.; Ari, M.D.; Morey, R.E.; Janan, F.A.; Hayes, J.M.; Hossain, M.A.; Brooks, W.A.; Levett, P.N. Leptospirosis during dengue outbreak, Bangladesh. Emerg. Infect. Dis. 2005, 11, 766–769. [Google Scholar] [CrossRef]
- Haake, D.A. Spirochetes; Elsevier: Amsterdam, The Netherlands, 2009; pp. 278–292. Available online: https://micropspbgmu.ru/downloads/Spirochetes.pdf (accessed on 20 April 2025).
- Zakeri, S.; Sepahian, N.; Afsharpad, M.; Esfandiari, B.; Ziapour, P.; Djadid, N.D. Molecular epidemiology of leptospirosis in northern Iran by nested polymerase chain reaction/restriction fragment length polymorphism and sequencing methods. Am. J. Trop. Med. Hyg. 2010, 82, 899–903. [Google Scholar] [CrossRef]
- Zakeri, S.; Khorami, N.; Ganji, Z.F.; Sepahian, N.; Malmasi, A.A.; Gouya, M.M.; Djadid, N.D. Leptospira wolffii, a potential new pathogenic Leptospira species detected in human, sheep and dog. Infect. Genet. Evol. 2010, 10, 273–277. [Google Scholar] [CrossRef]
- Balamurugan, V.; Gangadhar, N.L.; Mohandoss, N.; Thirumalesh, S.R.; Dhar, M.; Shome, R.; Krishnamoorthy, P.; Prabhudas, K.; Rahman, H. Characterization of leptospira isolates from animals and humans: Phylogenetic analysis identifies the prevalence of intermediate species in India. SpringerPlus 2013, 2, 362. [Google Scholar] [CrossRef]
- Philip, N.; Bahtiar Affendy, N.; Ramli, S.N.A.; Arif, M.; Raja, P.; Nagandran, E.; Renganathan, P.; Taib, N.M.; Masri, S.N.; Yuhana, M.Y.; et al. Leptospira interrogans and Leptospira kirschneri are the dominant Leptospira species causing human leptospirosis in Central Malaysia. PLoS Negl. Trop. Dis. 2020, 14, e0008197. [Google Scholar] [CrossRef]
- Chiriboga, J.; Barragan, V.; Arroyo, G.; Sosa, A.; Birdsell, D.N.; España, K.; Mora, A.; Espín, E.; Mejía, M.E.; Morales, M.; et al. High Prevalence of Intermediate Leptospira spp. DNA in Febrile Humans from Urban and Rural Ecuador. Emerg. Infect. Dis. 2015, 21, 2141–2147. [Google Scholar] [CrossRef]
- Chiani, Y.; Jacob, P.; Varni, V.; Landolt, N.; Schmeling, M.F.; Pujato, N.; Caimi, K.; Vanasco, B. Isolation and clinical sample typing of human leptospirosis cases in Argentina. Infect. Genet. Evol. 2016, 37, 245–251. [Google Scholar] [CrossRef]
- Gurung, S.; Tewari, D.; Sherpa, U.; Chhophel, T.P.; Siddique, A.I.; Sarmah, N.; Borkakoty, B. Fatal case of Leptospira wolffii infection in Sikkim, India: An autochthonous case from a new geographical region. Indian J. Med. Microbiol. 2025, 54, 100813. [Google Scholar] [CrossRef]
- Siddique, A.I.; Borkakoty, B.; Bali, N.K.; Sarmah, N.; Dutta, M.; Jakharia, A.; Rahman, M.; Bhowmick, I.P.; Kalimuthusamy, N. Outbreak of leptospirosis caused by Leptospira wolffii with HAV coinfections in lower Assam, Northeast India in 2024: Clinical impact and public health implications. Indian J. Med. Microbiol. 2025, 57, 100923. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Eo, K.Y.; Lee, W.S.; Kimura, J.; Yamamoto, N. DNA-based detection of Leptospira wolffii, Giardia intestinalis and Toxoplasma gondii in environmental feces of wild animals in Korea. J. Vet. Med. Sci. 2021, 83, 850–854. [Google Scholar] [CrossRef] [PubMed]
- Azali, M.A.; Yean Yean, C.; Harun, A.; Aminuddin Baki, N.N.; Ismail, N. Molecular Characterization of Leptospira spp. in Environmental Samples from North-Eastern Malaysia Revealed a Pathogenic Strain, Leptospira alstonii. J. Trop. Med. 2016, 2016, 2060241. [Google Scholar] [CrossRef] [PubMed]
- Pui, C.F.; Bilung, L.M.; Apun, K.; Su’ut, L. Diversity of Leptospira spp. in Rats and Environment from Urban Areas of Sarawak, Malaysia. J. Trop. Med. 2017, 2017, 3760674. [Google Scholar] [CrossRef]
- Ling, B.L.; Tay, Z.E.; Philip, N. Detection of Leptospira in environmental samples of wet markets and paddy fields in Penang, Malaysia. Trop. Biomed. 2025, 42, 51–57. [Google Scholar] [CrossRef]
- Chaiwattanarungruengpaisan, S.; Suwanpakdee, S.; Sangkachai, N.; Chamsai, T.; Taruyanon, K.; Thongdee, M. Potentially Pathogenic Leptospira Species Isolated from a Waterfall in Thailand. Jpn. J. Infect. Dis. 2018, 71, 65–67. [Google Scholar] [CrossRef]
- Krairojananan, P.; Wasuworawong, K.; Leepitakrat, S.; Monkanna, T.; Wanja, E.W.; Davidson, S.A.; Poole-Smith, B.K.; McCardle, P.W.; Mann, A.; Lindroth, E.J. Leptospirosis Risk Assessment in Rodent Populations and Environmental Reservoirs in Humanitarian Aid Settings in Thailand. Microorganisms 2024, 13, 29. [Google Scholar] [CrossRef]
- Sato, Y.; Hermawan, I.; Kakita, T.; Okano, S.; Imai, H.; Nagai, H.; Kimura, R.; Yamashiro, T.; Kajita, T.; Toma, C. Analysis of human clinical and environmental Leptospira to elucidate the eco-epidemiology of leptospirosis in Yaeyama, subtropical Japan. PLoS Negl. Trop. Dis. 2022, 16, e0010234. [Google Scholar] [CrossRef]
- Mendoza, M.V.; Rivera, W.L. Application of simplified MLST scheme for direct typing of clinical samples from human leptospirosis cases in a tertiary hospital in the Philippines. PLoS ONE 2021, 16, e0258891. [Google Scholar] [CrossRef]
- Thaipadungpanit, J.; Wuthiekanun, V.; Chierakul, W.; Smythe, L.D.; Petkanchanapong, W.; Limpaiboon, R.; Apiwatanaporn, A.; Slack, A.T.; Suputtamongkol, Y.; White, N.J.; et al. A dominant clone of Leptospira interrogans associated with an outbreak of human leptospirosis in Thailand. PLoS Negl. Trop. Dis. 2007, 1, e56. [Google Scholar] [CrossRef]
- Thipmontree, W.; Suputtamongkol, Y.; Tantibhedhyangkul, W.; Suttinont, C.; Wongswat, E.; Silpasakorn, S. Human leptospirosis trends: Northeast Thailand, 2001–2012. Int. J. Environ. Res. Public Health 2014, 11, 8542–8551. [Google Scholar] [CrossRef] [PubMed]
- Grillová, L.; Robinson, M.T.; Chanthongthip, A.; Vincent, A.T.; Nieves, C.; Oppelt, J.; Mariet, J.F.; Lorioux, C.; Vongsouvath, M.; Mayxay, M.; et al. Genetic diversity of Leptospira isolates in Lao PDR and genome analysis of an outbreak strain. PLoS Negl. Trop. Dis. 2021, 15, e0010076. [Google Scholar] [CrossRef] [PubMed]
- Mai, L.T.P.; Dung, L.P.; Mai, T.N.P.; Hanh, N.T.M.; Than, P.D.; Tran, V.D.; Quyet, N.T.; Hai, H.; Ngoc, D.B.; Hai, P.T.; et al. Characteristics of human leptospirosis in three different geographical and climatic zones of Vietnam: A hospital-based study. Int. J. Infect. Dis. 2022, 120, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Rafizah, A.A.; Aziah, B.D.; Azwany, Y.N.; Imran, M.K.; Rusli, A.M.; Nazri, S.M.; Nikman, A.M.; Nabilah, I.; Asma’, H.S.; Zahiruddin, W.M.; et al. A hospital-based study on seroprevalence of leptospirosis among febrile cases in northeastern Malaysia. Int. J. Infect. Dis. 2013, 17, e394–e397. [Google Scholar] [CrossRef]
- Philip, N.; Ahmed, K. Leptospirosis in Malaysia: Current status, insights, and future prospects. J. Physiol. Anthropol. 2023, 42, 30. [Google Scholar] [CrossRef]
- Chaudhry, R.; Das, A.; Premlatha, M.M.; Choudhary, A.; Chourasia, B.K.; Chandel, D.S.; Dey, A.B. Serological & molecular approaches for diagnosis of leptospirosis in a tertiary care hospital in north India: A 10-year study. Indian J. Med. Res. 2013, 137, 785–790. [Google Scholar] [PubMed]
- Pinto, G.V.; Senthilkumar, K.; Rai, P.; Kabekkodu, S.P.; Karunasagar, I.; Kumar, B.K. Identification of Dominant Leptospira Serogroups among Leptospirosis Cases and Their Clinical Outcomes: A Prospective Hospital-Based Study in Mangaluru, India. Am. J. Trop. Med. Hyg. 2024, 110, 1230–1236. [Google Scholar] [CrossRef]
- Jayasundara, D.; Senavirathna, I.; Warnasekara, J.; Gamage, C.; Siribaddana, S.; Kularatne, S.A.M.; Matthias, M.; Mariet, J.F.; Picardeau, M.; Agampodi, S.; et al. 12 Novel clonal groups of Leptospira infecting humans in multiple contrasting epidemiological contexts in Sri Lanka. PLoS Negl. Trop. Dis. 2021, 15, e0009272, Erratum in PLoS Negl. Trop. Dis. 2021, 15, e0009471. [Google Scholar] [CrossRef]
- Trott, D.J.; Abraham, S.; Adler, B. Antimicrobial Resistance in Leptospira, Brucella, and Other Rarely Investigated Veterinary and Zoonotic Pathogens. Microbiol. Spectr. 2018, 6, 10-1128. [Google Scholar] [CrossRef]
- Munoz-Zanzi, C.; Groene, E.; Morawski, B.M.; Bonner, K.; Costa, F.; Bertherat, E.; Schneider, M.C. A systematic literature review of leptospirosis outbreaks worldwide, 1970–2012. Rev. Panam. Salud. Publica 2020, 44, e78. [Google Scholar] [CrossRef] [PubMed]
- Arzouni, J.P.; Parola, P.; La Scola, B.; Postic, D.; Brouqui, P.; Raoult, D. Human infection caused by Leptospira fainei. Emerg. Infect. Dis. 2002, 8, 865–868. [Google Scholar] [CrossRef] [PubMed]
- Tsuboi, M.; Koizumi, N.; Hayakawa, K.; Kanagawa, S.; Ohmagari, N.; Kato, Y. Imported Leptospira licerasiae Infection in Traveler Returning to Japan from Brazil. Emerg. Infect. Dis. 2017, 23, 548–549. [Google Scholar] [CrossRef] [PubMed]
- Caraballo, L.; Rangel, Y.; Reyna-Bello, A.; Muñoz, M.; Figueroa-Espinosa, R.; Sanz-Rodriguez, C.E.; Guerrero, E.; Loureiro, C.L.; Liu, Q.; Takiff, H.E. Outbreak of Intermediate Species Leptospira venezuelensis Spread by Rodents to Cows and Humans in L. interrogans-Endemic Region, Venezuela. Emerg. Infect. Dis. 2024, 30, 1514–1522. [Google Scholar] [CrossRef]
- Lata, K.S.; Kumar, S.; Vindal, V.; Patel, S.; Das, J. A core and pan gene map of Leptospira genus and its interactions with human host. Microb. Pathog. 2022, 162, 105347. [Google Scholar] [CrossRef]
- de Abreu Fonseca, C.; Teixeira de Freitas, V.L.; Caló Romero, E.; Spinosa, C.; Arroyo Sanches, M.C.; da Silva, M.V.; Shikanai-Yasuda, M.A. Polymerase chain reaction in comparison with serological tests for early diagnosis of human leptospirosis. Trop. Med. Int. Health 2006, 11, 1699–1707. [Google Scholar] [CrossRef]
- Ko, A.I.; Goarant, C.; Picardeau, M. Leptospira: The dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat. Rev. Microbiol. 2009, 7, 736–747. [Google Scholar] [CrossRef]
- Chadsuthi, S.; Chalvet-Monfray, K.; Geawduanglek, S.; Wongnak, P.; Cappelle, J. Spatial-temporal patterns and risk factors for human leptospirosis in Thailand, 2012–2018. Sci. Rep. 2022, 12, 5066. [Google Scholar] [CrossRef]
- Gupta, N.; Wilson, W.; Ravindra, P. Leptospirosis in India: A systematic review and meta-analysis of clinical profile, treatment and outcomes. Infez. Med. 2023, 31, 290–305. [Google Scholar]
- Baharom, M.; Ahmad, N.; Hod, R.; Ja’afar, M.H.; Arsad, F.S.; Tangang, F.; Ismail, R.; Mohamed, N.; Mohd Radi, M.F.; Osman, Y. Environmental and Occupational Factors Associated with Leptospirosis: A Systematic Review. Heliyon 2023, 10, e23473. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).