Development of a Predictive Model for N-Dealkylation of Amine Contaminants Based on Machine Learning Methods
Abstract
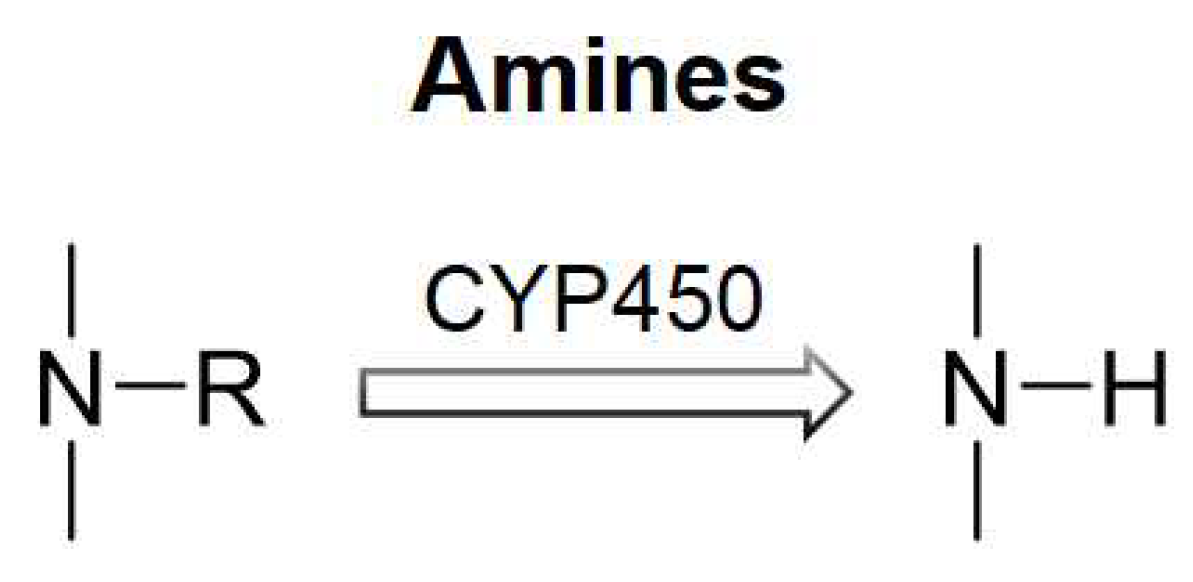
:1. Introduction
2. Materials and Methods
2.1. Dataset Establishment
2.2. Feature Selection
2.3. Machine Learning Algorithms Utilized
2.4. Evaluation of Model Performance
3. Results
3.1. Data Collection and Feature Selection
3.2. Single Machine Learning Prediction Model
3.3. Model Explanation
- SlogP_VSA2 defined using MoeType functions using electrostatic potential on the van der Waals surface of an organic spacer [25].
- AATSC2V defined using autocorrelation functions using averaged and centered Moreau–Broto autocorrelation of lag 2 weighted by vdw volume [25].
- ETA_dBeta defined using ETA functions using with the difference between contributions from sigma bonds and non-sigma bonds (pi-bonds) [25].
- ATSC1i defined using autocorrelation functions using averaged centered Broto–Moreau autocorrelation weighted by van der Waals volumes [35].
3.4. Voting Ensemble Learning (VEL) Approach for Final Predictions
3.5. Application Domain
3.6. Misclassification Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Trowbridge, A.; Walton, S.M.; Gaunt, M.J. New strategies for the transition-metal catalyzed synthesis of aliphatic amines. Chem. Rev. 2020, 120, 2613–2692. [Google Scholar] [CrossRef]
- Gulde, R.; Meier, U.; Schymanski, E.L.; Kohler, H.P.E.; Helbling, D.E.; Derrer, S.; Rentsch, D.; Fenner, K. Systematic exploration of biotransformation reactions of amine-containing micropollutants in activated sludge. Environ. Sci. Technol. 2016, 50, 2908–2920. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Wu, Y.; He, Q.; Sun, P.; Chen, Q.; Xia, C.; Huang, Y.; Yang, J.; Liu, M. Ubiquity of amino accelerators and antioxidants in road dust from multiple land types: Targeted and nontargeted analysis. Environ. Sci. Technol. 2023, 57, 10361–10372. [Google Scholar] [CrossRef]
- Deng, Q.; He, B.; Shen, M.; Ge, J.; Du, B.; Zeng, L. First Evidence of Hindered Amine Light Stabilizers As Abundant, Ubiquitous, Emerging Pollutants in Dust and Air Particles: A New Concern for Human Health. Environ. Sci. Technol. 2024, 58, 1349–1358. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.; Okazaki, O.; Seto, Y.; Macdonald, T. Radical cation intermediates in N-dealkylation reactions. Xenobiotica 1995, 25, 689–709. [Google Scholar] [CrossRef]
- Shaik, S.; Cohen, S.; Wang, Y.; Chen, H.; Kumar, D.; Thiel, W. P450 Enzymes: Their Structure, Reactivity, and Selectivity Modeled by QM/MM Calculations. Chem. Rev. 2010, 110, 949–1017. [Google Scholar] [CrossRef]
- Dang, N.L.; Hughes, T.B.; Miller, G.P.; Swamidass, S.J. Computationally assessing the bioactivation of drugs by N-dealkylation. Chem. Res. Toxicol. 2018, 31, 68–80. [Google Scholar] [CrossRef]
- Wong, D.T.; Bymaster, F.P.; Reid, L.R.; Mayle, D.A.; Krushinski, J.H.; Robertson, D.W. Norfluoxetine enantiomers as inhibitors of serotonin uptake in rat brain. Neuropsychopharmacology 1993, 8, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Iverson, S.L.; Uetrecht, J.P. Identification of a reactive metabolite of terbinafine: Insights into terbinafine-induced hepatotoxicity. Chem. Res. Toxicol. 2001, 14, 175–181. [Google Scholar] [CrossRef]
- Najmi, A.A.; Bischoff, R.; Permentier, H.P. N-dealkylation of amines. Molecules 2022, 27, 3293. [Google Scholar] [CrossRef]
- Koleva, Y.K.; Madden, J.C.; Cronin, M.T. Formation of categories from structure-activity relationships to Allow read-across for risk assessment: Toxicity of α, β-unsaturated carbonyl compounds. Chem. Res. Toxicol. 2008, 21, 2300–2312. [Google Scholar] [CrossRef] [PubMed]
- Gheni, S.A.; Ali, M.M.; Ta, G.C.; Harbin, H.J.; Awad, S.A. Toxicity, hazards, and safe handling of primary aromatic amines. ACS Chem. Health Saf. 2023, 31, 8–21. [Google Scholar] [CrossRef]
- Benigni, R.; Giuliani, A.; Franke, R.; Gruska, A. Quantitative structure-activity relationships of mutagenic and carcinogenic aromatic amines. Chem. Rev. 2000, 100, 3697–3714. [Google Scholar] [CrossRef] [PubMed]
- Raju, D.R.; Kumar, A.; Naveen, B.; Shetty, A.; Akshai, P.; Kumar, R.P.; Lalitha, R.; Sigamani, G. Extensive modelling and quantum chemical study of sterol C-22 desaturase mechanism: A commercially important cytochrome P450 family. Catal. Today 2022, 397, 50–62. [Google Scholar] [CrossRef]
- Léon, I.; Tasinato, N.; Spada, L.; Alonso, E.R.; Mata, S.; Balbi, A.; Puzzarini, C.; Alonso, J.L.; Barone, V. Looking for the Elusive Imine Tautomer of Creatinine: Different States of Aggregation Studied by Quantum Chemistry and Molecular Spectroscopy. ChemPlusChem 2021, 86, 1374–1386. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Zhang, H.; Ding, W.; Song, R.; Han, Y.; Yu, H.; Paneth, P. Theoretical Kinetic Isotope Effects in Establishing the Precise Biodegradation Mechanisms of Organic Pollutants. Environ. Sci. Technol. 2023, 57, 4915–4929. [Google Scholar] [CrossRef]
- Chai, L.; Zhang, H.; Song, R.; Yang, H.; Yu, H.; Paneth, P.; Kepp, K.P.; Akamatsu, M.; Ji, L. Precision biotransformation of emerging pollutants by human cytochrome P450 using computational–experimental synergy: A case study of tris (1, 3-dichloro-2-propyl) phosphate. Environ. Sci. Technol. 2021, 55, 14037–14050. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, x.; Song, R.; Ding, W.; Li, F.; Ji, L. Emerging metabolic profiles of sulfonamide antibiotics by cytochromes P450: A computational–experimental synergy study on emerging pollutants. Environ. Sci. Technol. 2023, 57, 5368–5379. [Google Scholar] [CrossRef]
- Zorn, K.M.; Foil, D.H.; Lane, T.R.; Russo, D.P.; Hillwalker, W.; Feifarek, D.J.; Jones, F.; Klaren, W.D.; Brinkman, A.M.; Ekins, S. Machine learning models for estrogen receptor bioactivity and endocrine disruption prediction. Environ. Sci. Technol. 2020, 54, 12202–12213. [Google Scholar] [CrossRef]
- Zhong, S.; Zhang, K.; Bagheri, M.; Burken, J.G.; Gu, A.; Li, B.; Ma, X.; Marrone, B.L.; Ren, Z.J.; Schrier, J.; et al. Machine learning: New ideas and tools in environmental science and engineering. Environ. Sci. Technol. 2021, 55, 12741–12754. [Google Scholar] [CrossRef]
- Cheng, S.; Yuan, S.; Wu, X.; Lei, T.; Ji, J.; Yin, Y.; Liu, Y.; Liu, C.; Zhang, Y.; Zhu, Y. Identification of chemicals based on locomotor tracks of Daphnia magna using deep learning. Environ. Sci. Technol. Lett. 2023, 10, 998–1003. [Google Scholar] [CrossRef]
- Adadi, A.; Berrada, M. Peeking inside the black-box: A survey on explainable artificial intelligence (XAI). IEEE Access 2018, 6, 52138–52160. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The human metabolome database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Meijer, J.; Lamoree, M.; Hamers, T.; Antignac, J.P.; Hutinet, S.; Debrauwer, L.; Covaci, A.; Huber, C.; Krauss, M.; Walker, D.I.; et al. An annotation database for chemicals of emerging concern in exposome research. Environ. Int. 2021, 152, 106511. [Google Scholar] [CrossRef]
- Moriwaki, H.; Tian, Y.S.; Kawashita, N.; Takagi, T. Mordred: A molecular descriptor calculator. J. Cheminform. 2018, 10, 1–14. [Google Scholar] [CrossRef]
- Hall, M.A.; Holmes, G. Benchmarking attribute selection techniques for discrete class data mining. IEEE Trans. Knowl. Data Eng. 2003, 15, 1437–1447. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.Y. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 2017, 30, 3149–3157. [Google Scholar]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K. Xgboost: Extreme Gradient Boosting; R Package Version 0.4-2; Scientific Research: Atlanta, GA, USA, 2015; Volume 1, pp. 1–4.
- Gardner, M.W.; Dorling, S. Artificial neural networks (the multilayer perceptron)—A review of applications in the atmospheric sciences. Atmos. Environ. 1998, 32, 2627–2636. [Google Scholar] [CrossRef]
- Peppes, N.; Daskalakis, E.; Alexakis, T.; Adamopoulou, E.; Demestichas, K. Performance of machine learning-based multi-model voting ensemble methods for network threat detection in agriculture 4.0. Sensors 2021, 21, 7475. [Google Scholar] [CrossRef]
- Jaworska, J.; Nikolova-Jeliazkova, N.; Aldenberg, T. QSAR applicability domain estimation by projection of the training set in descriptor space: A review. Altern. Lab. Anim. 2005, 33, 445–459. [Google Scholar] [CrossRef]
- Ryu, J.Y.; Kim, H.U.; Lee, S.Y. Deep learning improves prediction of drug–drug and drug–food interactions. Proc. Natl. Acad. Sci. USA 2018, 115, E4304–E4311. [Google Scholar] [CrossRef]
- Lundberg, S. A unified approach to interpreting model predictions. arXiv 2017, arXiv:1705.07874. [Google Scholar]
- Todeschini, R.; Consonni, V. Molecular Descriptors for Chemoinformatic; Wiley-VCH: Weinheim, Germany, 2009; pp. 1–2. [Google Scholar]
- Zhang, H.; Huang, C.H. Oxidative transformation of fluoroquinolone antibacterial agents and structurally related amines by manganese oxide. Environ. Sci. Technol. 2005, 39, 4474–4483. [Google Scholar] [CrossRef]
- Paul, T.; Miller, P.L.; Strathmann, T.J. Visible-light-mediated TiO2 photocatalysis of fluoroquinolone antibacterial agents. Environ. Sci. Technol. 2007, 41, 4720–4727. [Google Scholar] [CrossRef]
- Li, X.; Liu, G.; Wang, Z.; Zhang, L.; Liu, H.; Ai, H. Ensemble multiclassification model for aquatic toxicity of organic compounds. Aquat. Toxicol. 2023, 255, 106379. [Google Scholar] [CrossRef]
- Choi, J.; Lim, K.J.; Ji, B. Robust imputation method with context-aware voting ensemble model for management of water-quality data. Water Res. 2023, 243, 120369. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Cheng, S.; Ding, W.; Huang, J.; van Eldik, R.; Ji, L. Insight into chemically reactive metabolites of aliphatic amine pollutants: A de novo prediction strategy and case study of sertraline. Environ. Int. 2024, 186, 108636. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Cheng, S.; Ge, M.; Ji, L. Evidence for the formation of 6PPD-quinone from antioxidant 6PPD by cytochrome P450. J. Hazard. Mater. 2024, 480, 136273. [Google Scholar] [CrossRef]





| Name | Range | Mean Value | Type |
|---|---|---|---|
| ATSC1i | −42.10–11.98 | −9.82 | Autocorrelation |
| AATSC2v | −12.74–24.65 | 3.39 | Autocorrelation |
| ETA_eta_B | −0.01–2.33 | 0.28 | ExtendedTopochemicalAtom |
| ETA_dBeta | −18.00–4.50 | −2.47 | ExtendedTopochemicalAtom |
| CIC3 | 0.00–2.65 | 0.83 | InformationContent |
| SMR_VSA6 | 0.00–99.36 | 19.09 | MoeType |
| SlogP_VSA2 | 5.90–260.06 | 33.18 | MoeType |
| Machine Learning Algorithm | Hyperparameter |
|---|---|
| RF | n_estimators = 4, max_depth = 3 |
| GBDT | n_estimators = 5, max_depth = 7 |
| XGB | min_child_weight = 0.125, subsample = 0.8, colsample_bytree = 0.9, learning_rate = 0.01, n_estimators = 200 |
| MLP | hidden_layer_sizes = (80, 100, 50), learning_rate_init = 0.01, max_iter = 800 |
| Single Classifier | Accuracy | Sensitivity | Specificity | Precision | F1 Score | MCC |
|---|---|---|---|---|---|---|
| RF | 88.6% | 93.1% | 83.7% | 87.7% | 0.90 | 0.76 |
| GBDT | 87.3% | 98.5% | 72.4 | 82.6% | 0.89 | 0.75 |
| XGB | 91.7% | 99.2% | 81.6 | 90.0% | 0.93 | 0.83 |
| MLP | 86.4% | 88.5% | 83.7 | 87.8% | 0.88 | 0.72 |
| Single Classifier | Accuracy | Sensitivity | Specificity | Precision | F1 Score | MCC |
|---|---|---|---|---|---|---|
| RF | 77.6% | 84.0% | 72.7% | 70.0% | 0.76 | 0.56 |
| GBDT | 81.0% | 96.0% | 69.7% | 70.6% | 0.81 | 0.66 |
| XGB | 79.3% | 100.0% | 63.6% | 67.6% | 0.80 | 0.65 |
| MLP | 79.3% | 88.0% | 72.7% | 71.0% | 0.78 | 0.60 |
| Consensus Classifier | Accuracy | Sensitivity | Specificity | Precision | F1 Score | MCC |
|---|---|---|---|---|---|---|
| RF+XGB+GBDT | 90.4% | 99.2% | 78.6% | 86.0% | 0.92 | 0.86 |
| RF+XGB+MLP | 89.5% | 93.8% | 83.7% | 88.4% | 0.91 | 0.78 |
| RF+GBDT+MLP | 89.5% | 93.8% | 83.7% | 88.4% | 0.91 | 0.78 |
| XGB+MLP+GBDT | 89.5% | 96.2% | 80.6% | 86.8% | 0.91 | 0.78 |
| Consensus Classifier | Accuracy | Sensitivity | Specificity | Precision | F1 Score | MCC |
|---|---|---|---|---|---|---|
| RF+XGB+GBDT | 86.2% | 96.0% | 78.8% | 77.4% | 0.85 | 0.74 |
| RF+XGB+MLP | 84.5% | 96.0% | 75.8% | 75.0% | 0.84 | 0.71 |
| RF+GBDT+MLP | 84.5% | 96.0% | 75.8% | 75.0% | 0.84 | 0.71 |
| XGB+MLP+GBDT | 82.8% | 96.0% | 72.7% | 72.7% | 0.82 | 0.68 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, S.; Zhang, Q.; Min, H.; Jiang, W.; Liu, J.; Liu, C.; Wang, Z. Development of a Predictive Model for N-Dealkylation of Amine Contaminants Based on Machine Learning Methods. Toxics 2024, 12, 931. https://doi.org/10.3390/toxics12120931
Cheng S, Zhang Q, Min H, Jiang W, Liu J, Liu C, Wang Z. Development of a Predictive Model for N-Dealkylation of Amine Contaminants Based on Machine Learning Methods. Toxics. 2024; 12(12):931. https://doi.org/10.3390/toxics12120931
Chicago/Turabian StyleCheng, Shiyang, Qihang Zhang, Hao Min, Wenhui Jiang, Jueting Liu, Chunsheng Liu, and Zehua Wang. 2024. "Development of a Predictive Model for N-Dealkylation of Amine Contaminants Based on Machine Learning Methods" Toxics 12, no. 12: 931. https://doi.org/10.3390/toxics12120931
APA StyleCheng, S., Zhang, Q., Min, H., Jiang, W., Liu, J., Liu, C., & Wang, Z. (2024). Development of a Predictive Model for N-Dealkylation of Amine Contaminants Based on Machine Learning Methods. Toxics, 12(12), 931. https://doi.org/10.3390/toxics12120931








