Proteomics for Biomarker Discovery for Diagnosis and Prognosis of Kidney Transplantation Rejection
Abstract
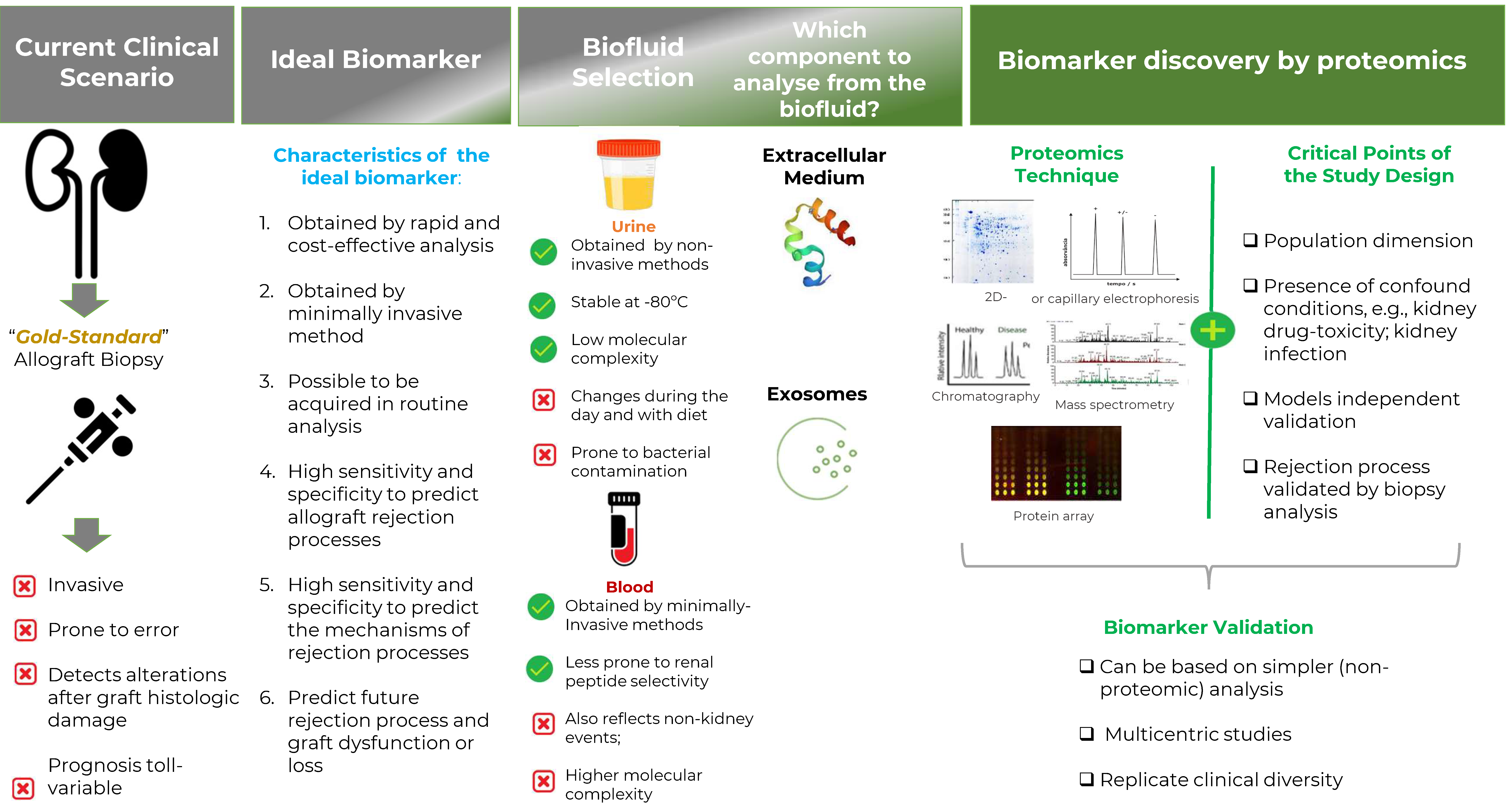
1. Introduction
2. Why Proteomics?
3. Which Biofluid to Analyze?
4. Biofluids Proteomics
4.1. Urinary and Blood Proteomics
| Biofluid Type Proteomic Technique | Population Dimension (It Is Indicated If an Independent Validation Set Was Used) | Prediction Models (Peptide Fragments/Proteins Used in the Model) | Ref |
|---|---|---|---|
| Urine (14 peptides previously discovered) | *No-A-TCMR 390, borderline A-TCMR 157, A-TCMR IA+B 36. A-TCMR IIA+IIB+ I 46 (3 countries) | AUC (A-TCMR) 0.67 (collagen a(I) and (III) chain fragments) | [75] |
| Urine LC-TOF MS/MS | *STA 14, A-ABMR 22 Validation set: *STA 18, A-ABMR 19., HC 12 | AUC (A-ABMR) 0.95, sensitivity 1.00, specificity 0.78 (epidermal growth factor, collagen alpha-1 (VI) chain, Nidogen-1) | [74] |
| Urine iTRAQ LC-MS/MS | *STA 117, AR 112, CAN 116, BKVN 51 Validation set: *STA 47, AR 42, CAN 46, BKVN 16 | AUC (AR) 0.93; AUC (CAN) 0.99; AUC (BKVN) 0.83 (AR: 11 peptides; CAN: 12 peptides; and BKVN: 12 peptides) | [79] |
| Urine LC-MS/MS | *STA 5, Sub-Cli-R 6, IFTA 6 Validation set: *STA 22, ScR 17, GN 15, Viral nephropathies 7, IFTA = 20, IFTAi 13; B-T 13 | AUC (matrix metalloproteinase-7: creatinine, inflamed vs. non-inflamed biopsies) 0.74 | [29] |
| Urine SELDI-TOF-MS | *STA 26, AR 26 Validation set: *STA 16, AR 16 | AUCs (alpha-1-microglobulin) 0.81 and (haptoglobin) 0.76 | [80] |
| Urine CE-MS | *STA 23, Subcli-TCMRC 16 Validation set: *STA 36, SubCli-R 18, Cli-R 10 | AUC (TCMRC) 0.91 (collagen α (I); α (III); matrix metalloproteinase-8) | [71] |
| Urine SELDI-TOF-MS/protein chip array | *STA 36, AR 55, ATN 10 | ATN vs. STA: sensitivity 1.0 and specificity 1.0; STA vs. AR: sensitivity 0.86 and specificity 0.85 (p < 0.001) (ATN vs. STA: 2655; 11,730; 13,134 Da. STA vs. AR: 2364; 33,344; 66,479 Da) | [81] |
| Urine LC-MS/MS and ELISA | *STA, AR 10, HC 20 Validation set: *STA 20, AR 20, HC 20 | AUC (CD44) 0.97; AUC (PEDF) 0.93; AUC (UMOD) 0.85 (MHC antigens, complement cascade, extracellular matrix proteins) | [54] |
| Urine MALDI-TOF MS | *STA 10, AR 10, BKVN 6 Validation set: *STA 10, AR 10, BKVN 4, NS 10, HC 10 | AUC (AR) 0.96 (40 peptides) | [82] |
| Urine LC-MALDI-TOF MS | *STA 8, C-ABMR 10, IFTA 8, HC 10 Validation set: *C-ABMR 8, IFTA 6 | AUC (C-ABMR) 1.00 (6 peptides—m/z:1539.8, 1540.03, 1542.1, 1575.48, 1587.86, and 1657.4) | [69] |
| Urine LC-MALDI-TOF MS | *STA 5, C-ABMR 10, IFTA 8, HC 9 Validation set: *STA 9, C-ABMR 11, IFTA 10, HC 9 | C-ABMR: sensitivities 0.70 and specificities 0.70 (m/z: 610.7, 638.0, 642,6, 645.6, and 1096.8) | [70] |
| Urine SELDI-TOF-MS/ Protein chip array | *STA 22, Sub-Cli-R 27 Validations set: *STA 14, SubCli-R 10 | Sensitivity 0.90 and specificity 0.71 (m/z: 2761, 10762, 11729, 11940) | [76] |
| Urine SELDI-TOF-MS | *STA 22, AR 18, ATN 5, dnG 5, HC 28, UTI 5 | AR vs. STA p < 0.0001 Detected (peaks I+II+III) 94% AR and 18% STA and 0% HC | [61] |
| Urine SELDI-TOF-MS | *STA 22, AR 23, HC 20 | sensitivity 0.905–0.913 and specificity 0.772–0.833 (2003.0, 2802.6, 4756.3, 5872.4, 6990.6, 19,018.8, 25,665.7 Da) | [55] |
| Urine SELDI-TOF-MS | *STA 15, AR 17 | Tree decision model: sensitivity 0.83 and specificity 1.00 (decision trees 3.4, 10.0 Kd) | [83] |
| Plasma LC-MS/MS | *STA 25, A-CR 6 | p < 0.05 (24 proteins) | [57] |
| Plasma plus Blood iTRAQ MALDI-TOF/TOF MS/MS | *AR 20, non-AR 20 | AUC (21 peptides) 0.57 AUC (90 probes gene) 0.71 | [84] |
| Plasma iTRAQ MALDI-TOF/TOF | *AR 11, non-AR 21 | AUC 0.86 (titin, kininogen-1, and lipopolysaccharide-binding protein) | [56] |
| Serum iTRAQ LC-ESI-MS/MS | *AR 3, HC 9 | Q ≤ 0.05 (109 proteins) | [85] |
| Serum MALDI-TOF MS | *STA 12, AR 12, CR 12, HC 13 | Identification 83% AR and 99% CR (AR: 18 peptides; CR: 6 peptides) | [77] |
- Define a specific proteomics technique since each technique will highlight a different set of proteins;
- Define a specific biofluid, as the proteomic is specific to the biofluid;
- High dimension of the population evaluated;
- Apply an independent validation data set to test prediction models;
- Consider a high diversity of confound conditions, including patients with and without rejection processes, including, e.g., kidney drug toxicity, ischemic/reperfusion injury, and infections, among other diseases;
- All samples should be classified according to a parallel and rigorous histological-based biopsy analysis;
- The prediction power of the model should be quantified by measures such as AUC, sensitivity, and specificity, among others.
4.2. PBMC Proteomics
4.3. Exosomes Proteomics
4.4. Multi-Omics Approach
5. Final Considerations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Lohéac, C.; Aubert, O.; Loupy, A.; Legendre, C. Étude Des Étiologies Spécifiques de Perte Des Greffons Rénaux: Place Du Rejet Médié Par Les Anticorps et Approche En Population. Néphrol. Thér. 2018, 14, S39–S50. [Google Scholar] [CrossRef] [PubMed]
- Schold, J.D.; Buccini, L.D.; Goldfarb, D.A.; Flechner, S.M.; Poggio, E.D. Association between Kidney Transplant Center Performance and the Survival Benefit of Transplantation Versus Dialysis. Clin. J. Am. Soc. Nephrol. 2014, 9, 1773–1780. [Google Scholar] [CrossRef] [PubMed]
- Chadban, S.J.; Ahn, C.; Axelrod, D.A.; Foster, B.J.; Kasiske, B.L.; Kher, V.; Kumar, D.; Oberbauer, R.; Pascual, J.; Pilmore, H.L.; et al. Summary of the Kidney Disease: Improving Global Outcomes (KDIGO) Clinical Practice Guideline on the Evaluation and Management of Candidates for Kidney Transplantation. Transplantation 2020, 104, 708–714. [Google Scholar] [CrossRef]
- Rosselli, D.; Rueda, J.; Diaz, C.E. Cost-Effectiveness of Kidney Transplantation Compared with Chronic Dialysis in End-Stage Renal Disease. Saudi J. Kidney Dis. Transplant. 2015, 26, 733–738. [Google Scholar] [CrossRef] [PubMed]
- Burton, H.; Iyamu Perisanidou, L.; Steenkamp, R.; Evans, R.; Mumford, L.; Evans, K.M.; Caskey, F.J.; Hilton, R. Causes of Renal Allograft Failure in the UK: Trends in UK Renal Registry and National Health Service Blood and Transplant Data from 2000 to 2013. Nephrol. Dial. Transplant. 2019, 34, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Leendert, C.P. Chronic Renal Transplant Loss. Kidney Int. 1995, 47, 1491–1499. [Google Scholar] [CrossRef]
- Hongera, G.; Fornaro, I.; Granado, C.; Tiercy, J.-M.; Hosli, I.; Schaub, S. Frequency and Determinants of Pregnancy-Induced Child-Specific Sensitization. Am. J. Transplant. 2013, 13, 746–753. [Google Scholar] [CrossRef]
- Pratschke, J.; Dragun, D.; Hauser, I.A.; Horn, S.; Mueller, T.F.; Schemmer, P.; Thaiss, F. Immunological Risk Assessment: The Key to Individualized Immunosuppression after Kidney Transplantation. Transplant. Rev. 2016, 30, 77–84. [Google Scholar] [CrossRef]
- Marfo, K.; Lu, A.; Ling, M.; Akalin, E. Desensitization Protocols and Their Outcome. Clin. J. Am. Soc. Nephrol. 2011, 6, 922–936. [Google Scholar] [CrossRef]
- Khodadadi, L.; Adib, M.; Pourazar, A. Immunoglobulin Class (IgG, IgM) Determination by Dithiothreitol in Sensitized Kidney Transplant Candidates. Transplant. Proc. 2006, 38, 2813–2815. [Google Scholar] [CrossRef]
- Nakorchevsky, A.; Hewel, J.A.; Kurian, S.M.; Mondala, T.S.; Campbell, D.; Head, S.R.; Marsh, C.L.; Yates, J.R.; Salomon, D.R. Molecular Mechanisms of Chronic Kidney Transplant Rejection via Large-Scale Proteogenomic Analysis of Tissue Biopsies. J. Am. Soc. Nephrol. 2010, 21, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Nankivell, B.J.; Kuypers, D.R. Diagnosis and Prevention of Chronic Kidney Allograft Loss. Lancet 2011, 378, 1428–1437. [Google Scholar] [CrossRef]
- Halloran, P.F.; Reeve, J.P.; Pereira, A.B.; Hidalgo, L.G.; Famulski, K.S. Antibody-Mediated Rejection, T Cell-Mediated Rejection, and the Injury-Repair Response: New Insights from the Genome Canada Studies of Kidney Transplant Biopsies. Kidney Int. 2014, 85, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Chong, A.S. Mechanisms of Organ Transplant Injury Mediated by B Cells and Antibodies: Implications for Antibody-mediated Rejection. Am. J. Transplant. 2020, 20, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Lefaucheur, C.; Viglietti, D.; Bentlejewski, C.; Duong van Huyen, J.-P.; Vernerey, D.; Aubert, O.; Verine, J.; Jouven, X.; Legendre, C.; Glotz, D.; et al. IgG Donor-Specific Anti-Human HLA Antibody Subclasses and Kidney Allograft Antibody-Mediated Injury. J. Am. Soc. Nephrol. 2016, 27, 293–304. [Google Scholar] [CrossRef]
- Terasaki, P.I.; Cai, J. Humoral Theory of Transplantation: Further Evidence. Curr. Opin. Immunol. 2005, 17, 541–545. [Google Scholar] [CrossRef]
- Merhi, B.; Bayliss, G.; Gohh, R.Y. Role for Urinary Biomarkers in Diagnosis of Acute Rejection in the Transplanted Kidney. World J. Transplant. 2015, 5, 251–261. [Google Scholar] [CrossRef]
- Ponticelli, C. The Mechanisms of Acute Transplant Rejection Revisited. J. Nephrol. 2012, 25, 150–158. [Google Scholar] [CrossRef]
- Randhawa, P. T-Cell-Mediated Rejection of the Kidney in the Era of Donor-Specific Antibodies. Curr. Opin. Organ Transplant. 2015, 20, 325–332. [Google Scholar] [CrossRef]
- Sarwal, M.; Chua, M.-S.; Kambham, N.; Hsieh, S.-C.; Satterwhite, T.; Masek, M.; Salvatierra, O. Molecular Heterogeneity in Acute Renal Allograft Rejection Identified by DNA Microarray Profiling. N. Engl. J. Med. 2003, 349, 125–138. [Google Scholar] [CrossRef]
- Gwinner, W. Renal Transplant Rejection Markers. World J. Urol. 2007, 25, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Halloran, P.F.; Pereira, A.B.; Chang, J.; Matas, A.; Picton, M.; De Freitas, D.; Bromberg, J.; Serõn, D.; Sellarés, J.; Einecke, G.; et al. Microarray Diagnosis of Antibody-Mediated Rejection in Kidney Transplant Biopsies: An International Prospective Study (INTERCOM). Am. J. Transplant. 2013, 13, 2865–2874. [Google Scholar] [CrossRef] [PubMed]
- Lasmar, M.F.; Dutra, R.S.; Nogueira-Machado, J.A.; Fabreti-Oliveira, R.A.; Siqueira, R.G.; Nascimento, E. Effects of Immunotherapy Induction on Outcome and Graft Survival of Kidney-Transplanted Patients with Different Immunological Risk of Rejection. BMC Nephrol. 2019, 20, 314. [Google Scholar] [CrossRef]
- Senev, A.; Lerut, E.; Van Sandt, V.; Coemans, M.; Callemeyn, J.; Sprangers, B.; Kuypers, D.; Emonds, M.; Naesens, M. Specificity, Strength, and Evolution of Pretransplant Donor-specific HLA Antibodies Determine Outcome after Kidney Transplantation. Am. J. Transplant. 2019, 19, 3100–3113. [Google Scholar] [CrossRef] [PubMed]
- Townamchai, N.; Safa, K.; Chandraker, A. Immunologic Monitoring in Kidney Transplant Recipients. Kidney Res. Clin. Pract. 2013, 32, 52–61. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ishida, H.; Hirai, T.; Kohei, N.; Yamaguchi, Y.; Tanabe, K. Significance of Qualitative and Quantitative Evaluations of Anti-HLA Antibodies in Kidney Transplantation. Transpl. Int. 2011, 24, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R. Donor-Specific Antibodies in Kidney Transplant Recipients. Clin. J. Am. Soc. Nephrol. 2017, 13, 82–192. [Google Scholar] [CrossRef]
- Kenta, I.; Takaaki, K. Molecular Mechanisms of Antibody-Mediated Rejection and Accommodation in Organ Transplantation. Nephron 2020, 144, 2–6. [Google Scholar] [CrossRef]
- Ho, J.; Rush, D.N.; Krokhin, O.; Antonovici, M.; Gao, A.; Bestland, J.; Wiebe, C.; Hiebert, B.; Rigatto, C.; Gibson, I.W.; et al. Elevated Urinary Matrix Metalloproteinase-7 Detects Underlying Renal Allograft Inflammation and Injury. Transplantation 2016, 100, 648–654. [Google Scholar] [CrossRef]
- Rush, D. Protocol Biopsies for Renal Transplantation. Saudi J. Kidney Dis. Transpl. 2010, 21, 219–226. [Google Scholar] [CrossRef]
- Yango, A.; Gohh, R.; Wang, L.J.; Morrissey, P.; Shih, M.; Lowery, K.; Charpentier, K.; Gautam, A.; Mendonca, C.; Kumar, S.; et al. The Utility of 6-Month Protocol Renal Biopsy under Modern Immunosuppression. Clin. Nephrol. 2008, 70, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Halloran, P.F.; Chang, J.; Famulski, K.; Hidalgo, L.G.; Salazar, I.D.R.; Merino Lopez, M.; Matas, A.; Picton, M.; de Freitas, D.; Bromberg, J.; et al. Disappearance of T Cell-Mediated Rejection Despite Continued Antibody-Mediated Rejection in Late Kidney Transplant Recipients. J. Am. Soc. Nephrol. 2015, 26, 1711–1720. [Google Scholar] [CrossRef] [PubMed]
- Hariharan, S. BK Virus Nephritis after Renal Transplantation. Kidney Int. 2006, 69, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Djamali, A.; Premasathian, N.; Pirsch, J.D. Outcomes in Kidney Transplantation. Semin. Nephrol. 2003, 23, 306–316. [Google Scholar] [CrossRef]
- Eikmans, M.; Gielis, E.M.; Ledeganck, K.J.; Yang, J.; Abramowicz, D.; Claas, F.F.J. Non-Invasive Biomarkers of Acute Rejection in Kidney Transplantation: Novel Targets and Strategies. Front. Med. 2019, 5, 358. [Google Scholar] [CrossRef]
- Erpicum, P.; Hanssen, O.; Weekers, L.; Lovinfosse, P.; Meunier, P.; Tshibanda, L.; Krzesinski, J.; Hustinx, R.; Jouret, F. Non-Invasive Approaches in the Diagnosis of Acute Rejection in Kidney Transplant Recipients, Part II: Omics Analyses of Urine and Blood Samples. Clin. Kidney J. 2017, 10, 106–115. [Google Scholar] [CrossRef][Green Version]
- Chakraborty, A.; Sarwal, M. Protein Biomarkers in Renal Transplantation. Expert Rev. Proteom. 2018, 15, 41–54. [Google Scholar] [CrossRef]
- Hricik, D.E.; Nickerson, P.; Formica, R.N.; Poggio, E.D.; Rush, D.; Newell, K.A.; Goebel, J.; Gibson, I.W.; Fairchild, R.L.; Riggs, M.; et al. Multicenter Validation of Urinary CXCL9 as a Risk-Stratifying Biomarker for Kidney Transplant Injury. Am. J. Transplant. 2013, 13, 2634–2644. [Google Scholar] [CrossRef]
- Hirt-Minkowski, P.; Amico, P.; Ho, J.; Gao, A.; Bestland, J.; Hopfer, H.; Steiger, J.; Dickenmann, M.; Burkhalter, F.; Rush, D.; et al. Detection of Clinical and Subclinical Tubulo-Interstitial Inflammation by the Urinary CXCL10 Chemokine in a Real-Life Setting. Am. J. Transplant. 2012, 12, 1811–1823. [Google Scholar] [CrossRef]
- Araújo, R.; Bento, L.F.N.; Fonseca, T.A.H.; Von Rekowski, C.P.; da Cunha, B.R.; Calado, C.R.C. Infection Biomarkers Based on Metabolomics. Metabolites 2022, 12, 92. [Google Scholar] [CrossRef]
- Lin, X.-C.; Sui, W.-G.; Qi, S.-W.; Tang, D.-E.; Cong, S.; Zou, G.-M.; Zhang, Y.; Li, H.; Chen, W.-B.; Cheng, Z.-Q.; et al. Quantitative Proteomic Profiling of Renal Tissue in Human Chronic Rejection Biopsy Samples After Renal Transplantation. Transplant. Proc. 2015, 47, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.-M.; Thijs, L.; Liu, Y.-P.; Zhang, Z.; Jacobs, L.; Koeck, T.; Zürbig, P.; Lichtinghagen, R.; Brand, K.; Kuznetsova, T.; et al. The Urinary Proteome as Correlate and Predictor of Renal Function in a Population Study. Nephrol. Dial. Transplant. 2014, 29, 2260–2268. [Google Scholar] [CrossRef]
- Raimondo, F.; Corbetta, S.; Chinello, C.; Pitto, M.; Magni, F. The Urinary Proteome and Peptidome of Renal Cell Carcinoma Patients: A Comparison of Different Techniques. Expert Rev. Proteom. 2014, 11, 503–514. [Google Scholar] [CrossRef]
- Kaysheva, A.L.; Kopylov, A.T.; Kushlinskii, N.E.; Gershtein, E.S.; Alferov, A.A.; Morozov, A.A.; Kazantseva, I.A.; Pleshakova, T.O.; Archakov, A.I.; Ivanov, Y.D. Comparative Analysis of Blood Plasma Proteome in Patients with Renal Cell Carcinoma. Bull. Exp. Biol. Med. 2019, 167, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Ricci, P.; Magalhães, P.; Krochmal, M.; Pejchinovski, M.; Daina, E.; Caruso, M.R.; Goea, L.; Belczacka, I.; Remuzzi, G.; Umbhauer, M.; et al. Urinary Proteome Signature of Renal Cysts and Diabetes Syndrome in Children. Sci. Rep. 2019, 9, 2225. [Google Scholar] [CrossRef]
- Matías-García, P.R.; Wilson, R.; Guo, Q.; Zaghlool, S.B.; Eales, J.M.; Xu, X.; Charchar, F.J.; Dormer, J.; Maalmi, H.; Schlosser, P.; et al. Plasma Proteomics of Renal Function: A Transethnic Meta-Analysis and Mendelian Randomization Study. J. Am. Soc. Nephrol. 2021, 32, 1747–1763. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zu, Q.; Lu, J.; Zhang, L.; Zhu, Q.; Sun, X.; Dong, J. Effects of Donor-Recipient Age Difference in Renal Transplantation, an Investigation on Renal Function and Fluid Proteome. Clin. Interv. Aging 2021, 16, 1457–1470. [Google Scholar] [CrossRef]
- Aslam, B.; Basit, M.; Nisar, M.A.; Khurshid, M.; Rasool, M.H. Proteomics: Technologies and Their Applications. J. Chromatogr. Sci. 2017, 55, 182–196. [Google Scholar] [CrossRef]
- Alharbi, R.A. Proteomics Approach and Techniques in Identification of Reliable Biomarkers for Diseases. Saudi J. Biol. Sci. 2020, 27, 968–974. [Google Scholar] [CrossRef]
- Xie, S.; Moya, C.; Bilgin, B.; Jayaraman, A.; Walton, S.P. Emerging Affinity-Based Techniques in Proteomics. Expert Rev. Proteom. 2009, 6, 573–583. [Google Scholar] [CrossRef]
- Flechner, S.M.; Kurian, S.M.; Head, S.R.; Sharp, S.M.; Whisenant, T.C.; Zhang, J.; Chismar, J.D.; Horvath, S.; Mondala, T.; Gilmartin, T.; et al. Kidney Transplant Rejection and Tissue Injury by Gene Profiling of Biopsies and Peripheral Blood Lymphocytes. Am. J. Transplant. 2004, 4, 1475–1489. [Google Scholar] [CrossRef] [PubMed]
- Pietrowska, M.; Wlosowicz, A.; Gawin, M.; Widlak, P. MS-Based Proteomic Analysis of Serum and Plasma: Problem of High Abundant Components and Lights and Shadows of Albumin Removal. In Advances in Experimental Medicine and Biology; Springer Nature: Cham, Switzerland, 2019; Volume 1073, pp. 57–76. ISBN 9783030122980. [Google Scholar]
- Tu, C.; Rudnick, P.A.; Martinez, M.Y.; Cheek, K.L.; Stein, S.E.; Slebos, R.J.C.; Liebler, D.C. Depletion of Abundant Plasma Proteins and Limitations of Plasma Proteomics. J. Proteome Res. 2010, 9, 4982–4991. [Google Scholar] [CrossRef] [PubMed]
- Sigdel, T.K.; Kaushal, A.; Gritsenko, M.; Norbeck, A.D.; Qian, W.; Xiao, W.; Camp, D.G.; Smith, R.D.; Sarwal, M.M. Shotgun Proteomics Identifies Proteins Specific for Acute Renal Transplant Rejection. Proteom. Clin. Appl. 2010, 4, 32–47. [Google Scholar] [CrossRef] [PubMed]
- O’Riordan, E. Bioinformatic Analysis of the Urine Proteome of Acute Allograft Rejection. J. Am. Soc. Nephrol. 2004, 15, 3240–3248. [Google Scholar] [CrossRef] [PubMed]
- Freue, G.V.C.; Sasaki, M.; Meredith, A.; Günther, O.P.; Bergman, A.; Takhar, M.; Mui, A.; Balshaw, R.F.; Ng, R.T.; Opushneva, N.; et al. Proteomic Signatures in Plasma during Early Acute Renal Allograft Rejection. Mol. Cell. Proteom. 2010, 9, 1954–1967. [Google Scholar] [CrossRef]
- Perez, J.D.; Sakata, M.M.; Colucci, J.A.; Spinelli, G.A.; Felipe, C.R.; Carvalho, V.M.; Cardozo, K.H.M.; Medina-Pestana, J.O.; Tedesco-Silva, H.; Schor, N.; et al. Plasma Proteomics for the Assessment of Acute Renal Transplant Rejection. Life Sci. 2016, 158, 111–120. [Google Scholar] [CrossRef]
- Kaisar, M.; van Dullemen, L.F.A.; Thézénas, M.-L.; Zeeshan Akhtar, M.; Huang, H.; Rendel, S.; Charles, P.D.; Fischer, R.; Ploeg, R.J.; Kessler, B.M. Plasma Degradome Affected by Variable Storage of Human Blood. Clin. Proteom. 2016, 13, 26. [Google Scholar] [CrossRef]
- Hepburn, S.; Cairns, D.A.; Jackson, D.; Craven, R.A.; Riley, B.; Hutchinson, M.; Wood, S.; Smith, M.W.; Thompson, D.; Banks, R.E. An Analysis of the Impact of Pre-Analytical Factors on the Urine Proteome: Sample Processing Time, Temperature, and Proteolysis. Proteom. Clin. Appl. 2015, 9, 507–521. [Google Scholar] [CrossRef]
- Ostroff, R.; Foreman, T.; Keeney, T.R.; Stratford, S.; Walker, J.J.; Zichi, D. The Stability of the Circulating Human Proteome to Variations in Sample Collection and Handling Procedures Measured with an Aptamer-Based Proteomics Array. J. Proteom. 2010, 73, 649–666. [Google Scholar] [CrossRef]
- Schaub, S.; Rush, D.; Wilkins, J.; Gibson, I.W.; Weiler, T.; Sangster, K.; Nicolle, L.; Karpinski, M.; Jeffery, J.; Nickerson, P. Proteomic-Based Detection of Urine Proteins Associated with Acute Renal Allograft Rejection. J. Am. Soc. Nephrol. 2004, 15, 219–227. [Google Scholar] [CrossRef]
- Nagaraj, N.; Mann, M. Quantitative Analysis of the Intra- and Inter-Individual Variability of the Normal Urinary Proteome. J. Proteome Res. 2011, 10, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Swensen, A.C.; He, J.; Fang, A.C.; Ye, Y.; Nicora, C.D.; Shi, T.; Liu, A.Y.; Sigdel, T.K.; Sarwal, M.M.; Qian, W.-J. A Comprehensive Urine Proteome Database Generated From Patients With Various Renal Conditions and Prostate Cancer. Front. Med. 2021, 8, 548212. [Google Scholar] [CrossRef] [PubMed]
- Habuka, M.; Fagerberg, L.; Hallström, B.M.; Kampf, C.; Edlund, K.; Sivertsson, Å.; Yamamoto, T.; Pontén, F.; Uhlén, M.; Odeberg, J. The Kidney Transcriptome and Proteome Defined by Transcriptomics and Antibody-Based Profiling. PLoS ONE 2014, 9, e116125. [Google Scholar] [CrossRef] [PubMed]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/humanproteome/kidney (accessed on 15 March 2019).
- He, T.; Pejchinovski, M.; Mullen, W.; Beige, J.; Mischak, H.; Jankowski, V. Peptides in Plasma, Urine, and Dialysate: Toward Unravelling Renal Peptide Handling. Proteom. Clin. Appl. 2021, 15, 2000029. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, P.; Pontillo, C.; Pejchinovski, M.; Siwy, J.; Krochmal, M.; Makridakis, M.; Carrick, E.; Klein, J.; Mullen, W.; Jankowski, J.; et al. Comparison of Urine and Plasma Peptidome Indicates Selectivity in Renal Peptide Handling. Proteom. Clin. Appl. 2018, 12, 1700163. [Google Scholar] [CrossRef]
- Savage, A.K.; Gutschow, M.V.; Chiang, T.; Henderson, K.; Green, R.; Chaudhari, M.; Swanson, E.; Heubeck, A.T.; Kondza, N.; Burley, K.C.; et al. Multimodal Analysis for Human Ex Vivo Studies Shows Extensive Molecular Changes from Delays in Blood Processing. iScience 2021, 24, 102404. [Google Scholar] [CrossRef]
- Quintana, L.F.; Solé-Gonzalez, A.; Kalko, S.G.; Bañon-Maneus, E.; Solé, M.; Diekmann, F.; Gutierrez-Dalmau, A.; Abian, J.; Campistol, J.M. Urine Proteomics to Detect Biomarkers for Chronic Allograft Dysfunction. J. Am. Soc. Nephrol. 2009, 20, 428–435. [Google Scholar] [CrossRef]
- Quintana, L.F.; Campistol, J.M.; Alcolea, M.P.; Bañon-Maneus, E.; Sol-González, A.; Cutillas, P.R. Application of Label-Free Quantitative Peptidomics for the Identification of Urinary Biomarkers of Kidney Chronic Allograft Dysfunction. Mol. Cell. Proteom. 2009, 8, 1658–1673. [Google Scholar] [CrossRef]
- Metzger, J.; Chatzikyrkou, C.; Broecker, V.; Schiffer, E.; Jaensch, L.; Iphoefer, A.; Mengel, M.; Mullen, W.; Mischak, H.; Haller, H.; et al. Diagnosis of Subclinical and Clinical Acute T-Cell-Mediated Rejection in Renal Transplant Patients by Urinary Proteome Analysis. Proteom. Clin. Appl. 2011, 5, 322–333. [Google Scholar] [CrossRef]
- Mao, Y.; Bai, J.; Chen, J.; Shou, Z.; He, Q.; Wu, J.; Chen, Y.; Cheng, Y. A Pilot Study of GC/MS-Based Serum Metabolic Profiling of Acute Rejection in Renal Transplantation. Transpl. Immunol. 2008, 19, 74–80. [Google Scholar] [CrossRef]
- Wittke, S.; Haubitz, M.; Walden, M.; Rohde, F.; Schwarz, A.; Mengel, M.; Mischak, H.; Haller, H.; Gwinner, W. Detection of Acute Tubulointerstitial Rejection by Proteomic Analysis of Urinary Samples in Renal Transplant Recipients. Am. J. Transplant. 2005, 5, 2479–2488. [Google Scholar] [CrossRef] [PubMed]
- Heidari, S.S.; Nafar, M.; Kalantari, S.; Tavilani, H.; Karimi, J.; Foster, L.; Moon, K.M.; Khodadadi, I. Urinary Epidermal Growth Factor Is a Novel Biomarker for Early Diagnosis of Antibody Mediated Kidney Allograft Rejection: A Urinary Proteomics Analysis. J. Proteom. 2021, 240, 104208. [Google Scholar] [CrossRef] [PubMed]
- Gwinner, W.; Karch, A.; Braesen, J.H.; Khalifa, A.A.; Metzger, J.; Naesens, M.; Van Loon, E.; Anglicheau, D.; Marquet, P.; Budde, K.; et al. Noninvasive Diagnosis of Acute Rejection in Renal Transplant Patients Using Mass Spectrometric Analysis of Urine Samples: A Multicenter Diagnostic Phase III Trial. Transplant. Direct 2022, 8, e1316. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Yu, J.; Chen, J.; Yang, H.; He, Q.; Shou, Z.; Wu, J.; Zheng, S. Diagnosis of Renal Allograft Subclinical Rejection by Urine Protein Fingerprint Analysis. Transpl. Immunol. 2008, 18, 255–259. [Google Scholar] [CrossRef]
- Sui, W.; Huang, L.; Dai, Y.; Chen, J.; Yan, Q.; Huang, H. Proteomic Profiling of Renal Allograft Rejection in Serum Using Magnetic Bead-Based Sample Fractionation and MALDI-TOF MS. Clin. Exp. Med. 2010, 10, 259–268. [Google Scholar] [CrossRef]
- Sigdel, T.K.; Salomonis, N.; Nicora, C.D.; Ryu, S.; He, J.; Dinh, V.; Orton, D.J.; Moore, R.J.; Hsieh, S.-C.; Dai, H.; et al. The Identification of Novel Potential Injury Mechanisms and Candidate Biomarkers in Renal Allograft Rejection by Quantitative Proteomics. Mol. Cell. Proteom. 2014, 13, 621–631. [Google Scholar] [CrossRef]
- Sigdel, T.K.; Gao, Y.; He, J.; Wang, A.; Nicora, C.D.; Fillmore, T.L.; Shi, T.; Webb-Robertson, B.-J.; Smith, R.D.; Qian, W.-J.; et al. Mining the Human Urine Proteome for Monitoring Renal Transplant Injury. Kidney Int. 2016, 89, 1244–1252. [Google Scholar] [CrossRef]
- Stubendorff, B.; Finke, S.; Walter, M.; Kniemeyer, O.; von Eggeling, F.; Gruschwitz, T.; Steiner, T.; Ott, U.; Wolf, G.; Wunderlich, H.; et al. Urine Protein Profiling Identified Alpha-1-Microglobulin and Haptoglobin as Biomarkers for Early Diagnosis of Acute Allograft Rejection Following Kidney Transplantation. World J. Urol. 2014, 32, 1619–1624. [Google Scholar] [CrossRef]
- Yang, H.; Mao, Y.; Yu, J.; Chen, J.; He, Q.; Shou, Z.; Wu, J.; Chen, Y.; Zheng, S. Diagnosis of C4d+ Renal Allograft Acute Humoral Rejection by Urine Protein Fingerprint Analysis. J. Int. Med. Res. 2010, 38, 176–186. [Google Scholar] [CrossRef]
- Ling, X.B.; Sigdel, T.K.; Lau, K.; Ying, L.; Lau, I.; Schilling, J.; Sarwal, M.M. Integrative Urinary Peptidomics in Renal Transplantation Identifies Biomarkers for Acute Rejection. J. Am. Soc. Nephrol. 2010, 21, 646–653. [Google Scholar] [CrossRef]
- Clarke, W.; Silverman, B.C.; Zhang, Z.; Chan, D.W.; Klein, A.S.; Molmenti, E.P. Characterization of Renal Allograft Rejection by Urinary Proteomic Analysis. Ann. Surg. 2003, 237, 660–665. [Google Scholar] [CrossRef] [PubMed]
- Günther, O.P.; Shin, H.; Ng, R.T.; McMaster, W.R.; McManus, B.M.; Keown, P.A.; Tebbutt, S.J.; Lê Cao, K.-A. Novel Multivariate Methods for Integration of Genomics and Proteomics Data: Applications in a Kidney Transplant Rejection Study. OMICS A J. Integr. Biol. 2014, 18, 682–695. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ou, M.; Lin, H.; Lai, L.; Chen, H.; Chen, J.; Sui, W.; Xue, W.; Zhang, R.; Gan, Q.; et al. Proteomic Analysis of Differentially Expressed Proteins in the Serum of Patients with Acute Renal Allograft Rejection Using ITRAQ Labelling Technology. Mol. Med. Rep. 2020, 22, 2329–2341. [Google Scholar] [CrossRef] [PubMed]
- Van Besouw, N.M.; Yan, L.; De Kuiper, R.; Klepper, M.; Reijerkerk, D.; Dieterich, M.; Roelen, D.L.; Claas, F.H.J.; Clahsen-Van Groningen, M.C.; Hesselink, D.A.; et al. The Number of Donor-Specific IL-21 Producing Cells before and after Transplantation Predicts Kidney Graft Rejection. Front. Immunol. 2019, 10, 748. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, K.; Yamamoto, T.; Inanaga, Y.; Hiramitsu, T.; Miwa, Y.; Murotani, K.; Narumi, S.; Watarai, Y.; Katayama, A.; Uchida, K.; et al. MiR-142-5p and MiR-486-5p as Biomarkers for Early Detection of Chronic Antibody-Mediated Rejection in Kidney Transplantation. Biomarkers 2017, 22, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Gui, B.; Tian, P.; Yao, G.; Fu, R.; Wang, L.; Ge, H.; Ou, Y. TIPE2, a Novel Biomarker for Clinical Chronic Kidney Allograft Rejection. Artif. Organs 2013, 37, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Schaenman, J.M.; Rossetti, M.; Sidwell, T.; Groysberg, V.; Sunga, G.; Korin, Y.; Liang, E.; Zhou, X.; Abdalla, B.; Lum, E.; et al. Increased T Cell Immunosenescence and Accelerated Maturation Phenotypes in Older Kidney Transplant Recipients. Hum. Immunol. 2018, 79, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Nazari, B.; Amirzargar, A.; Nikbin, B.; Nafar, M.; Ahmadpour, P.; Einollahi, B.; Pezeshki, M.L.; Khatami, S.M.R.; Ansaripour, B.; Nikuinejad, H.; et al. Comparison of the Th1, IFN-γ Secreting Cells and FoxP3 Expression between Patients with Stable Graft Function and Acute Rejection Post Kidney Transplantation. Iran. J. Allergy Asthma Immunol. 2013, 12, 262–268. [Google Scholar]
- Barabadi, M.; Shahbaz, S.K.; Foroughi, F.; Hosseinzadeh, M.; Nafar, M.; Yekaninejad, M.S.; Amirzargar, A. High Expression of FOXP3 MRNA in Blood and Urine as a Predictive Marker in Kidney Transplantation. Prog. Transplant. 2018, 28, 134–141. [Google Scholar] [CrossRef]
- Savaryn, J.P.; Toby, T.K.; Catherman, A.D.; Fellers, R.T.; LeDuc, R.D.; Thomas, P.M.; Friedewald, J.J.; Salomon, D.R.; Abecassis, M.M.; Kelleher, N.L. Comparative Top down Proteomics of Peripheral Blood Mononuclear Cells from Kidney Transplant Recipients with Normal Kidney Biopsies or Acute Rejection. Proteomics 2016, 16, 2048–2058. [Google Scholar] [CrossRef]
- Gonzalez-Nolasco, B.; Wang, M.; Prunevieille, A.; Benichou, G. Emerging Role of Exosomes in Allorecognition and Allograft Rejection. Curr. Opin. Organ Transplant. 2018, 23, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Fleissner, F.; Goerzig, Y.; Haverich, A.; Thum, T. Microvesicles as Novel Biomarkers and Therapeutic Targets in Transplantation Medicine. Am. J. Transplant. 2012, 12, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Shen, L.; Shi, H.; Pan, Z.; Wu, L.; Yan, Y.; Zhang, X.; Mao, F.; Qian, H.; Xu, W. Exosomes from Human Umbilical Cord Mesenchymal Stem Cells: Identification, Purification, and Biological Characteristics. Stem Cells Int. 2016, 2016, 1929536. [Google Scholar] [CrossRef] [PubMed]
- Feigerlova, E.; Battaglia-Hsu, S.-F.; Hauet, T.; Guéant, J.-L.; Feigerlová, E.; Battaglia-Hsu, S.-F.; Hauet, T.; Guéant, J.-L. Extracellular Vesicles as Immune Mediators in Response to Kidney Injury. Am. J. Physiol. Physiol. 2018, 314, F9–F21. [Google Scholar] [CrossRef]
- Hiemstra, T.F.; Charles, P.D.; Gracia, T.; Hester, S.S.; Gatto, L.; Al-Lamki, R.; Floto, R.A.; Su, Y.; Skepper, J.N.; Lilley, K.S.; et al. Human Urinary Exosomes as Innate Immune Effectors. J. Am. Soc. Nephrol. 2014, 25, 2017–2027. [Google Scholar] [CrossRef]
- Ezzelarab, M.B.; Raich-Regue, D.; Lu, L.; Zahorchak, A.F.; Perez-Gutierrez, A.; Humar, A.; Wijkstrom, M.; Minervini, M.; Wiseman, R.W.; Cooper, D.K.C.; et al. Renal Allograft Survival in Nonhuman Primates Infused With Donor Antigen-Pulsed Autologous Regulatory Dendritic Cells. Am. J. Transplant. 2017, 17, 1476–1489. [Google Scholar] [CrossRef]
- Aiello, S.; Rocchetta, F.; Longaretti, L.; Faravelli, S.; Todeschini, M.; Cassis, L.; Pezzuto, F.; Tomasoni, S.; Azzollini, N.; Mister, M.; et al. Extracellular Vesicles Derived from T Regulatory Cells Suppress T Cell Proliferation and Prolong Allograft Survival. Sci. Rep. 2017, 7, 11518. [Google Scholar] [CrossRef]
- Yu, X.; Huang, C.; Song, B.; Xiao, Y.; Fang, M.; Feng, J.; Wang, P. CD4+CD25+ Regulatory T Cells-Derived Exosomes Prolonged Kidney Allograft Survival in a Rat Model. Cell. Immunol. 2013, 285, 62–68. [Google Scholar] [CrossRef]
- Pang, X.-L.; Wang, Z.-G.; Liu, L.; Feng, Y.-H.; Wang, J.-X.; Xie, H.-C.; Yang, X.-L.; Li, J.-F.; Feng, G.-W. Immature Dendritic Cells Derived Exosomes Promotes Immune Tolerance by Regulating T Cell Differentiation in Renal Transplantation. Aging 2019, 11, 8911–8924. [Google Scholar] [CrossRef]
- Wu, X.; Wang, Z.; Wang, J.; Tian, X.; Cao, G.; Gu, Y.; Shao, F.; Yan, T. Exosomes Secreted by Mesenchymal Stem Cells Induce Immune Tolerance to Mouse Kidney Transplantation via Transporting LncRNA DANCR. Inflammation 2021. [Google Scholar] [CrossRef]
- Ramirez-Bajo, M.J.; Rovira, J.; Lazo-Rodriguez, M.; Banon-Maneus, E.; Tubita, V.; Moya-Rull, D.; Hierro-Garcia, N.; Ventura-Aguiar, P.; Oppenheimer, F.; Campistol, J.M.; et al. Impact of Mesenchymal Stromal Cells and Their Extracellular Vesicles in a Rat Model of Kidney Rejection. Front. Cell Dev. Biol. 2020, 8, 10. [Google Scholar] [CrossRef]
- Koch, M.; Lemke, A.; Lange, C. Extracellular Vesicles from MSC Modulate the Immune Response to Renal Allografts in a MHC Disparate Rat Model. Stem Cells Int. 2015, 2015, 486141. [Google Scholar] [CrossRef] [PubMed]
- Tower, C.M.; Reyes, M.; Nelson, K.; Leca, N.; Kieran, N.; Muczynski, K.; Jefferson, J.A.; Blosser, C.; Kukla, A.; Maurer, D.; et al. Plasma C4d+ Endothelial Microvesicles Increase in Acute Antibody-Mediated Rejection. Transplantation 2017, 101, 2235–2243. [Google Scholar] [CrossRef] [PubMed]
- Castellani, C.; Burrello, J.; Fedrigo, M.; Burrello, A.; Bolis, S.; Di Silvestre, D.; Tona, F.; Bottio, T.; Biemmi, V.; Toscano, G.; et al. Circulating Extracellular Vesicles as Non-Invasive Biomarker of Rejection in Heart Transplant. J. Hear. Lung Transplant. 2020, 39, 1136–1148. [Google Scholar] [CrossRef] [PubMed]
- Sigdel, T.K.; Ng, Y.W.; Lee, S.; Nicora, C.D.; Qian, W.-J.; Smith, R.D.; Camp, D.G.; Sarwal, M.M. Perturbations in the Urinary Exosome in Transplant Rejection. Front. Med. 2015, 1, 57. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, S.; Suazo, C.; Boltansky, A.; Ursu, M.; Carvajal, D.; Innocenti, G.; Vukusich, A.; Hurtado, M.; Villanueva, S.; Carreño, J.E.; et al. Urinary Exosomes as a Source of Kidney Dysfunction Biomarker in Renal Transplantation. Transplant. Proc. 2013, 45, 3719–3723. [Google Scholar] [CrossRef] [PubMed]
- Bruschi, M.; Santucci, L.; Ravera, S.; Candiano, G.; Bartolucci, M.; Calzia, D.; Lavarello, C.; Inglese, E.; Ramenghi, L.A.; Petretto, A.; et al. Human Urinary Exosome Proteome Unveils Its Aerobic Respiratory Ability. J. Proteom. 2016, 136, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Pisitkun, T.; Gandolfo, M.T.; Das, S.; Knepper, M.A.; Bagnasco, S.M. Application of Systems Biology Principles to Protein Biomarker Discovery: Urinary Exosomal Proteome in Renal Transplantation. Proteom. Clin. Appl. 2012, 6, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, P.A.; Pisitkun, T.; Hoffert, J.D.; Tchapyjnikov, D.; Star, R.A.; Kleta, R.; Wang, N.S.; Knepper, M.A. Large-Scale Proteomics and Phosphoproteomics of Urinary Exosomes. J. Am. Soc. Nephrol. 2009, 20, 363–379. [Google Scholar] [CrossRef]
- Al-Nedawi, K.; Haas-Neill, S.; Gangji, A.; Ribic, C.M.; Kapoor, A.; Margetts, P. Circulating Microvesicle Protein Is Associated with Renal Transplant Outcome. Transpl. Immunol. 2019, 55, 101210. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Lee, C.-H.; Kim, K.Y.; Jung, H.; Choi, J.-Y.; Cho, J.; Park, S.; Kim, Y.-L.; Baek, M.; Park, J.B.; et al. Novel Urinary Exosomal Biomarkers of Acute T Cell-Mediated Rejection in Kidney Transplant Recipients: A Cross-Sectional Study. PLoS ONE 2018, 13, e0204204. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.-Y.; Lee, C.-H.; Choi, J.-Y.; Cho, J.-H.; Park, S.-H.; Kim, Y.-L.; Moon, P.-G.; Baek, M.-C.; Berm Park, J.; Hoon Kim, Y.; et al. Potential Urinary Extracellular Vesicle Protein Biomarkers of Chronic Active Antibody-Mediated Rejection in Kidney Transplant Recipients. J. Chromatogr. B 2020, 1138, 121958. [Google Scholar] [CrossRef] [PubMed]
- Chari, R.; Coe, B.P.; Vucic, E.A.; Lockwood, W.W.; Lam, W.L. An Integrative Multi-Dimensional Genetic and Epigenetic Strategy to Identify Aberrant Genes and Pathways in Cancer. BMC Syst. Biol. 2010, 4, 67. [Google Scholar] [CrossRef]
- Mariani, M.; He, S.; McHugh, M.; Andreoli, M.; Pandya, D.; Sieber, S.; Wu, Z.; Fiedler, P.; Shahabi, S.; Ferlini, C. Integrated Multidimensional Analysis Is Required for Accurate Prognostic Biomarkers in Colorectal Cancer. PLoS ONE 2014, 9, e101065. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramalhete, L.M.; Araújo, R.; Ferreira, A.; Calado, C.R.C. Proteomics for Biomarker Discovery for Diagnosis and Prognosis of Kidney Transplantation Rejection. Proteomes 2022, 10, 24. https://doi.org/10.3390/proteomes10030024
Ramalhete LM, Araújo R, Ferreira A, Calado CRC. Proteomics for Biomarker Discovery for Diagnosis and Prognosis of Kidney Transplantation Rejection. Proteomes. 2022; 10(3):24. https://doi.org/10.3390/proteomes10030024
Chicago/Turabian StyleRamalhete, Luís M., Rúben Araújo, Aníbal Ferreira, and Cecília R. C. Calado. 2022. "Proteomics for Biomarker Discovery for Diagnosis and Prognosis of Kidney Transplantation Rejection" Proteomes 10, no. 3: 24. https://doi.org/10.3390/proteomes10030024
APA StyleRamalhete, L. M., Araújo, R., Ferreira, A., & Calado, C. R. C. (2022). Proteomics for Biomarker Discovery for Diagnosis and Prognosis of Kidney Transplantation Rejection. Proteomes, 10(3), 24. https://doi.org/10.3390/proteomes10030024








