The OsAP4-OsCATA/OsCATC Regulatory Module Orchestrates Drought Stress Adaptation in Rice Seedlings Through ROS Scavenging
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatic Analysis and Statistical Analysis
2.2. Plant Materials, Drought Stress Treatment, and Phenotypic Measurements
2.3. Measurement of Physiological and Biochemical Indices
2.4. Transcript Level Analysis
2.5. Subcellular Localization
2.6. Yeast Two-Hybrid (Y2H) Assay
2.7. Bimolecular Fluorescence Complementation (BiFC) Assay
2.8. Pull-Down Assay
2.9. Co-Immunoprecipitation (Co-IP) Assay
2.10. In Vivo Protein Degradation Assay
3. Results
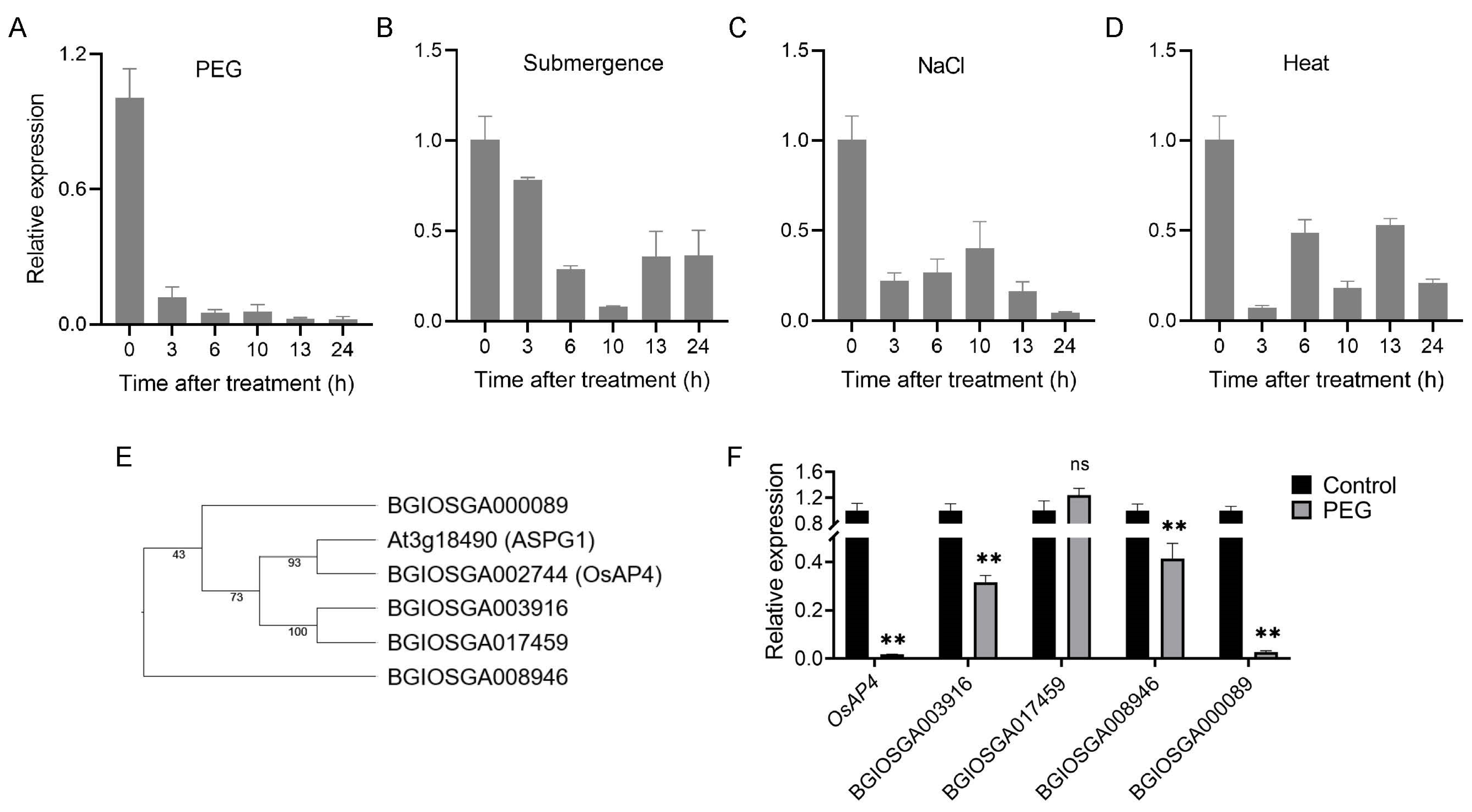
3.1. Expression Patterns of OsAP4
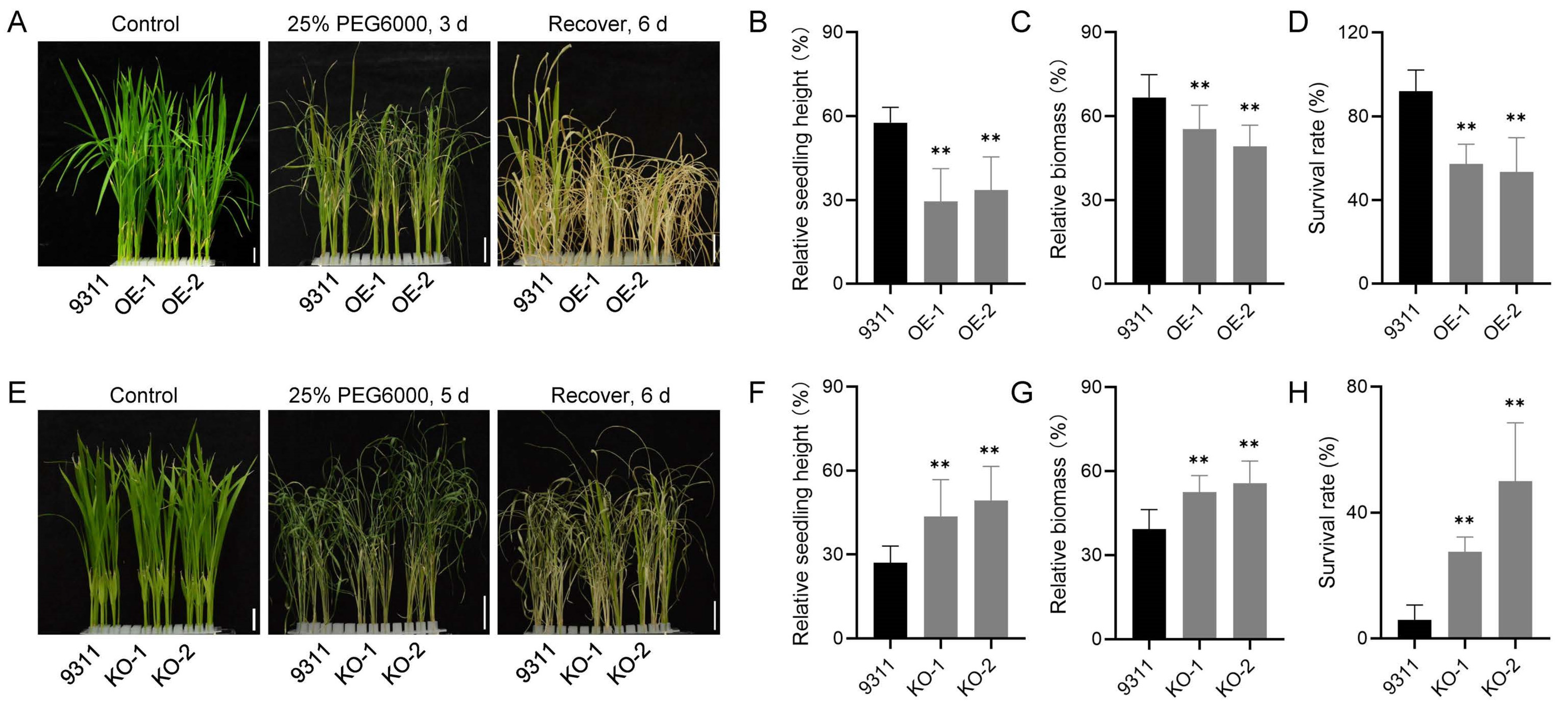
3.2. OsAP4 Negatively Regulates Drought Stress Tolerance in Rice
3.3. Identification of Proteins Interacting with OsAP4
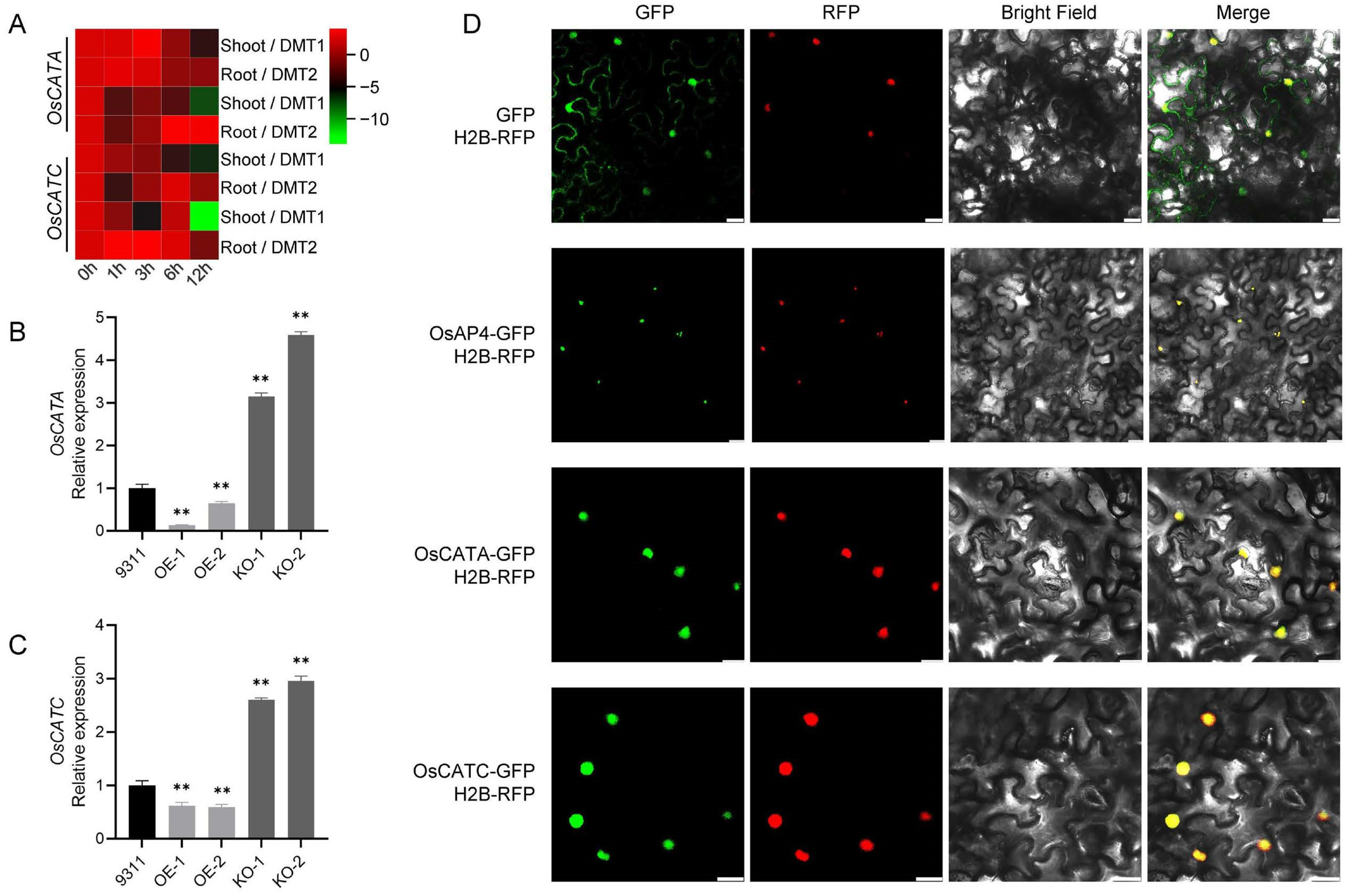
3.4. OsAP4 Interacts with and Destabilizes OsCATA/OsCATC
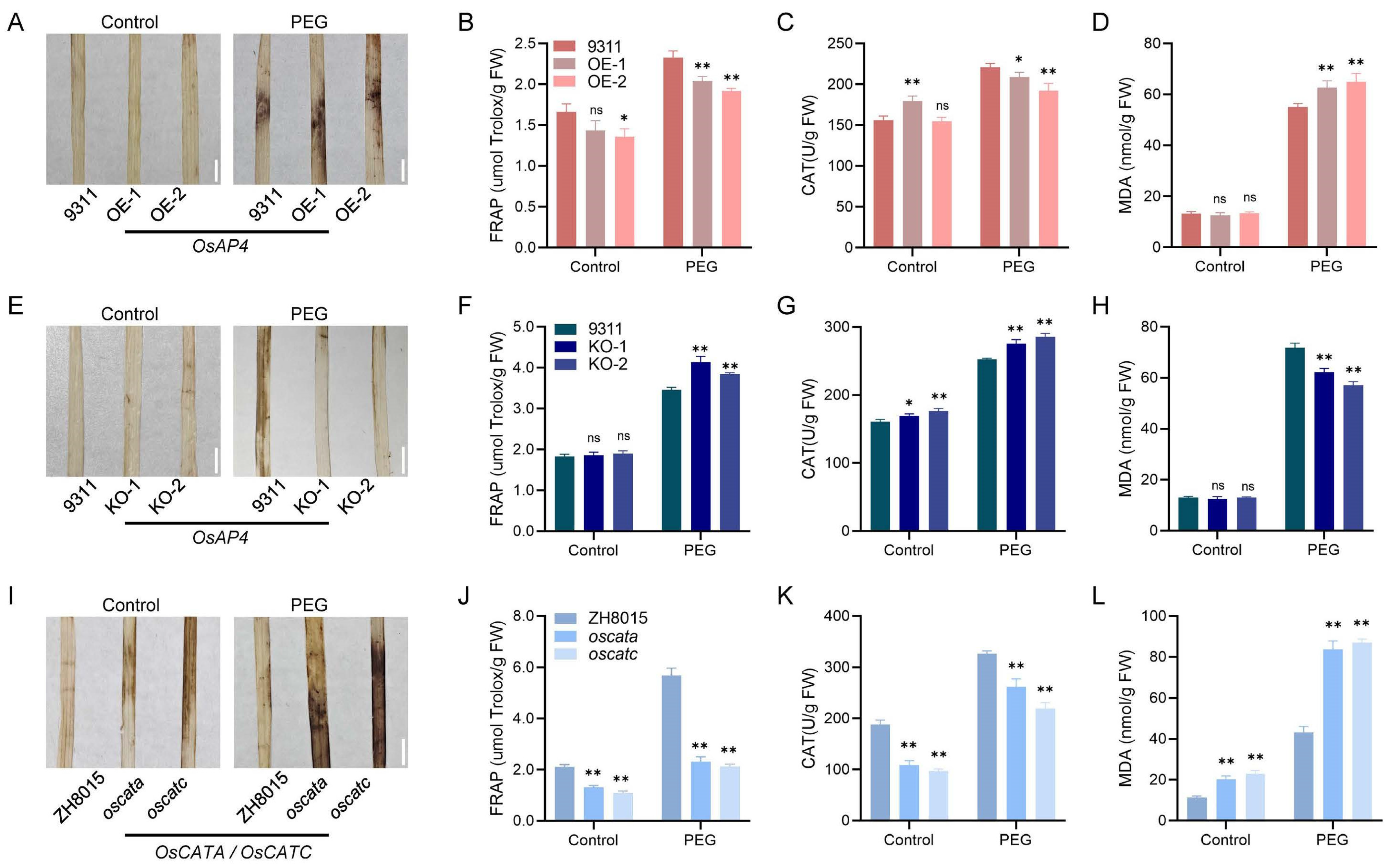
3.5. OsAP4 Controls ROS Scavenging Capacity
3.6. OsAP4 and OsCATA/OsCATC Expression in Response to ABA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Takahashi, K.; Athauda, S.; Matsumoto, K.; Rajapakshe, S.; Inoue, H. Nepenthesin, a unique member of a novel subfamily of aspartic peoteinases: Enzymatic and structural characteritics. Curr. Protein Pept. Sci. 2005, 6, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Mutlu, A.; Gal, S. Plant aspartic proteinases: Enzymes On the way to a function. Physiol. Plant. 1999, 105, 569–576. [Google Scholar] [CrossRef]
- Brik, A.; Wong, C. HIV-1 protease: Mechanism and drug discovery. Org. Biomol. Chem. 2002, 1, 5–14. [Google Scholar] [CrossRef]
- Dunn, B. Structure and mechanism of the pepsin-like family of aspartic peptidases. Chem. Rev. 2002, 102, 4431–4458. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huang, H.; Liu, Y.; Wu, Z.; Wang, F.; Fan, X.; Chen, P.; Wang, J. Optical control of protein functions via genetically encoded photocaged aspartic acids. J. Am. Chem. Soc. 2023, 145, 19218–19224. [Google Scholar] [CrossRef]
- Lin, L.; Wang, M.; Zeng, J.; Mao, Y.; Qin, R.; Deng, J.; Ouyang, X.; Hou, X.; Sun, C.; Wang, Y.; et al. Sequence variation of Candida albicans Sap2 enhances fungal pathogenicity via complement evasion and macrophage M2-like phenotype induction. Adv. Sci. 2023, 10, 2206713. [Google Scholar] [CrossRef]
- Kato, Y.; Yamamoto, Y.; Murakami, S.; Sato, F. Post-translational regulation of CND41 protease activity in senescent tobacco leaves. Planta 2005, 222, 643–651. [Google Scholar] [CrossRef]
- Wang, Y.; Garrido-Oter, R.; Wu, J.; Winkelmüller, T.M.; Agler, M.; Colby, T.; Nobori, T.; Kemen, E.; Tsuda, K. Site-specific cleavage of bacterial MucD by secreted proteases mediates antibacterial resistance in Arabidopsis. Nat. Commun. 2019, 10, 2853. [Google Scholar] [CrossRef]
- Li, W.; Li, P.; Deng, Y.; Zhang, Z.; Situ, J.; Huang, J.; Li, M.; Xi, P.; Jiang, Z.; Kong, G. Litchi aspartic protease LcAP1 enhances plant resistance via suppressing cell death triggered by the pectate lyase PlPeL8 from Peronophythora litchii. New Phytol. 2024, 242, 2682–2701. [Google Scholar] [CrossRef]
- Xia, Y.; Ma, Z.; Qiu, M.; Guo, B.; Zhang, Q.; Jiang, H.; Zhang, B.; Lin, Y.; Xuan, M.; Sun, L.; et al. N-glycosylation shields Phytophthora sojae apoplastic effector PsXEG1 from a specific host aspartic protease. Proc. Natl. Acad. Sci. USA 2020, 117, 27685–27693. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, C.; Li, S.; Zhang, B.; Luo, P.; Peng, X.; Zhao, P.; Dresselhaus, T.; Sun, M.-X. A female in vivo haploid-induction system via mutagenesis of egg cell-specific peptidases. Mol. Plant 2023, 16, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Niu, K.; Chang, C.; Zhang, M.; Guo, Y.; Yan, Y.; Sun, H.; Zhang, G.; Li, X.; Gong, Y.; Ding, C.; et al. Suppressing ASPARTIC PROTEASE 1 prolongs photosynthesis and increases wheat grain weight. Nat. Plants 2023, 9, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Guo, M.; Cheng, J.; Cheng, H.; Liu, X.; Ji, H.; Liu, G.; Cheng, Y.; Yang, C.; Turner, S. Aspartic proteases modulate programmed cell death and secondary cell wall synthesis during wood formation in poplar. J. Exp. Bot. 2022, 73, 6876–6890. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, H.; Li, X.; Tang, L.; Cai, X.; Liu, C.; Zhang, X.; Zhang, J. Aspartyl proteases identified as candidate genes of a fiber length QTL, qFLD05, that regulates fiber length in cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 2024, 137, 59. [Google Scholar] [CrossRef]
- Sebastián, D.; Fernando, F.; Raúl, D.; Gabriela, G. Overexpression of Arabidopsis aspartic protease APA1 gene confers drought tolerance. Plant Sci. 2020, 292, 110406. [Google Scholar] [CrossRef]
- Yao, X.; Xiong, W.; Ye, T.; Wu, Y. Overexpression of the aspartic protease ASPG1 gene confers drought avoidance in Arabidopsis. J. Exp. Bot. 2012, 63, 2579–2593. [Google Scholar] [CrossRef]
- Raimbault, A.-K.; Zuily-Fodil, Y.; Soler, A.; de Carvalho, M.H.C. A novel aspartic acid protease gene from pineapple fruit (Ananas comosus): Cloning, characterization and relation to postharvest chilling stress resistance. J. Plant Physiol. 2013, 170, 1536–1540. [Google Scholar] [CrossRef]
- Chen, J.; Ouyang, Y.; Wang, L.; Xie, W.; Zhang, Q. Aspartic proteases gene family in rice: Gene structure and expression, predicted protein features and phylogenetic relation. Gene 2009, 442, 108–118. [Google Scholar] [CrossRef]
- Chang, S.; Chen, Y.; Jia, S.; Li, Y.; Liu, K.; Lin, Z.; Wang, H.; Chu, Z.; Liu, J.; Xi, C.; et al. Auxin apical dominance governed by the OsAsp1-OsTIF1 complex determines distinctive rice caryopses development on different branches. PLOS Genet. 2020, 16, e1009157. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, X.; Cheng, K.; Jiang, Y.; Ouyang, Y.; Xu, C.; Li, X.; Xiao, J.; Zhang, Q. OsAP65, a rice aspartic protease, is essential for male fertility and plays a role in pollen germination and pollen tube growth. J. Exp. Bot. 2013, 64, 3351–3360. [Google Scholar] [CrossRef]
- Niu, N.; Liang, W.; Yang, X.; Jin, W.; Wilson, Z.A.; Hu, J.; Zhang, D. EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat. Commun. 2013, 4, 1445. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhou, L.; Lan, Y.; Li, X.; Wang, J.; Dong, J.; Guo, W.; Jing, D.; Liu, Q.; Zhang, S.; et al. An aspartic protease 47 causes quantitative recessive resistance to rice black-streaked dwarf virus disease and southern rice black-streaked dwarf virus disease. New Phytol. 2022, 233, 2520–2533. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.; Nakamura, H.; Ichikawa, H.; Miyao, A.; Hirochika, H.; Kobayashi, K.; Yamaoka, N.; Nishiguchi, M. Response of an aspartic protease gene OsAP77 to fungal, bacterial and viral infections in rice. Rice 2014, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.-W.; Li, J.-H.; Gao, X.; Wang, S.-J.; Yuan, T.-T.; Lu, Y.-T. Pathogen-induced methylglyoxal negatively regulates rice bacterial blight resistance by inhibiting OsCDR1 protease activity. Mol. Plant 2024, 17, 325–341. [Google Scholar] [CrossRef]
- Ji, Q.; Zhang, M.; Lu, J.; Wang, H.; Lin, B.; Liu, Q.; Chao, Q.; Zhang, Y.; Liu, C.; Gu, M.; et al. Molecular basis underlying the S5-dependent reproductive isolation and compatibility of indica/japonica rice hybrids. Plant Physiol. 2012, 158, 1319–1328. [Google Scholar] [CrossRef]
- Zhong, H.; Liu, S.; Meng, X.; Sun, T.; Deng, Y.; Kong, W.; Peng, Z.; Li, Y. Uncovering the genetic mechanisms regulating panicle architecture in rice with GPWAS and GWAS. BMC Genom. 2021, 22, 86. [Google Scholar]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one”bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Wang, B.; Xue, P.; Zhang, Y.; Zhan, X.; Wu, W.; Yu, P.; Chen, D.; Fu, J.; Hong, Y.; Shen, X.; et al. OsCPK12 phosphorylates OsCATA and OsCATC to regulate H2O2 homeostasis and improve oxidative stress tolerance in rice. Plant Commun. 2024, 5, 100780. [Google Scholar] [CrossRef]
- Kawahara, Y.; Oono, Y.; Wakimoto, H.; Ogata, J.; Kanamori, H.; Sasaki, H.; Mori, S.; Matsumoto, T.; Itoh, T. TENOR: Database for comprehensive mRNA-Seq experiments in rice. Plant Cell Physiol. 2016, 57, e7. [Google Scholar] [CrossRef]
- An, Z.; Yang, Z.; Zhou, Y.; Huo, S.; Zhang, S.; Wu, D.; Shu, X.; Wang, Y. OsJRL negatively regulates rice cold tolerance via interfering phenylalanine metabolism and flavonoid biosynthesis. Plant Cell Environ. 2024, 47, 4071–4085. [Google Scholar] [CrossRef]
- Li, N.; Lin, B.; Wang, H.; Li, X.; Yang, F.; Ding, X.; Yan, J.; Chu, Z. Natural variation in ZmFBL41 confers banded leaf and sheath blight resistance in maize. Nat. Genet. 2019, 51, 1540–1548. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Wang, K.; Chen, Z.; Cao, Y.; Gao, Q.; Li, Y.; Li, X.; Lu, H.; Du, H.; Lu, M.; et al. MBKbase for rice: An integrated omics knowledgebase for molecular breeding in rice. Nucleic Acids Res. 2020, 48, D1085–D1092. [Google Scholar] [CrossRef]
- Mhamdi, A.; Queval, G.; Chaouch, S.; Vanderauwera, S.; Van Breusegem, F.; Noctor, G. Catalase function in plants: A focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 2010, 61, 4197–4220. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Bao, X.; Sun, H.; Zhu, L.; Zhang, Y.; Zhang, K.; Bai, Z.; Zhu, J.; Liu, X.; Li, A.; et al. Optimizing root system architecture to improve cotton drought tolerance and minimize yield loss during mild drought stress. Field Crops Res. 2024, 308, 109305. [Google Scholar] [CrossRef]
- Wang, X.; Du, T.; Huang, J.; Peng, S.; Xiong, D. Leaf hydraulic vulnerability triggers the decline in stomatal and mesophyll conductance during drought in rice. J. Exp. Bot. 2018, 69, 4033–4045. [Google Scholar] [CrossRef] [PubMed]
- Peralta, D.A.; Araya, A.; Gomez-Casati, D.F.; Busi, M.V. Over-expression of SINAL7 increases biomass and drought tolerance, and also delays senescence in Arabidopsis. J. Biotechnol. 2018, 283, 11–21. [Google Scholar] [CrossRef]
- Jing, Y.; Pei, T.; Zhang, S.; Li, C.; Zhan, M.; Li, C.; Gong, X.; Mao, K.; Liu, C.; Ma, F. Overexpression of FERONIA receptor kinase MdMRLK2 regulates lignin accumulation and enhances water use efficiency in apple under long-term water deficit condition. Plant J. 2024, 119, 2638–2653. [Google Scholar] [CrossRef]
- Gao, H.; Cui, J.; Liu, S.; Wang, S.; Lian, Y.; Bai, Y.; Zhu, T.; Wu, H.; Wang, Y.; Yang, S.; et al. Natural variations of ZmSRO1d modulate the trade-off between drought resistance and yield by affecting ZmRBOHC-mediated stomatal ROS production in maize. Mol. Plant 2022, 15, 1558–1574. [Google Scholar] [CrossRef]
- Jing, X.-Q.; Shi, P.-T.; Zhang, R.; Zhou, M.-R.; Shalmani, A.; Wang, G.-F.; Liu, W.-T.; Li, W.-Q.; Chen, K.-M. Rice kinase OsMRLK63 contributes to drought tolerance by regulating reactive oxygen species production. Plant Physiol. 2024, 194, 2679–2696. [Google Scholar] [CrossRef]
- Joo, J.; Lee, Y.H.; Song, S.I. Rice CatA, CatB, and CatC are involved in environmental stress response, root growth, and photorespiration, respectively. J. Plant Biol. 2014, 57, 375–382. [Google Scholar] [CrossRef]
- Meng, Y.; Li, M.; Guo, Z.; Chen, J.; Wu, J.; Xia, Z. The transcription factor ZmbHLH105 confers cadmium tolerance by promoting abscisic acid biosynthesis in maize. J. Hazard. Mater. 2024, 480, 135826. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Li, L.; Liu, M.; Peng, D.; Wei, A.; Hou, B.; Lei, Y.; Li, X. TaFDL2-1A confers drought stress tolerance by promoting ABA biosynthesis, ABA responses, and ROS scavenging in transgenic wheat. Plant J. 2022, 112, 722–737. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Jin, L.; Sun, Y.; Zhan, C.; Tang, S.; Qin, T.; Liu, N.; Huang, J. OsNAC120 balances plant growth and drought tolerance by integrating GA and ABA signaling in rice. Plant Commun. 2024, 5, 100782. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, Y.; Xie, B.; Luo, X.; Li, Y. The OsAP4-OsCATA/OsCATC Regulatory Module Orchestrates Drought Stress Adaptation in Rice Seedlings Through ROS Scavenging. Plants 2025, 14, 2174. https://doi.org/10.3390/plants14142174
Jiang Y, Xie B, Luo X, Li Y. The OsAP4-OsCATA/OsCATC Regulatory Module Orchestrates Drought Stress Adaptation in Rice Seedlings Through ROS Scavenging. Plants. 2025; 14(14):2174. https://doi.org/10.3390/plants14142174
Chicago/Turabian StyleJiang, Yifei, Bin Xie, Xiong Luo, and Yangsheng Li. 2025. "The OsAP4-OsCATA/OsCATC Regulatory Module Orchestrates Drought Stress Adaptation in Rice Seedlings Through ROS Scavenging" Plants 14, no. 14: 2174. https://doi.org/10.3390/plants14142174
APA StyleJiang, Y., Xie, B., Luo, X., & Li, Y. (2025). The OsAP4-OsCATA/OsCATC Regulatory Module Orchestrates Drought Stress Adaptation in Rice Seedlings Through ROS Scavenging. Plants, 14(14), 2174. https://doi.org/10.3390/plants14142174





