Multi-Omics Research Accelerates the Clarification of the Formation Mechanism and the Influence of Leaf Color Variation in Tea (Camellia sinensis) Plants
Abstract
:1. Introduction
2. Multi-Omics Approaches Further Our Understanding of Leaf Color Variation in Tea
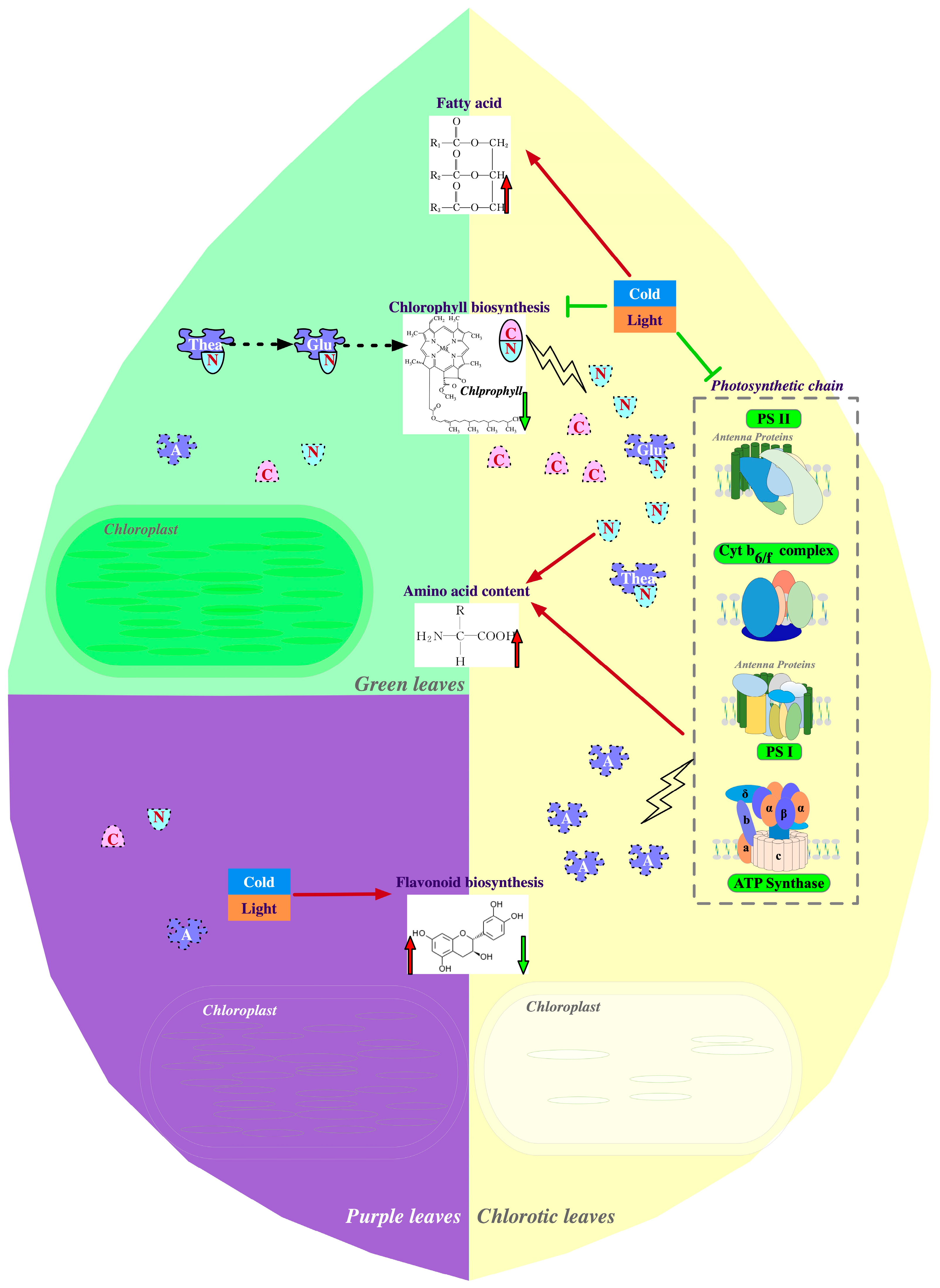
2.1. Physiological Mechanisms of Albinism or Etiolation in Tea
2.2. Molecular Mechanisms of Leaf Chlorosis in Tea Plant
2.3. Physiological Mechanisms of Purple Leaf Coloration in Tea
2.4. Molecular Mechanisms of Purple Coloration in Tea Leaves
3. Metabolic Reprogramming by Leaf Color Variations in Tea Plants
3.1. Influence of Chlorina Variations on Amino Acid Metabolism
3.2. Effects of Chlorina Variation on Flavonoid Metabolism
3.3. Effects of Chlorina Variations on Fatty Acid Metabolism
3.4. Metabolic Changes in Flavonoids Other Than Anthocyanins in Purple Tea
4. Conclusions and Future Prospects
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xia, E.H.; Tong, W.; Wu, Q.; Wei, S.; Zhao, J.; Zhang, Z.Z.; Wei, C.L.; Wan, X.C. Tea plant genomics: Achievements, challenges and perspectives. Hortic. Res. 2020, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.L.; Yang, H.; Wang, S.B.; Zhao, J.; Liu, C.; Gao, L.p.; Xia, E.H.; Lu, Y.; Tai, Y.L.; She, G.B.; et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. USA 2018, 115, E4151–E4158. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cheng, S.; Dai, J.G.; Wang, L.; Xu, Y.; Peng, X.Y.; Xie, X.F.; Peng, C. Molecular mechanisms and applications of tea polyphenols: A narrative review. J. Food Biochem. 2021, 45, e13910. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Y.; Liu, H.Y.; Wu, D.T.; Kenaan, A.; Geng, F.; Li, H.B.; Gunaratne, A.; Li, H.; Gan, R.Y. L-theanine: A unique functional amino acid in tea (Camellia sinensis L.) with multiple health benefits and food applications. Front. Nutr. 2022, 9, 853846. [Google Scholar] [CrossRef]
- Samynathan, R.; Thiruvengadam, M.; Nile, S.H.; Shariati, M.A.; Rebezov, M.; Mishra, R.K.; Venkidasamy, B.; Periyasamy, S.; Chung, I.-M.; Pateiro, M.; et al. Recent insights on tea metabolites, their biosynthesis and chemo-preventing effects: A review. Crit. Rev. Food Sci. Nutr. 2023, 63, 3130–3149. [Google Scholar] [CrossRef]
- Liu, D.D.; Mei, J.F.; Wang, J.Y.; Tang, R.J.; Chen, L.; Ma, C.L. Research Progress on Albino Trait of Tea Plant. China Tea 2020, 42, 24–35. [Google Scholar] [CrossRef]
- Wang, K.R.; Liang, Y.R.; Li, M.; Zhang, L.J. Backbone parents of albino tea plants and teir germplasm traits. J. Tea 2015, 41, 130–132,136. [Google Scholar] [CrossRef]
- Xu, Y.X.; Chen, W.; Ma, C.L.; Shen, S.Y.; Zhou, Y.Y.; Zhou, L.Q.; Chen, L. Proteome and acetyl-proteome profiling of Camellia sinensis cv. ‘Anji Baicha’ during periodic albinism reveals alterations in photosynthetic and secondary metabolite biosynthetic pathways. Front. Plant Sci. 2017, 8, 147. [Google Scholar] [CrossRef]
- Fan, Y.G.; Zhao, X.X.; Wang, H.Y.; Tian, Y.Y.; Xiang, Q.Z.; Zhanf, L.X. Effects of light intensity on metabolism of light-harvesting pigment and photosynthetic system in Camellia sinensis L. cultivar ‘Huangjinya’. Environ. Exp. Bot. 2019, 166, 103796. [Google Scholar] [CrossRef]
- Zhang, Q.F.; Li, C.L.; Jiao, Z.X.; Ruan, J.Y.; Liu, M.Y. Integration of metabolomics and transcriptomics reveal the mechanism underlying accumulation of flavonols in albino tea leaves. Molecules 2022, 27, 5792. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Huang, J.N.; Liu, S.Q.; Li, J.; Yang, X.H.; Liu, Y.S.; Liu, Z.H. Proteomic analysis of young leaves at three developmental stages in an albino tea cultivar. Proteome Sci. 2011, 9, 44. [Google Scholar] [CrossRef]
- Ma, Q.P.; Li, H.; Zou, Z.W.; Arkorful, E.; Lv, Q.R.; Zhou, Q.Q.; Chen, X.; Sun, K.; Li, X.H. Transcriptomic analyses identify albino-associated genes of a novel albino tea germplasm ‘Huabai 1′. Hortic. Res. 2018, 5, 54. [Google Scholar] [CrossRef]
- Jiang, X.F.; Zhao, H.; Guo, F.; Shi, X.P.; Ye, C.; Yang, P.X.; Liu, B.Y.; Ni, D.J. Transcriptomic analysis reveals mechanism of light-sensitive albinism in tea plant Camellia sinensis ‘Huangjinju’. BMC Plant Biol. 2020, 20, 216. [Google Scholar] [CrossRef]
- Liu, S.C.; Xu, Y.F.; Liu, Y.B.; Zhao, X.; Wei, J.; Lin, K.Q.; Liu, Y.; Yan, D.H. Metabolomic and transcriptomic analyses reveal the biosynthetic mechanisms of pigments and main taste compounds in an albino tea cultivar. Physiol. Plant 2023, 175, e13933. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.J.; Kong, X.G.; Zhang, Y.; Wang, S.Y.; Zhou, H.W.; Fang, D.S.; Yue, W.J.; Chen, C.S. Metabolomic profiling in combination with data association analysis provide insights about potential metabolic regulation networks among non-volatile and volatile metabolites in Camellia sinensis cv Baijiguan. Plants 2022, 11, 2557. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.C.; Wang, P.J.; Chen, X.J.; Yue, C.; Guo, Y.C.; Yang, J.F.; Sun, Y.; Ye, N.X. Integrated transcriptomics and metabolomics provide novel insight into changes in specialized metabolites in an albino tea cultivar (Camellia sinensis (L.) O. Kuntz). Plant Physiol. Biochem. 2021, 160, 27–36. [Google Scholar] [CrossRef]
- Ma, C.Y.; Cao, J.X.; Li, J.K.; Zhou, B.; Tang, J.C.; Miao, A.Q. Phenotypic, histological and proteomic analyses reveal multiple differences associated with chloroplast development in yellow and variegated variants from Camellia sinensis. Sci. Rep. 2016, 6, 33369. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.Z.; Zhang, C.Y.; Lu, C.; Wang, M.H.; Xie, N.C.; Chen, J.J.; Li, Y.F.; Chen, J.H.; Shen, C.W. Disruption of photomorphogenesis leads to abnormal chloroplast development and leaf variegation in Camellia sinensis. Front. Plant Sci. 2021, 12, 720800. [Google Scholar] [CrossRef]
- Li, X.X.; Li, Z.Y.; Zhu, W.; Wang, Y.Q.; Liang, Y.R.; Wang, K.R.; Ye, J.H.; Lu, J.L.; Zheng, X.Q. Anthocyanin metabolism and its differential regulation in purple tea (Camellia sinensis). Plant Physiol. Biochem. 2023, 201, 107875. [Google Scholar] [CrossRef]
- Wang, L.; Pan, D.; Liang, M.; Abubakar, Y.S.; Li, J.; Lin, J.; Chen, S.P.; Chen, W. Regulation of anthocyanin biosynthesis in purple leaves of Zijuan Tea (Camellia sinensis var. kitamura). Int. J. Mol. Sci. 2017, 18, 833. [Google Scholar] [CrossRef]
- Tan, L.Q.; Yang, C.J.; Zhou, B.; Wang, L.B.; Zou, Y.; Chen, W.; Xia, T.; Tang, Q. Inheritance and quantitative trait loci analyses of the anthocyanins and catechins of Camellia sinensis cultivar ‘Ziyan’ with dark-purple leaves. Physiol. Plant 2020, 170, 109–119. [Google Scholar] [CrossRef]
- Chen, X.J.; Wang, P.J.; Wei, M.X.; Lin, X.Y.; Gu, M.Y.; Fang, W.P.; Zheng, Y.C.; Zhao, F.; Jin, S.; Ye, N.X. Lipidomics analysis unravels changes from flavor precursors in different processing treatments of purple-leaf tea. J. Sci. Food Agric. 2022, 102, 3730–3741. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.; Wu, S.Y.; Li, H.K.; Tian, J.L. Health benefits of anthocyanin-containing foods, beverages, and supplements have unpredictable relation to gastrointestinal microbiota: A systematic review and meta-analysis of random clinical trials. Nutr. Res. 2023, 116, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, P.J.; Kong, W.L.; Chai, K.; Zhang, S.C.; Yu, J.X.; Wang, Y.B.; Jiang, M.W.; Lei, W.L.; Chen, X.; et al. Gene mining and genomics-assisted breeding empowered by the pangenome of tea plant Camellia sinensis. Nat. Plants 2023, 9, 1986–1999. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.T.; Chen, S.; Shi, L.Q.; Gong, D.P.; Zhang, S.C.; Zhao, Q.; Zhan, D.L.; Vasseur, L.; Wang, Y.B.; Yu, J.X.; et al. Haplotype-resolved genome assembly provides insights into evolutionary history of the tea plant Camellia sinensis. Nat. Genet. 2021, 53, 1250–1259. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.C.; Feng, H.; Chang, Y.X.; Ma, C.L.; Wang, L.Y.; Hao, X.Y.; Li, A.L.; Cheng, H.; Wang, L.; Cui, P.; et al. Population sequencing enhances understanding of tea plant evolution. Nat. Commun. 2020, 11, 4447. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.Y.; Zhang, Y.J.; Qiu, H.J.; Guo, Y.F.; Wan, H.L.; Zhang, X.L.; Scossa, F.; Alseekh, S.; Zhang, Q.H.; Wang, P.; et al. Genome assembly of wild tea tree DASZ reveals pedigree and selection history of tea varieties. Nat. Commun. 2020, 11, 3719. [Google Scholar] [CrossRef]
- Wang, P.; Grimm, B. Connecting chlorophyll metabolism with accumulation of the photosynthetic apparatus. Trends Plant Sci. 2021, 26, 484–495. [Google Scholar] [CrossRef]
- Wang, J.Y.; Chen, J.D.; Wang, S.L.; Chen, L.; Ma, C.L.; Yao, M.Z. Repressed Gene Expression of photosynthetic antenna proteins associated with yellow leaf variation as revealed by bulked segregant RNA-seq in tea plant Camellia sinensis. J. Agric. Food Chem. 2020, 68, 8068–8079. [Google Scholar] [CrossRef]
- Mei, X.; Zhang, K.K.; Lin, Y.G.; Su, H.F.; Lin, C.Y.; Chen, B.Y.; Yang, H.J.; Zhang, L.Y. Metabolic and Transcriptomic profiling reveals etiolated mechanism in huangyu tea (Camellia sinensis) leaves. Int. J. Mol. Sci. 2022, 23, 15044. [Google Scholar] [CrossRef]
- Li, C.F.; Yao, M.Z.; Ma, C.L.; Ma, J.Q.; Jin, J.Q.; Chen, L. Differential metabolic profiles during the albescent stages of ‘Anji Baicha’ (Camellia sinensis). PLoS ONE 2015, 10, e0139996. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.F.; Liu, M.Y.; Ruan, J.Y. Integrated transcriptome and metabolic analyses reveals novel insights into free amino acid metabolism in Huangjinya tea cultivar. Front. Plant Sci. 2017, 8, 291. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.F.; Han, Z.X.; Feng, L.; Gao, L.P.; Gao, M.J.; Gruber, M.Y.; Zhang, Z.L.; Xia, T.; Wan, X.C.; Wei, S. Metabolic flux redirection and transcriptomic reprogramming in the albino tea cultivar ‘Yu-Jin-Xiang’ with an emphasis on catechin production. Sci. Rep. 2017, 7, 45062. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.J.; Chen, Z.D.; Sun, W.J.; Deng, T.T.; Chen, M.J. De novo sequencing of the leaf transcriptome reveals complex light-responsive regulatory networks in Camellia sinensis cv. Baijiguan. Front. Plant Sci. 2016, 7, 332. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.B.; Li, J.L.; Wang, H.L.; Liu, H.J.; Yu, Z.H.; Zhao, Z. Whole-transcriptome sequencing reveals a cerna regulatory network associated with the process of periodic albinism under low temperature in Baiye No. 1 (Camellia sinensis). Int. J. Mol. Sci. 2023, 24, 7162. [Google Scholar] [CrossRef]
- Fang, D.; Shi, Y.Z.; Liu, M.Y.; Fan, K.; Zhang, Q.F.; Ruan, J.Y. iTRAQ-based quantitative proteomics analysis reveals the mechanism underlying the weakening of carbon metabolism in chlorotic tea leaves. Int. J. Mol. Sci. 2018, 19, 3943. [Google Scholar] [CrossRef]
- Zhang, Q.F.; Liu, M.Y.; Ruan, J.Y. Metabolomics analysis reveals the metabolic and functional roles of flavonoids in light-sensitive tea leaves. BMC Plant Biol. 2017, 17, 64. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Liu, H.R.; Wang, J.Y.; Li, Y.Y.; Liu, D.D.; Ye, Y.Y.; Huang, R.; Li, S.J.; Chen, L.; Chen, J.D.; et al. A key mutation in magnesium chelatase I subunit leads to a chlorophyll-deficient mutant of tea (Camellia sinensis). J. Exp. Bot. 2023, erad430. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Liu, G.Z.; Chen, J.J.; Xie, N.C.; Huang, J.A.; Shen, C.W. Translational landscape and metabolic characteristics of the etiolated tea plant (Camellia sinensis). Sci. Hortic. 2022, 303, 111193. [Google Scholar] [CrossRef]
- Xie, N.C.; Zhang, C.Y.; Zhou, P.Q.; Gao, X.Z.; Wang, M.H.; Tian, S.H.; Lu, C.; Wang, K.B.; Shen, C.W. Transcriptomic analyses reveal variegation-induced metabolic changes leading to high L-theanine levels in albino sectors of variegated tea (Camellia sinensis). Plant Physiol. Biochem. 2021, 169, 29–39. [Google Scholar] [CrossRef]
- Liu, Y.F.; Pang, D.D.; Jiang, H.B.; Chen, C.L.; Sun, Y.N.; Tian, Y.P.; Xu, Y.; Song, W.X.; Chen, L.B. Identifying key genes involved in yellow leaf variation in ‘Menghai Huangye’ based on biochemical and transcriptomic analysis. Funct. Integr. Genom. 2022, 22, 251–260. [Google Scholar] [CrossRef]
- Wang, L.; Yue, C.; Cao, H.L.; Zhou, Y.H.; Zeng, J.M.; Yang, Y.J.; Wang, X.C. Biochemical and transcriptome analyses of a novel chlorophyll-deficient chlorina tea plant cultivar. BMC Plant Biol. 2014, 14, 352. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Zhang, C.Y.; Ma, C.L.; Chen, L.; Yao, M.Z. Transcriptome and biochemical analyses of a chlorophyll-deficient bud mutant of tea plant (Camellia sinensis). Int. J. Mol. Sci. 2023, 24, 15070. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.J.; Gu, H.Y.; Ma, L.G.; Peng, Y.B.; Deng, X.W.; Chen, Z.L.; Qu, L.J. Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 2007, 17, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Hertle, A.P.; García-Cerdán, J.G.; Armbruster, U.; Shih, R.; Lee, J.J.; Wong, W.; Niyogi, K.K. A Sec14 domain protein is required for photoautotrophic growth and chloroplast vesicle formation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2020, 117, 9101–9111. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.X.; Shen, C.J.; Ma, J.Q.; Chen, W.; Mao, J.; Zhou, Y.Y.; Chen, L. Quantitative succinyl-proteome profiling of Camellia sinensis cv. ‘Anji Baicha’ during periodic albinism. Sci. Rep. 2017, 7, 1873. [Google Scholar] [CrossRef] [PubMed]
- Song, L.B.; Ma, Q.P.; Zou, Z.W.; Sun, K.; Yao, Y.T.; Tao, J.H.; Kaleri, N.A.; Li, X.H. Molecular link between leaf coloration and gene expression of flavonoid and carotenoid biosynthesis in Camellia sinensis cultivar ‘Huangjinya’. Front. Plant Sci. 2017, 8, 803. [Google Scholar] [CrossRef]
- Lu, M.Q.; Han, J.Y.; Zhu, B.Y.; Jia, H.Y.; Yang, T.Y.; Wang, R.J.; Deng, W.W.; Zhang, Z.Z. Significantly increased amino acid accumulation in a novel albino branch of the tea plant (Camellia sinensis). Planta 2019, 249, 363–376. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.X.; Yang, L.; Lei, Y.S.; Ju, R.N.; Miao, S.G.; Jin, S.H. Integrated transcriptome and amino acid profile analyses reveal novel insights into differential accumulation of theanine in green and yellow tea cultivars. Tree Physiol. 2022, 42, 1501–1516. [Google Scholar] [CrossRef]
- Yamashita, H.; Kambe, Y.; Ohshio, M.; Kunihiro, A.; Tanaka, Y.; Suzuki, T.; Nakamura, Y.; Morita, A.; Ikka, T. Integrated metabolome and transcriptome analyses reveal etiolation-induced metabolic changes leading to high amino acid contents in a light-sensitive Japanese albino tea cultivar. Front. Plant Sci. 2021, 11, 611140. [Google Scholar] [CrossRef]
- Liu, X.Y.; Cao, J.J.; Cheng, X.; Zhu, W.F.; Sun, Y.; Wan, X.C.; Liu, L.L. CsRVE1 promotes seasonal greening of albino Camellia sinensis cv. Huangkui by activating chlorophyll biosynthesis. Tree Physiol. 2023, 43, 1432–1443. [Google Scholar] [CrossRef]
- Lin, X.Y.; Wang, P.J.; Yang, R.X.; Zheng, Y.C.; Chen, X.M.; Zhang, L.; Xian, S.S.; Xing, Y.N. The albino mechanism of a new high theanine tea cultivar Fuhuang 1. Sci. Agric. Sin. 2022, 55, 22. [Google Scholar] [CrossRef]
- Lin, X.Y.; Shao, S.X.; Wang, P.J.; Yang, R.X.; Zheng, Y.C.; Chen, X.M.; Zhang, L.; Ye, N.X. The albino mechanism of a new high theanine tea cultivar Fuhuang 2. Chin. J. Biotechnol. 2022, 38, 3956–3972. [Google Scholar] [CrossRef]
- Sandmann, G. Antioxidant protection from UV- and light-stress related to carotenoid structures. Antioxidants 2019, 8, 219. [Google Scholar] [CrossRef] [PubMed]
- Di Mascio, P.; Kaiser, S.; Sies, H. Lycopene as the most efficient biological carotenoid singlet oxygen quencher. Arch. Biochem. Biophys. 1989, 274, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Boey Peng, L.; Akihiko, N.; Junji, T.; Kazunobu, T.; Tetsuya, S.; Kozo, T. Antioxidant activity of xanthophylls on peroxyl radical-mediated phospholipid peroxidation. Biochim. Biophys. Acta (BBA)—Lipids Lipid Metab. 1992, 1126, 178–184. [Google Scholar] [CrossRef]
- Woodall, A.A.; Britton, G.; Jackson, M.J. Carotenoids and protection of phospholipids in solution or in liposomes against oxidation by peroxyl radicals: Relationship between carotenoid structure and protective ability. Biochim. Biophys. Acta (BBA)—Gen. Subj. 1997, 1336, 575–586. [Google Scholar] [CrossRef]
- Du, H.Y.; Qi, M.Z.; Cui, X.P.; Cui, Y.M.; Yang, H.; Zhang, J.Y.; Ma, Y.J.; Zhang, S.S.; Zhang, X.; Yu, D.Y. Proteomic and functional analysis of soybean chlorophyll-deficient mutant cd1 and the underlying gene encoding the CHLI subunit of Mg-chelatase. Mol. Breed. 2018, 38, 71. [Google Scholar] [CrossRef]
- Satou, M.; Enoki, H.; Oikawa, A.; Ohta, D.; Saito, K.; Hachiya, T.; Sakakibara, H.; Kusano, M.; Fukushima, A.; Saito, K.; et al. Integrated analysis of transcriptome and metabolome of Arabidopsis albino or pale green mutants with disrupted nuclear-encoded chloroplast proteins. Plant Mol. Biol. 2014, 85, 411–428. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, L.L.; Cai, M.H.; Zhu, S.S.; Zhao, J.Y.; Zheng, T.H.; Xu, X.Y.; Zeng, Z.Q.; Niu, J.; Jiang, L.; et al. A point mutation of magnesium chelatase OsCHLI gene dampens the interaction between CHLI and CHLD subunits in rice. Plant Mol. Biol. Rep. 2015, 33, 1975–1987. [Google Scholar] [CrossRef]
- Hansson, A.; Kannangara, C.G.; von Wettstein, D.; Hansson, M. Molecular basis for semidominance of missense mutations in the XANTHA-H (42-kDa) subunit of magnesium chelatase. Proc. Natl. Acad. Sci. USA 1999, 96, 1744–1749. [Google Scholar] [CrossRef] [PubMed]
- Schön, A.; Krupp, G.; Gough, S.; Berry-Lowe, S.; Kannangara, C.G.; Söll, D. The RNA required in the first step of chlorophyll biosynthesis is a chloroplast glutamate tRNA. Nature 1986, 322, 281–284. [Google Scholar] [CrossRef] [PubMed]
- Teng, K.; Tan, P.H.; Guan, J.; Dong, D.; Liu, L.U.; Guo, Y.D.; Guo, W.E.; Yuesen, Y.; Fan, X.F.; Wu, J.Y. Functional characterization of the chlorophyll b reductase gene NYC1 associated with chlorophyll degradation and photosynthesis in Zoysia japonica. Environ. Exp. Bot. 2021, 191, 104607. [Google Scholar] [CrossRef]
- Sato, R.; Ito, H.; Tanaka, A. Chlorophyll b degradation by chlorophyll b reductase under high-light conditions. Photosynth. Res. 2015, 126, 249–259. [Google Scholar] [CrossRef]
- Niu, G.Q.; Guo, Q.; Wang, J.; Zhao, S.; He, Y.K.; Liu, L. Structural basis for plant lutein biosynthesis from α-carotene. Proc. Natl. Acad. Sci. USA 2020, 117, 14150–14157. [Google Scholar] [CrossRef] [PubMed]
- Pružinská, A.; Tanner, G.; Anders, I.; Roca, M.; Hörtensteiner, S. Chlorophyll breakdown: Pheophorbide a oxygenase is a Rieske-type iron–sulfur protein, encoded by the accelerated cell death 1 gene. Proc. Natl. Acad. Sci. USA 2003, 100, 15259–15264. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chen, Z.T.; Sun, Q.X.; Liu, J.X. Proteome-wide analyses reveal diverse functions of protein acetylation and succinylation modifications in fast growing stolons of bermudagrass (Cynodon dactylon L.). BMC Plant Biol. 2022, 22, 503. [Google Scholar] [CrossRef]
- Kobayashi, K.; Masuda, T. Chapter Five—Transcriptional control for the chlorophyll metabolism. In Advances in Botanical Research; Grimm, B., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 91, pp. 133–161. [Google Scholar]
- Sathasivam, R.; Radhakrishnan, R.; Kim, J.K.; Park, S.U. An update on biosynthesis and regulation of carotenoids in plants. S. Afr. J. Bot. 2021, 140, 290–302. [Google Scholar] [CrossRef]
- Engelken, J.; Brinkmann, H.; Adamska, I. Taxonomic distribution and origins of the extended LHC (light-harvesting complex) antenna protein superfamily. BMC Evol. Biol. 2010, 10, 233. [Google Scholar] [CrossRef]
- Takabayashi, A.; Kurihara, K.; Kuwano, M.; Kasahara, Y.; Tanaka, R.; Tanaka, A. The oligomeric states of the photosystems and the light-harvesting complexes in the Chl b-less mutant. Plant Cell Physiol. 2011, 52, 2103–2114. [Google Scholar] [CrossRef] [PubMed]
- Pietrzykowska, M.; Suorsa, M.; Semchonok, D.A.; Tikkanen, M.; Boekema, E.J.; Aro, E.-M.; Jansson, S. The light-harvesting chlorophyll a/b binding proteins Lhcb1 and Lhcb2 play complementary roles during state transitions in Arabidopsis. Plant Cell 2014, 26, 3646–3660. [Google Scholar] [CrossRef]
- Wietrzynski, W.; Engel, B.D. Chlorophyll biogenesis sees the light. Nat. Plants 2021, 7, 380–381. [Google Scholar] [CrossRef]
- Fu, C.X.; Tang, X.Y.; Rui, R.L.; Chen, Y.F.; Zhong, W.G. Pigment photobleach and its component changes in indica varieties leaves under high light. Chin. J. Rice Sci. 1993, 7, 83–87. [Google Scholar]
- Nick, S.; Meurer, J.; Soll, J.; Ankele, E. Nucleus-Encoded Light-harvesting chlorophyll a/b proteins are imported normally into chlorophyll b-free chloroplasts of Arabidopsis. Mol. Plant 2013, 6, 860–871. [Google Scholar] [CrossRef]
- Kindgren, P.; Norén, L.; Barajas López, J.d.D.; Shaikhali, J.; Strand, Å. Interplay between HEAT SHOCK PROTEIN 90 and HY5 controls PhANG expression in response to the GUN5 plastid signal. Mol. Plant 2012, 5, 901–913. [Google Scholar] [CrossRef]
- Sun, B.M.; Zhu, Z.S.; Cao, P.R.; Chen, H.; Chen, C.M.; Zhou, X.; Mao, Y.H.; Lei, J.J.; Jiang, Y.P.; Meng, W.; et al. Purple foliage coloration in tea (Camellia sinensis L.) arises from activation of the R2R3-MYB transcription factor CsAN1. Sci. Rep. 2016, 6, 32534. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.Z.; Zou, Z.W.; Zhang, X.Z.; Zhou, L.; Wang, Y.H.; Fang, W.P.; Zhu, X.J. Metabolic analyses reveal different mechanisms of leaf color change in two purple-leaf tea plant (Camellia sinensis L.) cultivars. Hortic. Res. 2018, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.Q.; Sun, W.J.; Lai, Z.X. Differential expression of genes in purple-shoot tea tender leaves and mature leaves during leaf growth. J. Sci. Food Agric. 2016, 96, 1982–1989. [Google Scholar] [CrossRef]
- Zhang, Q.F.; Hu, J.H.; Liu, M.Y.; Shi, Y.Z.; De Vos, R.C.H.; Ruan, J.Y. Stimulated biosynthesis of delphinidin-related anthocyanins in tea shoots reducing the quality of green tea in summer. J. Sci. Food Agric. 2020, 100, 1505–1514. [Google Scholar] [CrossRef]
- Maritim, T.K.; Korir, R.K.; Nyabundi, K.W.; Wachira, F.N.; Kamunya, S.M.; Muoki, R.C. Molecular regulation of anthocyanin discoloration under water stress and high solar irradiance in pluckable shoots of purple tea cultivar. Planta 2021, 254, 85. [Google Scholar] [CrossRef]
- Tan, L.Q.; Zhang, P.; Cui, D.; Yang, X.; Zhang, D.Y.; Yang, Y.; Chen, W.; Tang, D.D.; Tang, Q.; Li, P.W. Multi-omics analysis revealed anthocyanin accumulation differences in purple tea plants ‘Ziyan’, ‘Zijuan’ and their dark-purple hybrid. Sci. Hortic. 2023, 321, 112275. [Google Scholar] [CrossRef]
- Chen, X.J.; Wang, P.J.; Zheng, Y.C.; Gu, M.Y.; Lin, X.Y.; Wang, S.Y.; Jin, S.; Ye, N.X. Comparison of metabolome and transcriptome of flavonoid biosynthesis pathway in a purple-leaf tea germplasm Jinmingzao and a green-leaf tea germplasm huangdan reveals their relationship with genetic mechanisms of color formation. Int. J. Mol. Sci. 2020, 21, 4167. [Google Scholar] [CrossRef]
- Cai, J.; Lv, L.T.; Zeng, X.F.; Zhang, F.; Chen, Y.L.; Tian, W.L.; Li, J.R.; Li, X.Y.; Li, Y. Integrative analysis of metabolomics and transcriptomics reveals molecular mechanisms of anthocyanin metabolism in the Zikui tea plant (Camellia sinensis cv. Zikui). Int. J. Mol. Sci. 2022, 23, 4780. [Google Scholar] [CrossRef]
- Shi, J.; Simal-Gandara, J.; Mei, J.F.; Ma, W.J.; Peng, Q.H.; Shi, Y.L.; Xu, Q.; Lin, Z.; Lv, H.P. Insight into the pigmented anthocyanins and the major potential co-pigmented flavonoids in purple-coloured leaf teas. Food Chem. 2021, 363, 130278. [Google Scholar] [CrossRef]
- Li, M.W.; Shen, Y.; Ling, T.J.; Ho, C.T.; Li, D.X.; Guo, H.M.; Xie, Z.W. Analysis of Differentiated chemical components between Zijuan purple tea and Yunkang green tea by UHPLC-Orbitrap-MS/MS combined with chemometrics. Foods 2021, 10, 1070. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.W.; Shi, X.Y.; Duan, S.M.; Nian, B.; Chen, L.J.; Zhang, G.H.; Lv, C.Y.; Ma, Y.; Zhao, M. Multiomics analysis of the mechanisms behind flavonoid differences between purple and green tender shoots of Camellia sinensis var. assamica. G3-Genes Genom. Genet. 2022, 13, jkac297. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.Y.; Duan, J.H.; Lei, Y.; Kang, Y.K.; Luo, Y.; Chen, Y.Y.; Ding, D.; Li, S.J. Metabolomic and transcriptomic analyses reveal a MYB gene, CsAN1, involved in anthocyanins accumulation separation in F1 between ‘Zijuan’ (Camellia sinensis var. assamica) and ‘Fudingdabaicha’ (C. sinensis var. sinensis) tea plants. Front. Plant Sci. 2022, 13, 1008588. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Xie, H.; Liu, S.R.; Zhu, J.Y.; Zhao, S.Q.; Wei, C.L. Metabolites and transcriptional profiling analysis reveal the molecular mechanisms of the anthocyanin metabolism in the “Zijuan” tea plant (Camellia sinensis var. assamica). J. Agric. Food Chem. 2021, 69, 414–427. [Google Scholar] [CrossRef]
- Li, W.; Tan, L.Q.; Zou, Y.; Tan, X.Q.; Huang, J.C.; Chen, W.; Tang, Q. The Effects of ultraviolet A/B treatments on anthocyanin accumulation and gene expression in dark-purple tea cultivar ‘Ziyan’ (Camellia sinensis). Moleculars 2020, 25, 354. [Google Scholar] [CrossRef]
- Zhu, M.Z.; Zhou, F.; Ran, L.S.; Li, Y.L.; Tan, B.; Wang, K.B.; Huang, J.A.; Liu, Z.H. Metabolic profiling and gene expression analyses of purple-leaf formation in tea cultivars (Camellia sinensis var. sinensis and var. assamica). Front. Plant Sci. 2021, 12, 606962. [Google Scholar] [CrossRef]
- Kumari, M.; Thakur, S.; Kumar, A.; Joshi, R.; Kumar, P.; Shankar, R.; Kumar, R. Regulation of color transition in purple tea (Camellia sinensis). Planta 2019, 251, 35. [Google Scholar] [CrossRef]
- Li, Z.; Ahammed, G.J. Plant stress response and adaptation via anthocyanins: A review. Plant Stress 2023, 10, 100230. [Google Scholar] [CrossRef]
- Schaart, J.G.; Dubos, C.; Romero De La Fuente, I.; van Houwelingen, A.M.M.L.; de Vos, R.C.H.; Jonker, H.H.; Xu, W.; Routaboul, J.-M.; Lepiniec, L.; Bovy, A.G. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol. 2013, 197, 454–467. [Google Scholar] [CrossRef]
- Cui, D.L.; Zhao, S.X.; Xu, H.N.; Allan, A.C.; Zhang, X.D.; Fan, L.; Chen, L.M.; Su, J.; Shu, Q.; Li, K.Z. The interaction of MYB, bHLH and WD40 transcription factors in red pear (Pyrus pyrifolia) peel. Plant Mol. Biol. 2021, 106, 407–417. [Google Scholar] [CrossRef]
- Wei, K.; Zhang, Y.Z.; Wu, L.Y.; Li, H.L.; Ruan, L.; Bai, P.X.; Zhang, C.C.; Zhang, F.; Xu, L.Y.; Wang, L.Y.; et al. Gene expression analysis of bud and leaf color in tea. Plant Physiol. Biochem. 2016, 107, 310–318. [Google Scholar] [CrossRef]
- Wang, X.W.; Liu, B.Y.; Zhao, Q.S.; Sun, X.M.; Li, Y.Y.; Duan, Z.F.; Miao, X.L.; Luo, S.; Li, J.B. Genomic variance and transcriptional comparisons reveal the mechanisms of leaf color affecting palatability and stressed defense in tea plant. Genes 2019, 10, 929. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.J.; Shi, X.Y.; Nian, B.; Duan, S.M.; Jiang, B.; Wang, X.H.; Lv, C.Y.; Zhang, G.H.; Ma, Y.; Zhao, M. Alternative splicing regulation of anthocyanin biosynthesis in Camellia sinensis var. assamica unveiled by PacBio Iso-Seq. G3-Genes Genom. Genet. 2020, 10, 2713–2723. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.Q.; Chen, Z.D.; Lee, J.W.; Li, X.H.; Sun, W.J. Proteomic analysis of tea plants (Camellia sinensis) with purple young shoots during leaf development. PLoS ONE 2017, 12, e0177816. [Google Scholar] [CrossRef] [PubMed]
- Wilmouth, R.C.; Turnbull, J.J.; Welford, R.W.D.; Clifton, I.J.; Prescott, A.G.; Schofield, C.J. Structure and mechanism of anthocyanidin synthase from Arabidopsis thaliana. Structure 2002, 10, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Marrs, K.A.; Alfenito, M.R.; Lloyd, A.M.; Walbot, V. A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 1995, 375, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Braidot, E.; Zancani, M.; Petrussa, E.; Peresson, C.; Bertolini, A.; Patui, S.; Macrì, F.; Vianello, A. Transport and accumulation of flavonoids in grapevine (Vitis vinifera L.). Plant. Signal. Behav. 2008, 3, 626–632. [Google Scholar] [CrossRef]
- Zeng, C.Z.; Lin, H.Y.; Liu, Z.X.; Liu, Z.H. Analysis of young shoots of ‘Anji Baicha’ (Camellia sinensis) at three developmental stages using nontargeted LC-MS-Based metabolomics. J. Food Sci. 2019, 84, 1746–1757. [Google Scholar] [CrossRef]
- Huang, R.; Wang, J.Y.; Yao, M.Z.; Ma, C.L.; Chen, L. Quantitative trait loci mapping for free amino acid content using an albino population and SNP markers provides insight into the genetic improvement of tea plants. Hortic. Res. 2022, 9, uhab029. [Google Scholar] [CrossRef]
- Yu, Y.; Kou, X.B.; Gao, R.S.; Chen, X.F.; Zhao, Z.; Mei, H.L.; Li, J.J.; Jeyaraj, A.; Thangaraj, K.; Periakaruppan, R.; et al. Glutamine synthetases play a vital role in high accumulation of theanine in tender shoots of albino tea germplasm “Huabai 1”. J. Agric. Food Chem. 2021, 69, 13904–13915. [Google Scholar] [CrossRef]
- Li, C.F.; Xu, Y.X.; Ma, J.Q.; Jin, J.Q.; Huang, D.J.; Yao, M.Z.; Ma, C.L.; Chen, L. Biochemical and transcriptomic analyses reveal different metabolite biosynthesis profiles among three color and developmental stages in ‘Anji Baicha’ (Camellia sinensis). BMC Plant Biol. 2016, 16, 195. [Google Scholar] [CrossRef]
- Fu, X.M.; Cheng, S.H.; Liao, Y.Y.; Xu, X.L.; Wang, X.C.; Hao, X.Y.; Xu, P.; Dong, F.; Yang, Z.Y. Characterization of l-theanine hydrolase in vitro and subcellular distribution of its specific product ethylamine in tea (Camellia sinensis). J. Agric. Food Chem. 2020, 68, 10842–10851. [Google Scholar] [CrossRef]
- Liao, H.S.; Chung, Y.H.; Hsieh, M.H. Glutamate: A multifunctional amino acid in plants. Plant Sci. 2022, 318, 111238. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.; Wang, T.F.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant flavonoids: Classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef] [PubMed]
- Tattini, M.; Galardi, C.; Pinelli, P.; Massai, R.; Remorini, D.; Agati, G. Differential accumulation of flavonoids and hydroxycinnamates in leaves of Ligustrum vulgare under excess light and drought stress. New Phytol. 2004, 163, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Wada, H.; Kobayashi, K. Role of Galactolipids in plastid differentiation before and after light exposure. Plants 2019, 8, 357. [Google Scholar] [CrossRef]
- Pye, V.E.; Christensen, C.E.; Dyer, J.H.; Arent, S.; Henriksen, A. Peroxisomal plant 3-ketoacyl-coa thiolase structure and activity are regulated by a sensitive redox switch. J. Biol. Chem. 2010, 285, 24078–24088. [Google Scholar] [CrossRef] [PubMed]
- Arent, S.; Pye, V.E.; Henriksen, A. Structure and function of plant acyl-CoA oxidases. Plant Physiol. Biochem. 2008, 46, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.U. Lipid Metabolism in Plants. Plants 2020, 9, 871. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.W.; Lin, Y.T.; Li, H.M. Increased ratio of galactolipid MGDG: DGDG induces jasmonic acid overproduction and changes chloroplast shape. New Phytol. 2020, 228, 1327–1335. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, R.; Wang, G.; Li, M.; Roth, M.; Welti, R.; Wang, X. Differential changes in galactolipid and phospholipid species in soybean leaves and roots under nitrogen deficiency and after nodulation. Phytochemistry 2013, 96, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.Y.; Lam, S.M.; Zuo, J.H.; Yuan, S.Z.; Lv, J.Y.; Shi, J.Y.; Gao, L.P.; Chen, B.; Sui, Y.; Shui, G.H.; et al. Lipidomics reveals the difference of membrane lipid catabolism between chilling injury sensitive and non-sensitive green bell pepper in response to chilling. Postharvest Biol. Technol. 2021, 182, 111714. [Google Scholar] [CrossRef]
- Zhou, Z.; Chen, M.j.; Wu, Q.j.; Zeng, W.; Chen, Z.d.; Sun, W.j. Combined analysis of lipidomics and transcriptomics revealed the key pathways and genes of lipids in light-sensitive albino tea plant (Camellia sinensis cv. Baijiguan). Front. Plant Sci. 2022, 13. [Google Scholar] [CrossRef]
- Yang, X.Y.; Lu, M.Q.; Wang, Y.F.; Wang, Y.R.; Liu, Z.J.; Chen, S. Response mechanism of plants to drought stress. Horticulturae 2021, 7, 50. [Google Scholar] [CrossRef]
- Bhanot, V.; Fadanavis, S.V.; Panwar, J. Revisiting the architecture, biosynthesis and functional aspects of the plant cuticle: There is more scope. Environ. Exp. Bot. 2021, 183, 104364. [Google Scholar] [CrossRef]
- Saini, P.; Bhatia, S.; Mahajan, M.; Kaushik, A.; Sahu, S.K.; Kumar, A.; Satbhai, S.B.; Patel, M.K.; Saxena, S.; Chaurasia, O.P.; et al. ELONGATED HYPOCOTYL5 negatively regulates DECREASE WAX BIOSYNTHESIS to increase survival during UV-B stress. Plant Physiol. 2020, 184, 2091–2106. [Google Scholar] [CrossRef]
- Song, S.S.; Tao, Y.; Gao, L.H.; Liang, H.L.; Tang, D.S.; Lin, J.; Wang, Y.C.; Gmitter, F.G.; Li, C.F. An integrated metabolome and transcriptome analysis reveal the regulation mechanisms of flavonoid biosynthesis in a purple tea plant cultivar. Front. Plant Sci. 2022, 13, 880227. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Chen, X.Y.; Chen, J.H.; Zheng, P.; Liu, S.Q.; Tan, X.D.; Sun, B.M. Virus-induced gene silencing in the tea plant (Camellia sinensis). Plants 2023, 12, 3162. [Google Scholar] [CrossRef] [PubMed]
- Li, G.D.; Li, Y.; Yao, X.Z.; Lu, L.T. Establishment of a virus-induced gene-silencing (VIGS) system in tea plant and its use in the functional analysis of CsTCS1. Int. J. Mol. Sci. 2023, 24, 392. [Google Scholar] [CrossRef] [PubMed]
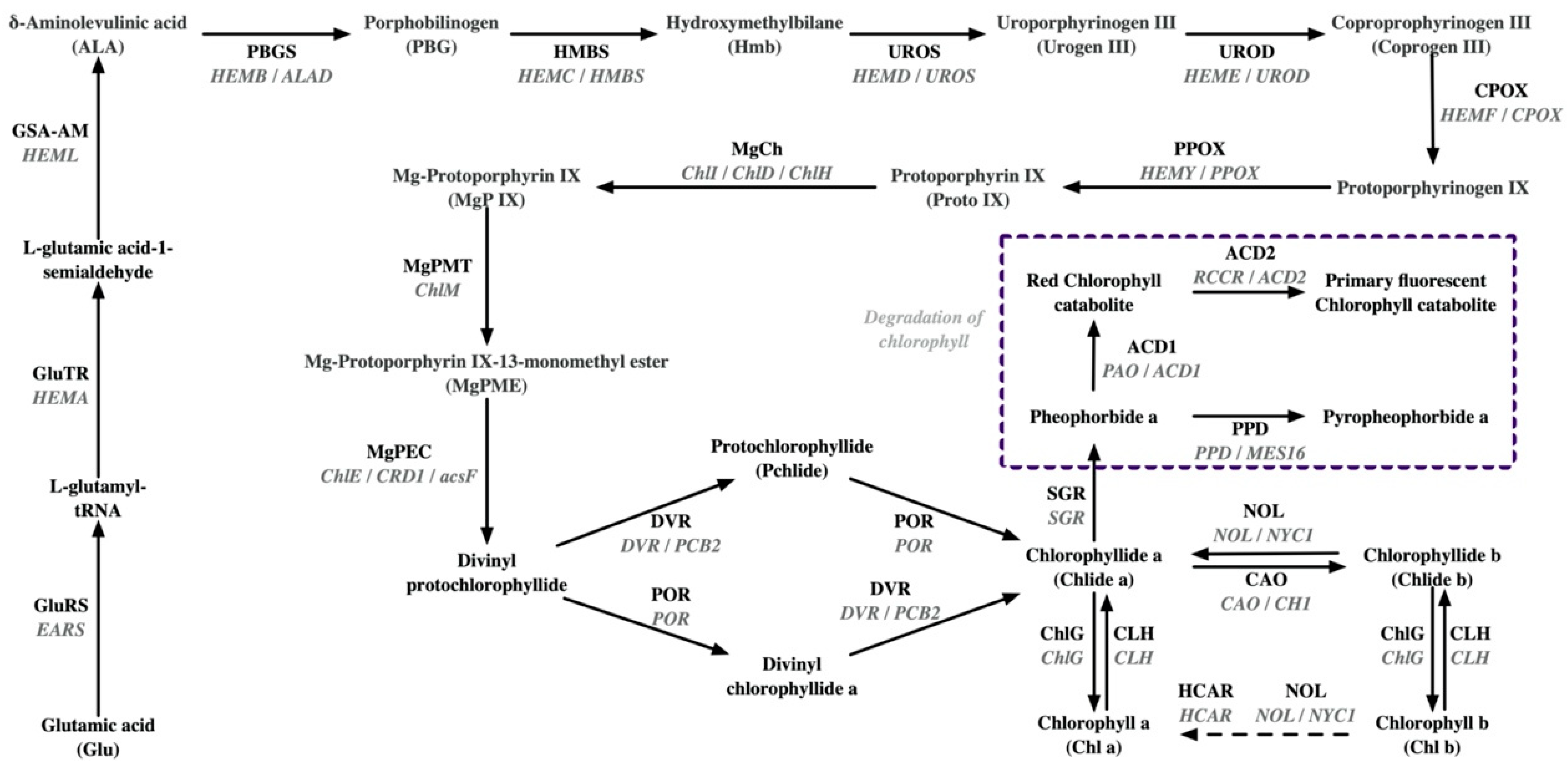

| Tea Varieties | Omics Approaches | Potential Molecular Mechanisms | References |
|---|---|---|---|
| Anjibaicha (Alternative names: Baiye No1, White leaf No.1, and Anji white 1) | Succinyl-proteome | Photosynthetic chain: The succinylation levels of PsbS and light-harvesting complex LHCA4 are down-regulated; the succinylation level of LHCB4 is up-regulated. | [46] |
| Proteome and acetyl-proteome | Photosynthetic chain: lower abundance of LHCB1, LHCB2, LHCB3, LHCB4, LHCB5, LHCB7, LHCA1, LHCA2, LHCA3, LHCA4, PsbC, PsbD, PsbO, PsbP, PsbQ, PsbR, PsbS, Psb27, PsaE, PsaG, PsaL, PsaN, PetA, PetC, PetE, PetF, PetH, ATPA, ATPB, ATPD, ATPE, ATPG; a lower acetylation level of LHCA1. | [8] | |
| Pangenome | Chlorophyll synthesis: The GluRS/EARS gene in Anjibaicha showed a loss of anti-codon recognition domains; SVs were revealed in ChlD. Chlorophyll degradation: 1 bp deletion is revealed in NOL/NYC1. | [24] | |
| Whole-transcriptome | Chlorophyll synthesis: down-regulated expression of POR (two alleles), CLH1, PBGD/HMBS/HEMC; up-regulated expression of COX15. Photosynthetic chain: down-regulated expression of CAB7, CAB21, LHCA4, CAB40 (three alleles), CAB13, LHCB5, PetA (two alleles), PsbA (TEA_001460), PsaB, PsbP1, ATPA, PsaH, ATPI, PetB, Psb28; up-regulated expression of PsbA (TEA_001460). | [35] | |
| Huangjinya | Proteome | Chlorophyll synthesis: higher abundance of GluRS/EARS, MgCh/ChlH, and HMBS; lower abundance of POR. Photosynthetic chain: lower abundance of Photosystem Q(B) protein. | [36] |
| Metabolome and proteome | Chlorophyll synthesis: higher abundance of GluRS/EARS and UROD/HEME; lower abundance of POR and CAO. Chlorophyll degradation: higher abundance of PAO/ACD1 Photosynthetic chain: lower abundance of LHCA1, LHCA3, LHCB1, LHCB2, LHCB3, LHCB4, LHCB5, LHCB6, PsaD, PsaF, PsaL, PsaN, PsbA, PsbD, PsbE, PsbO, PsbP, PsbQ; higher abundance of PsbS, FNR, ATPB. Carotenoid synthesis: lower abundance of ZEP; higher abundance of PSY. | [9] | |
| Pangenome | Chlorophyll synthesis: The GluRS/EARS gene in Huangjinya showed a loss of anti-codon recognition domains, which may inhibit chlorophyll synthesis; SVs were detected in CAO, CHLP, and GluTR. Carotenoid synthesis: mutated amino acids in CYP97A3HJY and elevated expression of the CYP97A3HJY allele. | [24] | |
| Transcriptome | Carotenoid synthesis: down-regulated expression of PSY, PDS, ZDS, LCYE, LCYB, CHY, ZEP, and VDE. | [47] | |
| Baijiguan | BSR-seq | Photosynthetic chain: co-down-regulated expression of LHCA3, three LHCB1 alleles (TEA001863, TEA001868, TEA030368), two LHCB3 alleles (TEA017256, TEA021966), and LHCB4 in bulked groups and parents. | [29] |
| Genome: Genotyping by sequencing and BSA-seq | Chlorophyll synthesis: a non-synonymous polymorphism (G1199A) in the magnesium chelatase I subunit (CsChlI). | [38] | |
| HY1 | Proteome | Photosynthetic chain: lower abundance of LHCA3, LHCA4, LHCB1, LHCB2, LHCB6, PsbC, PsBO, PsbS, PsaB, PsaC, PsaD, PsaF, PetA, PetH. | [17] |
| HY2 | Proteome | Photosynthetic chain: lower abundance of LHCB2, PsbA, PsbD, PsbC, PsbB, PsbQ, PsaA, PsaB, PsaD, PsaH, PetB, PetA, beta F-type ATPase. | [17] |
| HY | Transcriptome | Chlorophyll degradation: the activation of SGR and CLH. | [43] |
| Xiangfeihuangye | Transcriptome, translatome, and metabolome | Chlorophyll synthesis: up-regulation of HY5 in EL inhibited the expression of GluTR/HEMA and POR. | [39] |
| Huabai 1 | Transcriptome | Photosynthetic chain: down-regulated expression of light-harvesting complex II (LHCII) chlorophyll-a/b-binding protein. Chlorophyll degradation: up-regulated expression of SGR. | [12] |
| Albinistic branch of Huangshan | Transcriptome | Chlorophyll synthesis: down-regulated expression of four CHLP alleles (TEA027589, TEA019124, BGI_novel_G007262, TEA016514) and one POR allele (TEA014780); up-regulated expression of one CHLP allele (TEA009538), one POR allele (TEA027994), and one CLH allele (TEA027808). Photosynthetic chain: down-regulated expression of three LHCB1 alleles (TEA019232, TEA030366, TEA030368), PsbC, five PsbB alleles (TEA028468, TEA011113, TEA032780, TEA018797, BGI_novel_G013475), PsbP, Psb28, PsaA, five ATPD alleles (TEA030038, TEA004696, TEA002611, BGI_novel_G006800, BGI_novel_004911), two ATPA alleles (TEA019276, BGI_novel_G009498), ATPE. | [48] |
| Huangjinju | Transcriptome | Chlorophyll synthesis: up-regulated expression of POR. Photosynthetic chain: down-regulated expression of LHCA2, LHCA4, LHCB1, LHCB2, LHCB6. | [13] |
| Yanlingyinbiancha | Transcriptome | Chlorophyll synthesis: down-regulated expression of UROS/HEMD, PPOX, ChlH/GUN5, MgPEC/CRD1, DVR/PCB2, and CAO. Chlorophyll degradation: down-regulated expression of NOL/NYC1, HCAR, CLH1, and ACD2. Photosynthetic chain: fifty-five DEGs involved in photosynthetic complexes were found to be down-regulated. Carotenoid synthesis: down-regulated expression of Z-ISO, ZDS, ZEP, LUT2, NCED4. | [18] |
| Yanling Huayecha | Transcriptome | Chlorophyll synthesis: down-regulated expression of PPOX. Photosynthetic chain: down-regulated expression of LHCB6 and FdC2. Thylakoid membrane structure: down-regulated expression of SCY1. | [40] |
| Menghai Huangye | Transcriptome | Chlorophyll synthesis: four genes related to chlorophyll synthesis (HEME2 and POR). Photosynthetic chain: ten genes related to photosynthesis (LHCA and LHCB) are down-regulated. | [41] |
| Zhonghuang 3 | Transcriptome | Chlorophyll metabolism: down-regulated expression of GluTR/HEMA3 and CLH4. | [49] |
| Zhonghuang 3 | Transcriptome | Chlorophyll synthesis: down-regulated expression of GluTR/HEMA, GSA-AM/HEML, UROD/HEME, HEMF/CPOX, DVR (CSS0009780), and CHLP; up-regulated expression of PBGS/HEMB, DVR (CSS0011936), and CLH. Chlorophyll degradation: down-regulated expression of NOL/NYC1 (CSS0031926) and SGR (CSS0030812); up-regulated expression of NOL/NYC1 (CSS0015127), SGR (CSS0050352), and SGRL (CSS0004139 and CSS0036450). Carotenoid synthesis: up-regulated expression of Z-ISO, CRTISO (CSS0027469, CSS0033902, and CSS0044870), NCED1, NCED2; down-regulated expression of LCYB and NXS. | [14] |
| Zhonghuang 2 | Transcriptome | Transcripts encoding enzymes such as those functioning in early enzymatic steps, from the formation of glutamate 1-semialdehyde to protoporphyrin IX, showed lower levels. Critical enzymes for converting Mg-protoporphyrin IX into chlorophyll were also inhibited. | [42] |
| Koganemidori | Transcriptome | Chlorophyll synthesis: down-regulated expression of POR, CAO, and ChlG. Chlorophyll degradation: up-regulated expression of CLH. Transcriptional regulation: two homologs of GLK were significantly down-regulated. | [50] |
| Huangjinshuixian | Transcriptome and metabolome | Chlorophyll degradation: down-regulated expression of SGR. Carotenoid synthesis: the expression of DXS and GGPPS was significantly down-regulated. Transcriptional regulation: PIFs related to chlorophyll biosynthesis were significantly suppressed. | [16] |
| Huangyu | Transcriptome and metabolome | Chlorophyll synthesis: down-regulated expression of UROD/HEME, MgCh/ChlH, and CAO. Chlorophyll degradation: up-regulated expression of CLH. Photosynthetic chain: down-regulated expression of three LHCII genes (CSS0013089, CSS0017825, and CSS0039893) | [30] |
| Huangkui | Transcriptome | Transcriptional regulation: the transcriptional expression of CsRVE1 increased during seasonal greening and was tightly correlated with increases in the expression of genes involved in light harvesting (LHCB) and chlorophyll biosynthesis (MgCh/ChlH, GluTR/HEMA1, and CAO). | [51] |
| Fuhuang 1 | Transcriptome | Chlorophyll synthesis: down-regulated expression of CAO. Chlorophyll degradation: down-regulated expression of NOL/NYC1 and SGR. Photosynthetic chain: down-regulated expression of LHCA2, LHCA4, LHCB1, LHCB3. Carotenoid synthesis: down-regulated expression of LCYE, ZEP, NCED; up-regulated expression of PSY, PDS, VDE. | [52] |
| Fuhuang 2 | Transcriptome | Chlorophyll synthesis: down-regulated expression of GluTR/HEMA. Chlorophyll degradation: down-regulated expression of CLH. Photosynthetic chain: down-regulated expression of PsbB, PetC, ATPF1B, LCHBs, LCHAs. Carotenoid synthesis: down-regulated expression of NCED. | [53] |
| Tea Varieties | Sampling Location | Measurement Technique | Anthocyanin Composition | References |
|---|---|---|---|---|
| Zijuan | Tea garden of South China Agricultural University, Guangzhou, China | HPLC | Major anthocyanin compositions: cyanidin-3-O-galactoside and delphinidin-3-O-galactoside. | [77] |
| Tea garden of the Institute of Tea Science, Yunnan Province Academy of Agricultural Sciences (Menghai, China) | Non-targeted metabolomics approach: UHPLC– Orbitrap–MS/MS | Cyanidin 3-diglucoside 5-glucoside, cyanidin 3-O-(6-O-p-coumaroyl) glucoside, cyanidin 3-sambubioside, cyanidin 3-(6″-acetylglucoside)-5-glucoside, delphinidin 3-(6-p-coumaroyl) galactoside, delphinidin-3-O-arabinoside, pelargonidin 3-sophoroside 5-glucoside, pelargonidin 3-coumarylglucoside-5-acetylglucoside, pelargonidin 3-rhamnoside 5-glucoside; compared with Yunkang, the contents of cyanidin 3-diglucoside 5-glucoside and pelargonidin 3-sophoroside 5-glucoside are most increased in Zijuan. | [86] | |
| Pu’er City Institute of Tea Science, Yunnan Province | UPLC–ESI–MS/MS metabolomic analysis | Specific metabolites: petunidin 3-O-glucoside, peonidin 3-O-glucoside chloride, peonidin 3-O-glucoside, peonidin O-hexoside, malvidin 3-O-glucoside (oenin), petunidin 3,5-O-diglucoside. Marker metabolites: cyanidin 3-O-galactoside, cyanidin 3-O-glucoside (Kuromanin), delphinidin 3-O-glucoside (Mirtillin), pelargonidin 3-O-glucoside | [87] | |
| Changsha, Hunan, China | UPLC–ESI–MS/MS metabolomic analysis | Major anthocyanin compositions: cyanidin-3-ogalactoside, delphinidin-3-O-galactoside, and petunidin-3-O-galactoside | [88] | |
| Dechang Fabrication Base of Shucheng County in Anhui Province, China | LC−TOF–MS | Cyanidin-3-O-galactoside, Cyanidin 3-O-(6-O-p-coumaroyl) galactoside, Delphinidin 3-O-(6-O-p-coumaroyl) galactoside, Delphinidin-3-O-galactoside. | [89] | |
| Zijuan Ziyan and Chuanzi (ZZ) | Muchuan County, Sichuan Province, China | Targeted UPLC– ESI–MS/MS analysis | A total of 22 anthocyanins with a content ≥1 μg/g (DW) were detected in Chuanzi, Ziyan, and/or Zijuan and these included 6 cyanidins, 7 delphinidins, 5 pelargonidins, 2 peonidins, and 2 petunidins. In addition, 23 anthocyanins with a concentration of <1 μg/g were also detected. | [82] |
| Ziyan | Planted in plastic pots | HPLC | Delphinidin, cyanidin, and pelargonidin. | [90] |
| Hongyecha, Zijuan, 9803, Hongyafoshou | Changsha, Hunan, China | UPLC–DAD–QTOF–MS | Cyanidin-(E)-p-coumaroylgalactoside, cyanidin-3-O-galactoside, delphinidin-3-O-galactoside, delphinidin-(Z)-p-coumaroylgalactoside, delphinidin-(E)-p-coumaroylgalactoside, pelargonidin-O-hexose, and pelargonidin-O-dihexose. | [91] |
| Jinmingzao | Tea plantation of Wuqu in Fuan City, Fujian Province, China | Widely targeted metabolomics: UPLC–ESI–MS/MS | Cyanidin 3-O-glucoside, cyanidin 3-O-galactoside, cyanidin 3-rutinoside, cyanidin chloride, delphinidin 3-O-glucoside, peonidin 3-O-glucoside chloride (most affected). | [83] |
| Zikui | South Campus of Guizhou University, Huaxi District, Guiyang City, Guizhou Province, China | ESI–QTRAP–MS/MS | Cyanidin 3-O-galactoside, cyanidin 3-O-glucosid, petunidin 3-O-glucoside. | [84] |
| Longjing43 | Tea Research Institute, Chinese Academy of Agricultural Sciences, Hangzhou, China | LC–MS/MS | Delphinidin-hexose-coumaroyl showed the greatest increase. | [80] |
| TRFK 306 | Tea Research Institute (TRI), Kericho County, Kenya | HPLC | Malvidin 3-glucoside, peonidin 3-glucoside, pelargonidin 3,5-O-diglucoside, cyanidin 3-O-glucoside, cyanidin 3-O-galactoside, cyanidin 3-O-rutinoside. | [81] |
| 9 tea cultivars possessing purple leaves | Wuxi Institute of Tea Varieties in Wuxi City, Jiangsu Province, China | Widely targeted metabolomics: UPLC–ESI–MS/ MS | Thirty-three anthocyanins were identified, and delphinidin 3-O-galactoside and cyanidin 3-O-galactoside were found to be the most abundant in PTLs. | [85] |
| Unknown | Experimental tea farm (IHBT-269) of CSIR—Institute of Himalayan Bioresource Technology, HP, India | UHPLC | 3-O-alpha-l-arabinopyranosylproantho cyanidin A5′ and 3,3′-Di-O-galloylprocyanidin B. | [92] |
| Tea Varieties | Omics Approaches | Potential Molecular Mechanisms | References |
|---|---|---|---|
| Zijuan | Transcriptome | Transcriptional regulation: Activation of the R2R3-MYB transcription factor (TF) anthocyanin1 (CsAN1) and the bHLH TF CsGL3; CsAN1 interacts with bHLH TFs (CsGL3 and CsEGL3) and recruits a WD-repeat protein CsTTG1 to form the MYB-bHLH-WDR (MBW) complex that regulates anthocyanin accumulation. Late biosynthetic genes (LBGs): activation of CsF3′H, CsF3′5′H, CsDFR1, CsDFR2, CsANS1/LDOX1, CsANS2/LDOX2, and CsANS3/LDOX3. Metabolic substrate competition: activation of CsLAR1, CsLAR2, and CsLAR3, which encode enzymes for catechin biosynthesis, was highly expressed in red foliage. | [77] |
| Transcriptome, proteome | Phenylpropanoid metabolism: significantly increased expression of three PALs (CSA016076, 022024, 022025); significantly decreased expression of 4CL (CSA001434). Early biosynthesis genes (EBGs): significantly increased expression of CHS (CSA029775); significantly decreased expression of CHI (CSA008261). LBGs: significantly increased expression of two DFRs (CSA003949, XLOC_010242), one ANS/LDOX (CSA011508), six UGT75L12/13 (CSA005544, 005545, 010001, 036671, 036672, 029026), and two UGT94P1 (CSA007394, 008750); significantly decreased expression of F3′5′H (CSA031792), ANS/LDOX (CSA035767), two UGT75L12s (CSA008693, 028873), and two UGT94P1s (CSA005965, 026000). Metabolic substrate competition: significantly increased expression of two LARs (CSA014943, XLOC_016774). | [87] | |
| Transcriptome | Phenylpropanoid metabolism: activation of C4H. LBGs: activation of ANS/LDOX, UGT. Chlorophyll degradation: activation of CLH1. | [97] | |
| Full-length transcriptome | Alternative splicing (AS) events identified in transcriptional regulation (MYB113-1), phenylpropanoid metabolism (C4H1, PAL2), LBGs (UDP75L122), and metabolic substrate competition (FLS1). | [98] | |
| Proteome | EBGs: increased abundance of CHS and CHI. LBGs: increased abundance of DFR, ANS/LDOX, and UGT. Anthocyanin transportation: increased abundance of ABC transporter B8. | [20] | |
| Transcriptome | Transcriptional regulation: Most of the members belonging to the MYB, WRKY, AP2, GRF, bZIP, and MYC groups had a higher expression in Zijuan. LBGs: significantly increased expression of F3′5′H (CSS0022212.1), ANS/LDOX (CSS0010687.1), 3GT (anthocyanidin 3-O-glucosyltransferase, CSS0024320.1), 3AT (cyanidin-3-O-glucoside 6″-O-acyltransferase, CSS0015285.1). Metabolic substrate competition: significantly decreased expression of LAR (CSS0009063.1). Anthocyanin degradation: polyphenol oxidase (PPO, CSS0002951.1), showed negative correlation with the three anthocyanins, especially delphinidin and delargonidin. | [89] | |
| Chuanzi (ZZ) | Transcriptome | Transcriptional regulation: significantly increased expression of the well-known MYB transcription factor CsAN1/CsMYB75 (CSS0030514). LBGs: significantly increased expression of CsANSs/LDOXs (CSS0010687, CSS0018498 and CSS0046216), CsUGT94P1 (CSS0011196), and the anthocyanin O-methyltransferase gene (CsAOMT, CSS0015915). Anthocyanin transportation: significantly increased expression of CsGSTF1 (CSS0022086) and three other GST candidate genes (CSS0031248, CSS0026690, and CSS0018634) tightly linked to CsGSTF1. Metabolic substrate competition: down-regulated expression of LARs (CSS0028235 and CSS0009063) and ANRs (CSS0005927, and CSS0033195). | [82] |
| Zijuan, Jinguanyin and Jinmingzao | Pangenome | Read depth of the LTR insertion region in the promoter of CsMYB114 among a set of representative purple-leaf cultivars (‘ZJ’, ‘JMZ’, and ‘JGY’) and tea cultivars with green leaves (‘FDDB’, ‘BHZ’, and ‘GH3H’) | [24] |
| Ziyan | Transcriptome | Transcriptional regulation: UV-A induces the expression of the regulatory gene TT8; UV-AB induces the expression of the regulatory genes EGL1 and TT2. LBGs: UV-A induces the expression of F3H, F3′5′H, DFR, and ANS/LDOX; UV-AB induces the expression of F3′5′H, DFR, ANS/LDOX, and UGT. Metabolic substrate competition: UV radiation repressed the expression levels of LAR, ANR, and FLS, resulting in reduced ANR activity and a metabolic flux shift towards anthocyanin biosynthesis. | [90] |
| Wuyiqizhong18 | cDNA-AFLP | EBGs: increased expression of CHS. LBGs: increased expression of AT (TDF #3341_2f) and UGT (TDF #2421_1d and TDF #2411_1f). | [79] |
| Proteome | EBGs: increased abundance of CHS and CHI. Metabolic substrate competition: increased abundance of FLS. | [99] | |
| Jinmingzao | Transcriptome | Phenylpropanoid metabolism: activation of PAL, C4H, and 4CL. LBGs: activation of DFR, ANS/LDOX, and UGT (TEA004632 and TEA004632) genes. | [83] |
| Longjing43 | Transcriptome | Transcriptional regulation: activation of MYB75. LBGs: activation of ANS/LDOX and 3-GT. Anthocyanin transportation: activation of genes involved in anthocyanin transportation (GST, glutathione S-transferase). | [96] |
| Transcriptome | Phenylpropanoid metabolism: activation of PAL and C4H by high temperature and/or light levels in summer. EBGs: activation of CHI and CHS by high temperature and/or light levels in summer. LBGs: activation of ANR, ANS/LDOX, and DFR by high temperature and/or light levels in summer. Metabolic substrate competition: activation of FLS and LAR by high temperature and/or light levels in summer. | [80] | |
| TRFK 306 | Transcriptome | Transcriptional regulation: transcripts encoding pathway regulators of the MYB–bHLH–WD40 (MBW) complex were repressed, possibly contributing to the suppression of late biosynthetic genes of the pathway during the dry season. Anthocyanin transportation: suppression of anthocyanin transport genes could be linked to reduced accumulation of anthocyanin in the vacuole during the dry season. | [81] |
| Zikui | Transcriptome | Transcriptional regulation: CsMYB90 showed strong correlations with petunidin 3-O-glucoside, cyanidin 3-O-galactoside, and cyanidin 3-O-glucosid. LBGs: activation of two F3′H genes and two ANS/LDOX genes. Anthocyanin degradation: three negatively correlated PPO (polyphenol oxidase) genes with anthocyanin accumulation. | [84] |
| Hongyecha, Zijuan, 9803, Hongyafoshou | Transcriptome | Phenylpropanoid metabolism: activation of 4CL. LBGs: activation of ANS/LDOX and UGT. | [88] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, Y.-G.; Zhao, T.-T.; Xiang, Q.-Z.; Han, X.-Y.; Yang, S.-S.; Zhang, L.-X.; Ren, L.-J. Multi-Omics Research Accelerates the Clarification of the Formation Mechanism and the Influence of Leaf Color Variation in Tea (Camellia sinensis) Plants. Plants 2024, 13, 426. https://doi.org/10.3390/plants13030426
Fan Y-G, Zhao T-T, Xiang Q-Z, Han X-Y, Yang S-S, Zhang L-X, Ren L-J. Multi-Omics Research Accelerates the Clarification of the Formation Mechanism and the Influence of Leaf Color Variation in Tea (Camellia sinensis) Plants. Plants. 2024; 13(3):426. https://doi.org/10.3390/plants13030426
Chicago/Turabian StyleFan, Yan-Gen, Ting-Ting Zhao, Qin-Zeng Xiang, Xiao-Yang Han, Shu-Sen Yang, Li-Xia Zhang, and Li-Jun Ren. 2024. "Multi-Omics Research Accelerates the Clarification of the Formation Mechanism and the Influence of Leaf Color Variation in Tea (Camellia sinensis) Plants" Plants 13, no. 3: 426. https://doi.org/10.3390/plants13030426





