Mechanisms Affecting the Biosynthesis and Incorporation Rate of Selenocysteine
Abstract
:1. Introduction
2. The Biosynthesis Mechanism of Sec and the Role of Its Internal Influencing Factors
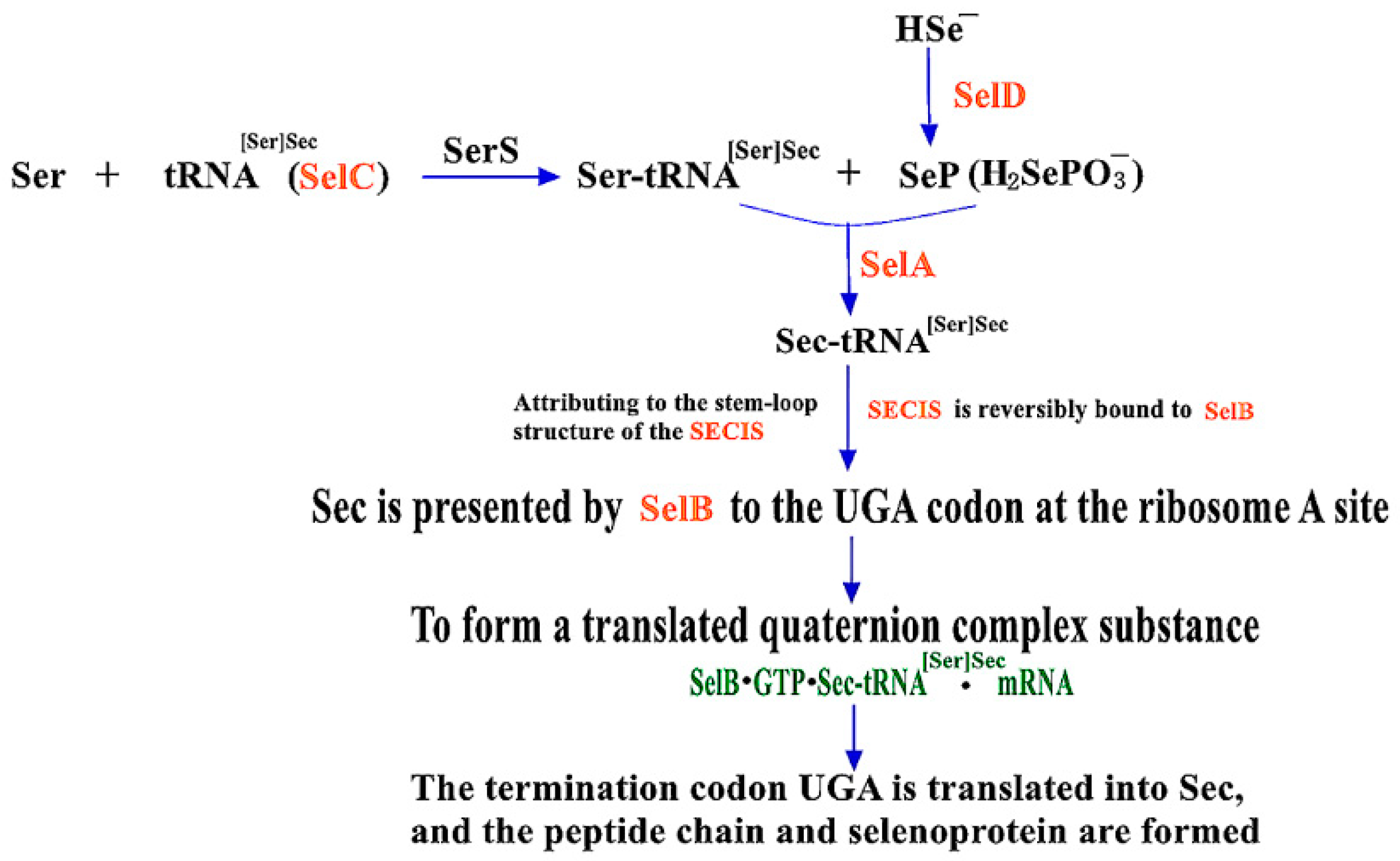
2.1. The Synthesis Mechanism of Sec in Prokaryotes and the Role of Main Influencing Factors
2.1.1. Mechanisms of Sec Synthesis in Prokaryotes
2.1.2. The Role of Four Gene Products in SBIP
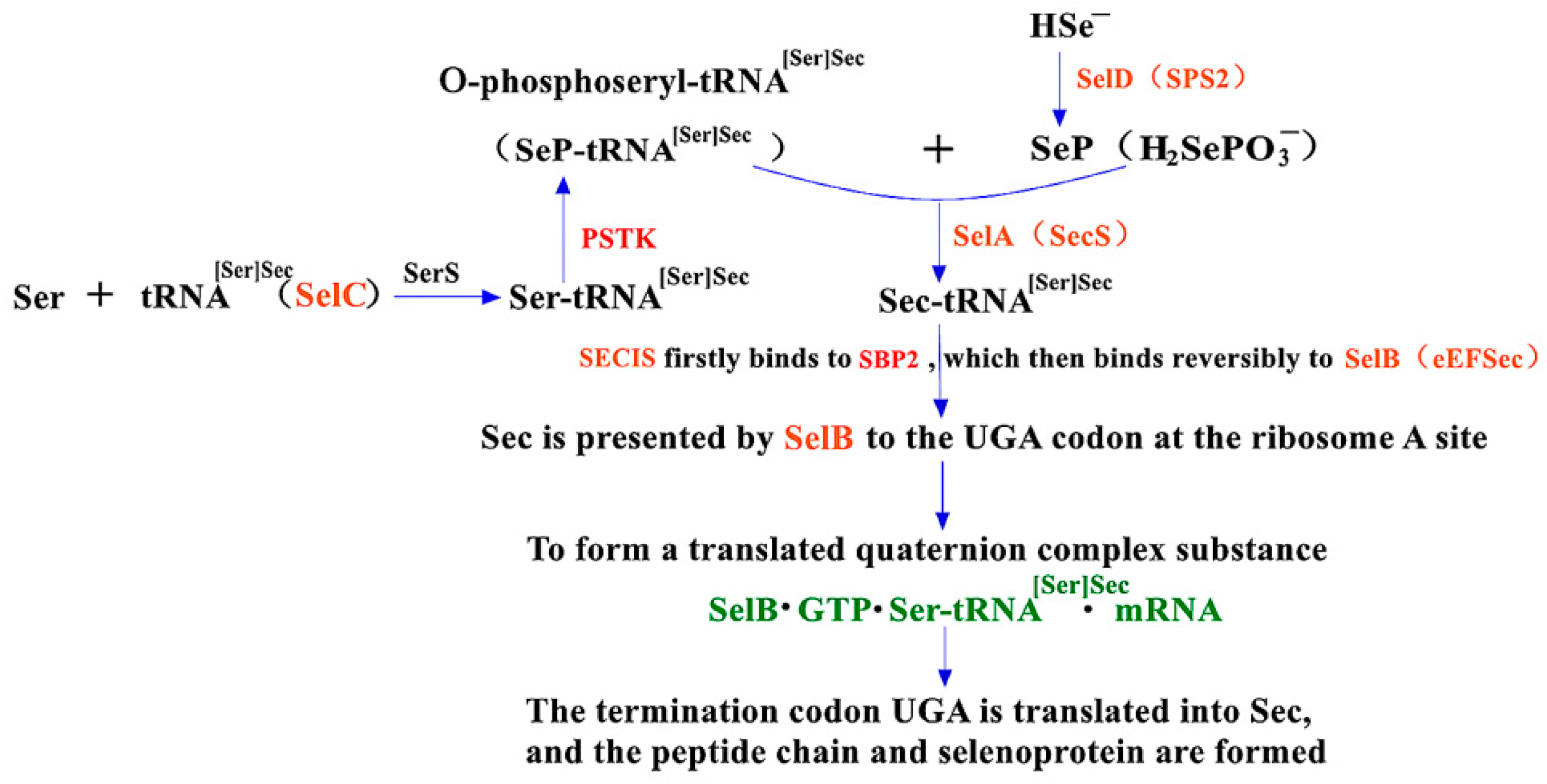
2.2. The Synthesis Mechanism of Sec in Eukaryotes and the Role of PSTK and SBP2 within It
3. Elements on mRNA and Their Functional Mechanisms
3.1. The Cis-Acting Element SECIS on mRNA and Its Functional Mechanism
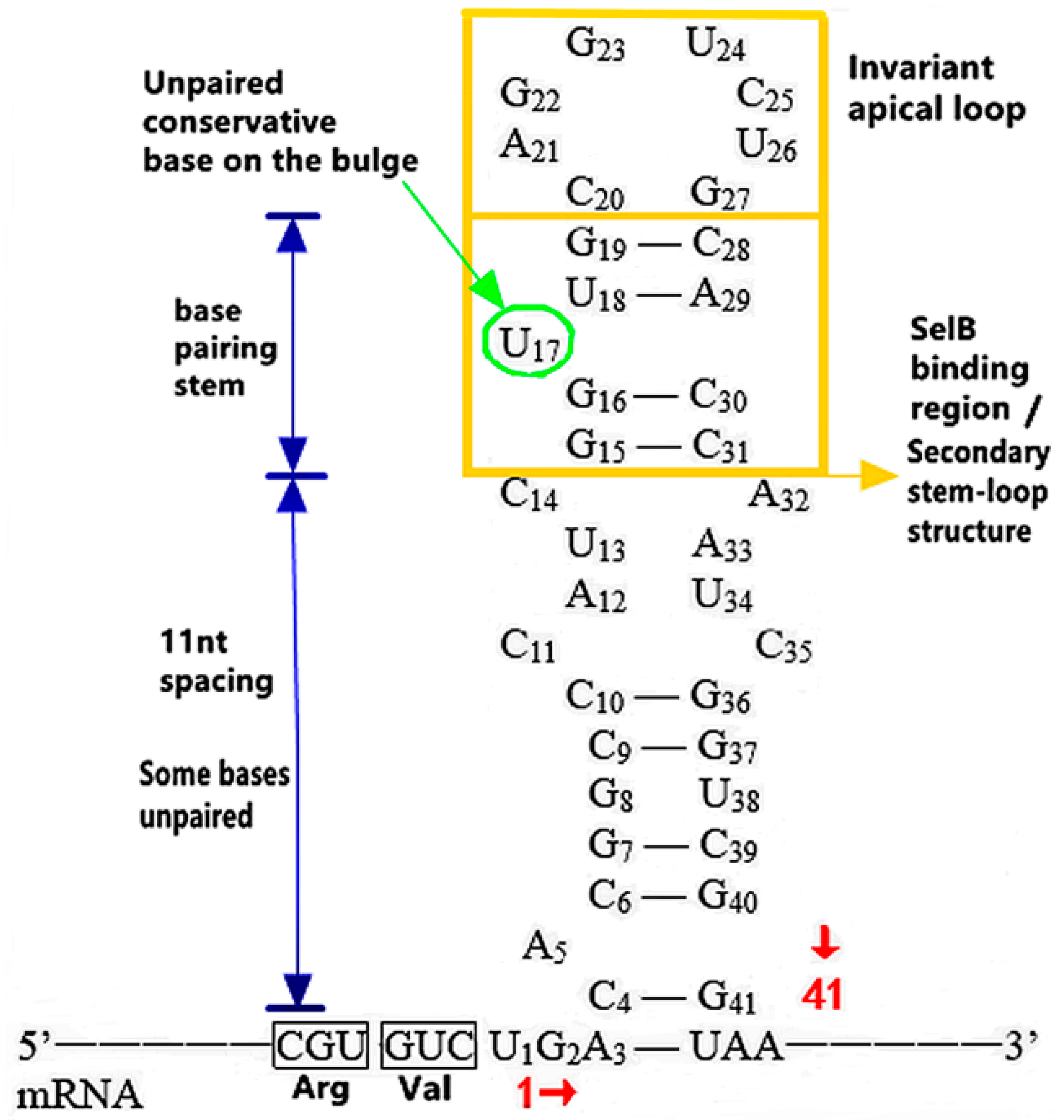
3.1.1. The Functional Mechanism of SECIS in Prokaryotes
The Distribution and Introduction Function of SECIS
The Action of Structure, Sequence and Location of SECIS
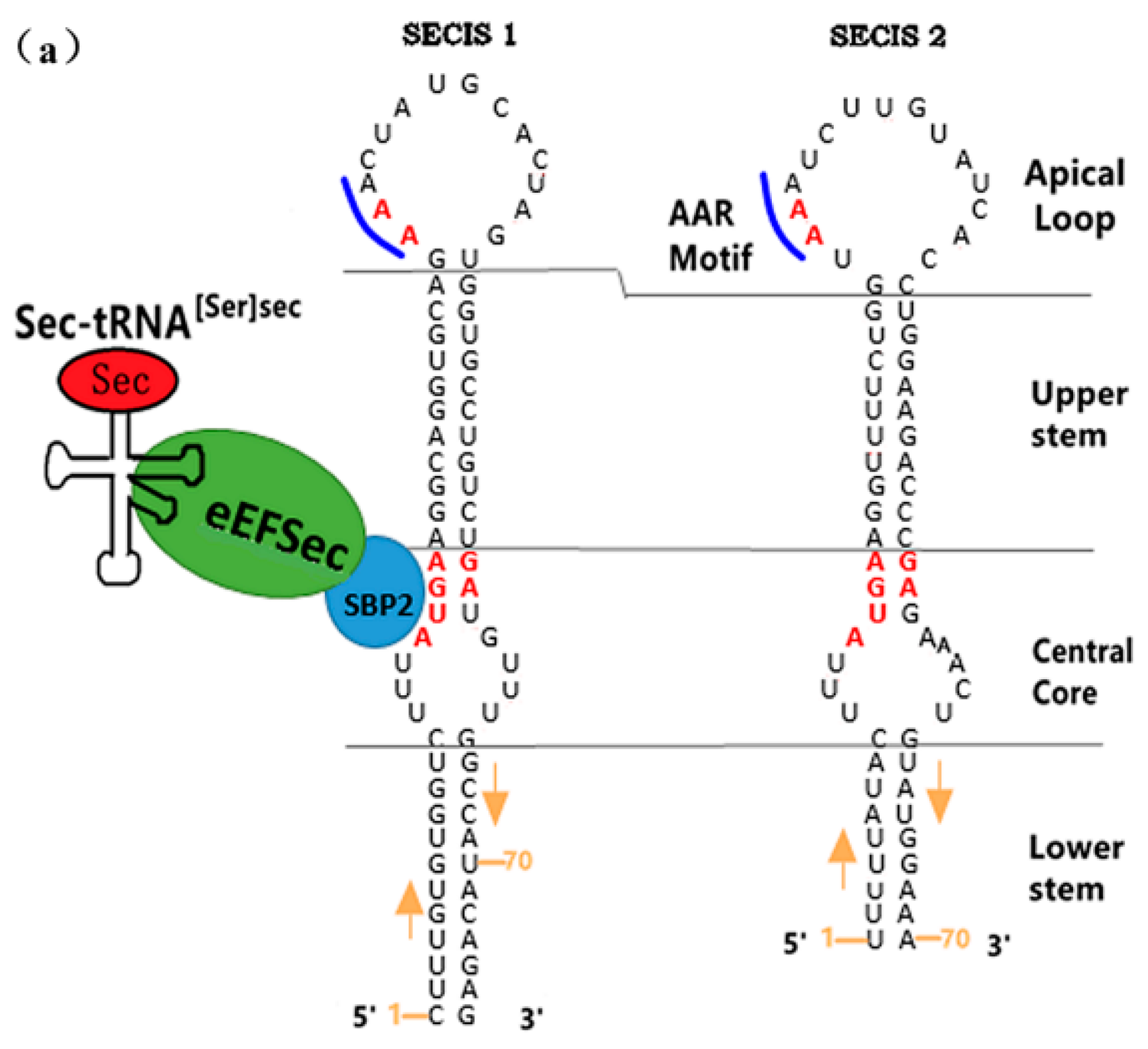
3.1.2. The Functional Mechanism of SECIS in Eukaryotes
The SECIS’s Distribution and Its Relative Distance from UGA
The Sequence Specificity of SECIS
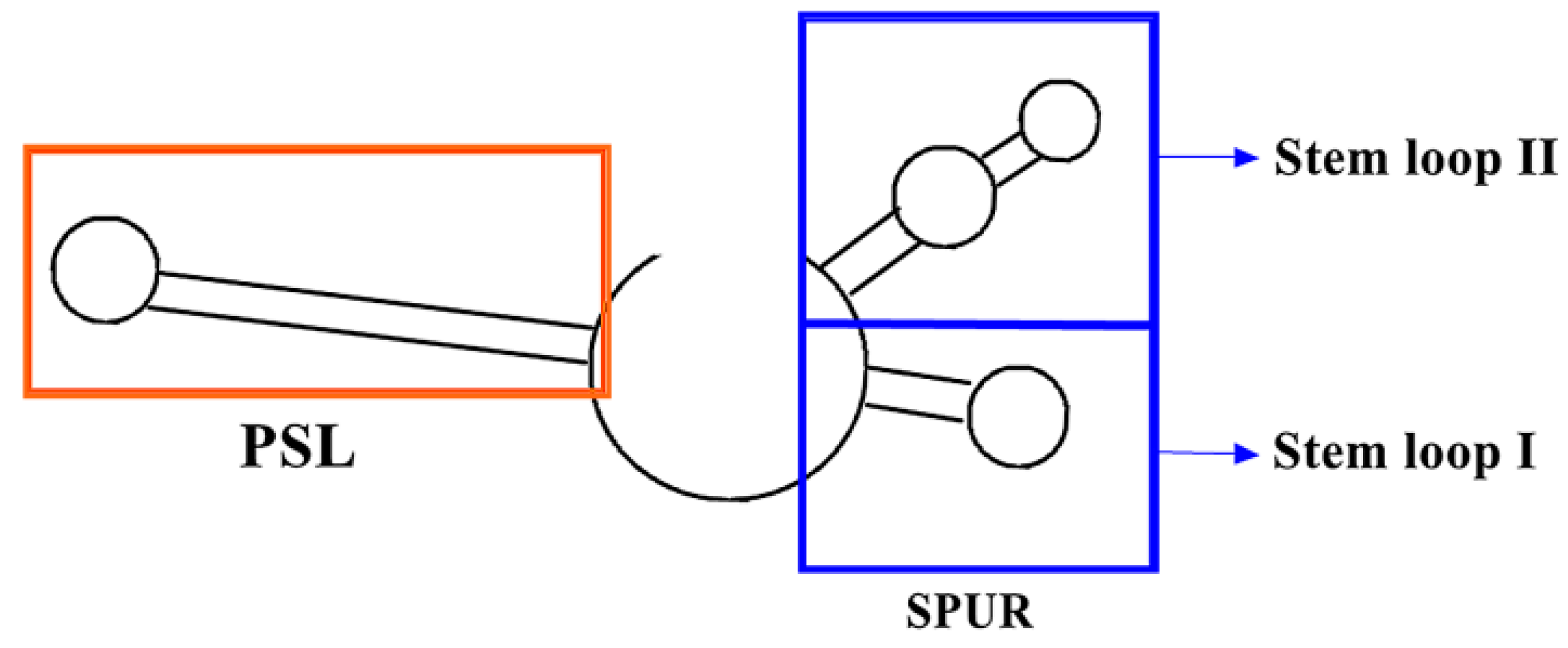
3.2. PSL (Proximal Stem Loop) and SPUR (SelS Positive UGA Recoding) on mRNA and Their Functional Mechanisms
3.3. Translation Competitor RF2 on mRNA and Its Functional Mechanism
4. The Specificity of UGA Codon
5. The Structure–Activity Relationship and Action Mechanism of Four SBIP Factors
5.1. Structure–Activity Relationship and Action Mechanism of a Synthase Factor—SelA
5.2. The Structure–Activity Relationship and Action Mechanism of a Specific Translation Elongation Factor—SelB
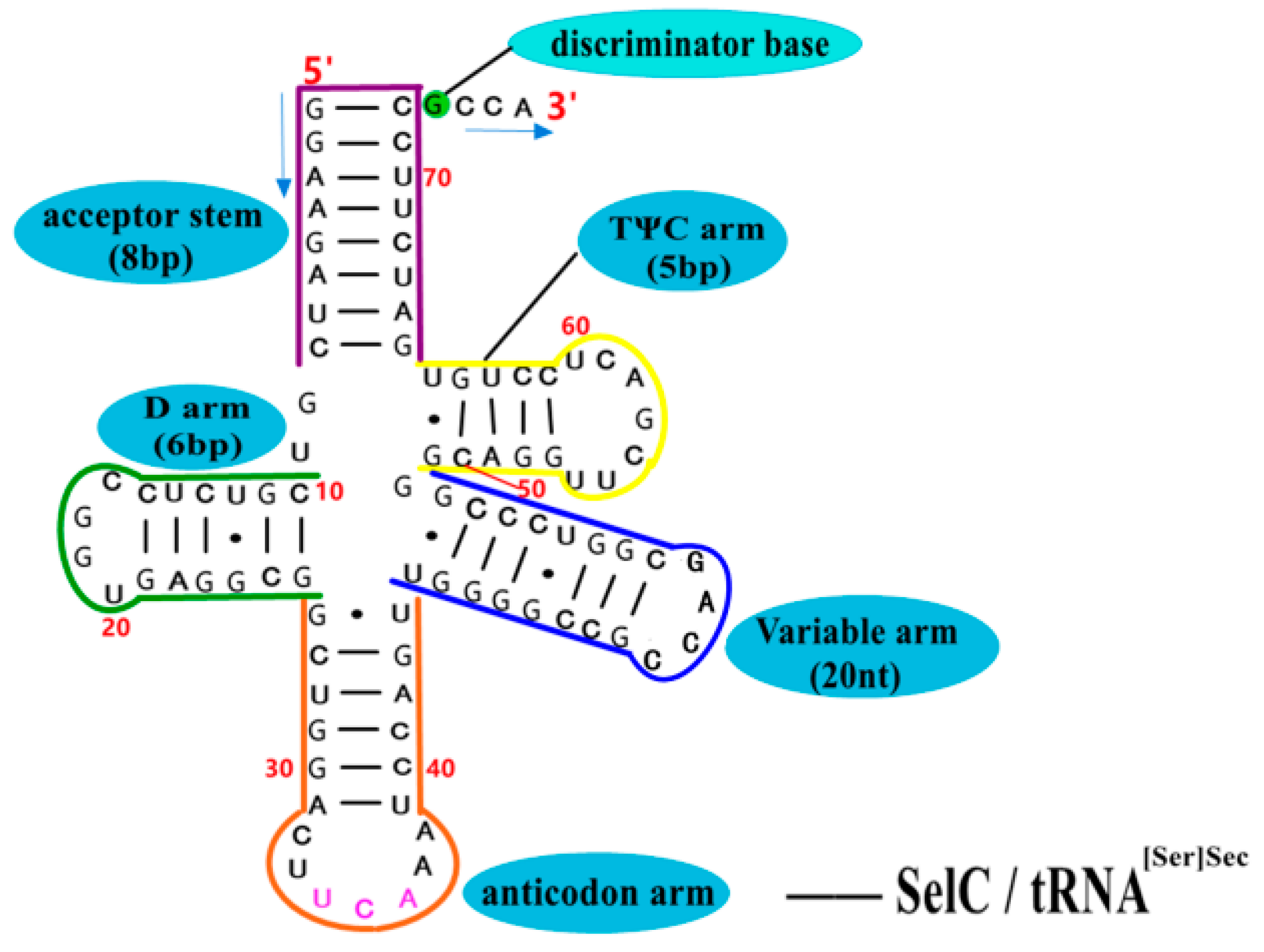
5.3. Structure–Activity Relationship and Action Mechanism of Transporter—SelC
5.4. Structure–Activity Relationship and Action Mechanism of Synthase Factor—Binding Protein SelD
6. Operating Mechanism of Two Key Enzyme Systems for Inorganic Selenium Source Flow before Sec Synthesis
6.1. Operating Mechanism of Transporter System
6.2. Operating Mechanism of Reductase System
7. Conclusions and Discussion
7.1. Some Other mRNA Elements Affecting the Incorporation of Sec
7.2. The Effect of Some Inherent Factors on Selenoprotein Expression
7.3. The Exploration and Thinking of New Mechanism
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Atkins, J.F.; Gesteland, R.F. The twenty-first amino acid. Nature 2000, 407, 463–465. [Google Scholar] [CrossRef]
- Ling, C.; Wang, E.D. Pyrrolysine: The 22nd amino acid. Prog. Biochem. Biophys. 2005, 32, 490–494. [Google Scholar]
- He, J.W.; Du, X.; Cai, J. Progress in biosynthesis of selenocysteine. China Brew. 2019, 38, 11–15. [Google Scholar]
- Wang, X.M.; Cao, J.; Wang, G.L. Research progress of organic selenium speciation analysis in food. J. Food Saf. Qual. 2019, 10, 7875–7881. [Google Scholar]
- Pinsent, J. The need for selenite and molybdate in the formation of formic dehydrogenase by members of the Coli-aerogenes group of bacteria. Biochem. J. 1954, 57, 10–16. [Google Scholar] [CrossRef] [Green Version]
- Chung, C.Z.; Miller, C.; Söll, D.; Krahn, N. Introducing Selenocysteine into Recombinant Proteins in Escherichia coli. Curr. Protoc. 2021, 1, e54. [Google Scholar] [CrossRef]
- Calomme, M.; Hu, J.; Van-Den-Branden, K.; Vanden-Berghe, D.A. Seleno-lactobacillus. An organic selenium source. Biol. Trace Elem. Res. 1995, 47, 379–383. [Google Scholar] [CrossRef]
- Mörschbächer, A.P.; Dullius, A.; Dullius, C.H.; Bandt, C.R.; Kuhn, D.; Brietzke, D.T.; Kuffel, F.J.M.; Etgeton, H.P.; Altmayer, T.; Gonçalves, T.E.; et al. Assessment of selenium bioaccumulation in lactic acid bacteria. J. Dairy Sci. 2018, 102, 10626–10635. [Google Scholar] [CrossRef] [Green Version]
- Hiroya, A.; Yoshihiro, S. Selenium Utilization Strategy by Microalgae. Molecules 2009, 14, 4880–4891. [Google Scholar]
- Li, G.; Wu, C.L.; Zhou, M.; Cui, Z.Y.; Cheng, X.F. Optimization of selenium-rich culture conditions for photosynthetic bacteria. Guangdong Feed 2015, 24, 31–33. [Google Scholar]
- Zhang, Y.; Gladyshev, V.N. An algorithm for identification of bacterial selenocysteine insertion sequence elements and selenoprotein genes. Bioinformatics 2005, 21, 2580–2589. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.H.; Mu, Y.; Li, W.J.; Yan, G.L.; Luo, G.M. The progress in mechanism of selenoprotein biosynthesis. J. Biochem. Biophys. 2002, 34, 395–399. [Google Scholar]
- Zhang, M.N.; Jiang, L.; Zhang, Y. Advance on Bioinformatic Analysis of Selenium Metabolic Network and Selenoproteomes. Curr. Biotechnol. 2017, 7, 537–543. [Google Scholar]
- Fernandes, J.; Hu, X.; Smith, M.R.; Go, Y.M.; Jones, D.P. Selenium at the redox interface of the genome, metabolome and exposome. Free Radic. Biol. Med. 2018, 127, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Ringquist, S.; Schneider, D.; Gibson, T.; Baron, C.; Böck, A.; Gold, L. Recognition of the mRNA selenocysteine insertion sequence by the specialized translational elongation factor SelB. Genes Dev. 1994, 8, 376–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vitor, H.B.S.; Adriano, D.F.F.; Luis, G.M.B.; Jessica, F.S.; Edson, C.J.; Marinonio, L.C.; Bibiana, M.D.S.; Mario, S.P.; Mario, D.O.N.; Otavio, H.T. The specific elongation factor to selenocysteine incorporation in Escherichia coli: Unique tRNASec recognition and its interactions. J. Mol. Biol. 2021, 433, 167279. [Google Scholar]
- Gursinsky, T.; Groebe, D.; Schierhorn, A.; Jaeger, J.; Andreesen, J.R.; Sohling, B. Factors and Selenocysteine Insertion Sequence Requirements for the Synthesis of Selenoproteins from a Gram-Positive Anaerobe in Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 1385–1393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grundner-Culemann, E.; Martin, G.W.; Harney, J.W.; Berry, M.J. Two distinct SECIS structures capable of directing selenocysteine incorporation in eukaryotes. RNA 1999, 5, 625–635. [Google Scholar] [CrossRef] [Green Version]
- Simonovic, M.; Puppala, A.K. On elongation factor eEFSec, its role and mechanism during selenium incorporation into nascent selenoproteins. Biochim. Biophys. ACTA-Gen. 2018, 1862, 2463–2472. [Google Scholar] [CrossRef]
- Fagegaltier, D.; Hubert, N.; Yamada, K.; Mizutani, T.; Carbon, P.; Krol, A. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 2000, 19, 4796–4805. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Gladyshev, V.N. Comparative genomics of trace elements: Emerging dynamic view of trace element utilization and function. Chem. Rev. 2009, 109, 4828–4861. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes, M.J.; Peterson, D.; Vicari, A.; Cocks, B.; Copeland, N.G.; Gilbert, D.J.; Jenkins, N.A.; Ferrick, D.A.; Kastelein, R.A.; Bazan, J.F.; et al. Identification of a novel selD homolog from Eukaryotes, Bacteria, and Archaea: Is there an autoregulatory mechanism in selenocysteine metabolism? Proc. Natl. Acad. Sci. USA 1996, 93, 15086–15091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.M.; Carlson, B.A.; Mix, H.; Zhang, Y.; Saira, K.; Glass, R.S.; Berry, M.J.; Hatfield, D.L. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007, 5, e4. [Google Scholar] [CrossRef] [Green Version]
- Itoh, Y.; Sekine, S.-I.; Yokoyama, S. Crystallization and preliminary X-ray crystallographic analysis of Aquifex aeolicus SelA, a bacterial selenocysteine synthase. ACTA Crystallogr. Sect. F Struct. Biol. Commun. 2012, 68, 1128–1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zinoni, F.; Heider, J.; Böck, A. Features of the formate dehydrogenase mRNA necessary for decoding of the UGA codon as selenocysteine. Proc. Natl. Acad. Sci. USA 1990, 87, 4660–4664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fourmy, D.; Guittet, E.; Yoshizawa, S. Structure of prokaryotic SECIS mRNA hairpin and its interaction with elongation factor SelB. J. Mol. Biol. 2002, 324, 137–150. [Google Scholar] [CrossRef]
- Liu, Z.S.; Reches, M.; Groisman, I.; Engelberg-Kulka, H. The nature of the minimal ‘selenocysteine insertion sequence’ (SECIS) in Escherichia coli. Nucleic Acids Res. 1998, 26, 896–902. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.S.; Reches, M.; Engelberg-Kulka, H. A sequence in the Escherichia coli fdhF “selenocysteine insertion sequence” (SECIS) operates in the absence of selenium. J. Mol. Biol. 1999, 294, 1073–1086. [Google Scholar] [CrossRef]
- Hazebrouck, S.; Camoin, L.; Faltin, Z.; Strosberg, A.; Eshdat, Y. Substituting selenocysteine for catalytic cysteine 41 enhances enzymatic activity of plant phospholipid hydroperoxide glutathione peroxidase expressed in Escherichia coli. J. Biol. Chem. 2000, 275, 28715–28721. [Google Scholar] [CrossRef] [Green Version]
- Fagegaltier, D.; Lescure, A.; Walczak, R.; Carbon, P.; Krol, A. Structural analysis of new local features in SECIS RNA hairpins. Nucleic Acids Res. 2000, 28, 2679–2689. [Google Scholar] [CrossRef] [Green Version]
- Hill, K.E.; Lloyd, R.S.; Burk, R.F. Conserved nucleotide sequences in the open reading frame and 3′ untranslated region of selenoprotein P mRNA. Proc. Natl. Acad. Sci. USA 1993, 90, 537–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berry, M.J.; Banu, L.; Chen, Y.Y.; Mandel, S.J.; Kieffer, J.D.; Harney, J.W.; Larsen, P.R. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature 1991, 353, 273–276. [Google Scholar] [CrossRef]
- Huang, K.X.; Xu, H.B. Chemistry and Biochemistry of Selenium and Its Applications in Life Sciences, 2nd ed.; Huazhong University of Science and Technology Press: Wuhan, China, 2009; pp. 106–142. ISBN 9787560953939. [Google Scholar]
- Martin, G.W.; Harney, J.W.; Berry, M.J. Selenocysteine incorporation in eukaryotes: Insights into mechanism and efficiency from sequence, structure, and spacing proximity studies of the type 1 deiodinase SECIS element. RNA 1996, 2, 171–182. [Google Scholar] [PubMed]
- Howard, M.T.; Copeland, P.R. New Directions for Understanding the Codon Redefinition Required for Selenocysteine Incorporation. Biol. Trace Elem. Res. 2019, 192, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Shetty, S.P.; Sturts, R.; Vetick, M.; Copeland, P.R. Processive incorporation of multiple selenocysteine residues is driven by a novel feature of the selenocysteine insertion sequence. J. Biol. Chem. 2018, 293, 19377–19386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shetty, S.P.; Shah, R.; Copeland, P.R. Regulation of Selenocysteine Incorporation into the Selenium Transport Protein, Selenoprotein, P. J. Biol. Chem. 2014, 289, 25317–25326. [Google Scholar] [CrossRef] [Green Version]
- Stoytcheva, Z.; Tujebajeva, R.M.; Harney, J.W.; Berry, M.J. Efficient incorporation of multiple selenocysteines involves an inefficient decoding step serving as a potential translational checkpoint and ribosome bottleneck. Mol. Cell. Biol. 2006, 26, 9177–9184. [Google Scholar] [CrossRef] [Green Version]
- Burk, R.F.; Hill, K.E.; Motley, A.K.; Hu, M.; Austin, L.M. Deletion of selenoprotein P disrupts selenium homeostasis in the mouse. FASEB J. 2004, 18, A849. [Google Scholar]
- Caban, K.; Kinzy, S.A.; Copeland, P.R. The L7Ae RNA binding motif is a multifunctional domain required for the ribosome-dependent Sec incorporation activity of Sec insertion sequence binding protein 2. Mol. Cell. Biol. 2007, 27, 6350–6360. [Google Scholar] [CrossRef] [Green Version]
- Klein, D.J.; Schmeing, T.M.; Moore, P.B.; Steitz, J.A. The kink-turn: A new RNA secondary structure motif. EMBO J. 2001, 20, 4214–4221. [Google Scholar] [CrossRef]
- Gonzalez-Flores, J.N.; Gupta, N.; DeMong, L.W.; Copeland, P.R. The selenocysteine-specific elongation factor contains a novel and multi-functional domain. J. Biol. Chem. 2012, 287, 38936–38945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.B.; Huang, K.X.; Qu, X.H.; Gao, Z.H.; Liu, Q.; Chen, R.S.; Xuan, Z.Y. Identification of selenocysteine insertion sequence (SECIS) element in eukaryotic selenoproteins by RNA Draw program. Chin. Sci. Bull. 2001, 46, 1159–1161. [Google Scholar] [CrossRef]
- Croteau, W.; Davey, J.C.; Galton, V.A.; StGermain, D.L. Cloning of the mammalian type II iodothyronine deiodinase. A selenoprotein differentially expressed and regulated in human and rat brain and other tissues. J. Clin. Investig. 1996, 98, 405–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buettner, C.; Harney, J.W.; Berry, M.J. The Caenorhabditis elegans homologue of thioredoxin reductase contains a selenocysteine insertion sequence (SECIS) element that differs from mammalian SECIS elements but directs selenocysteine incorporation. J. Biol. Chem. 1999, 274, 21598–21602. [Google Scholar] [CrossRef] [Green Version]
- Cockman, E.M.; Narayan, V.; Willard, B.; Shetty, S.P.; Copeland, P.R.; Driscoll, D.M. Identification of the Selenoprotein S Positive UGA Recoding (SPUR) element and its position-dependent activity. RNA Biol. 2019, 16, 1682–1696. [Google Scholar] [CrossRef] [PubMed]
- Budiman, M.E.; Bubenik, J.L.; Miniard, A.C.; Middleton, L.M.; Gerber, C.A.; Cash, A.; Driscoll, D.M. Eukaryotic initiation factor 4a3 is a selenium-regulated RNA-binding protein that selectively inhibits selenocysteine incorporation. Mol. Cell 2009, 35, 479–489. [Google Scholar] [CrossRef] [Green Version]
- Bubenik, J.L.; Ladd, A.N.; Gerber, C.A.; Budiman, M.E.; Driscoll, D.M. Known turnover and translation regulatory RNA-binding proteins interact with the 3′ UTR of SECIS-binding protein 2. RNA Biol. 2009, 6, 73–83. [Google Scholar] [CrossRef] [Green Version]
- Kotini, S.B.; Peske, F.; Rodnina, M.V. Partitioning between recoding and termination at a stop codon-selenocysteine insertion sequence. Nucleic Acids Res. 2015, 43, 6426–6438. [Google Scholar] [CrossRef] [Green Version]
- Hatfield, D.L.; Gladyshev, V.N. How Selenium Has Altered Our Understanding of the Genetic Code. Mol. Cell. Biol. 2002, 22, 3565–3576. [Google Scholar] [CrossRef] [Green Version]
- Itoh, Y.; Brocker, M.J.; Sekine, S.; Soll, D.; Yokoyama, S. Dimer-Dimer Interaction of the Bacterial Selenocysteine Synthase SelA Promotes Functional Active-Site Formation and Catalytic Specificity. J. Mol. Biol. 2014, 426, 1723–1735. [Google Scholar] [CrossRef] [Green Version]
- Araiso, Y.; Palioura, S.; Ishitani, R.; Sherrer, R.L.; O’Donoghue, P.; Yuan, J.; Oshikane, H.; Domae, N.; DeFranco, J.; Söll, D.; et al. Structural insights into RNA-dependent eukaryal and archaeal selenocysteine formation. Nucleic Acids Res. 2008, 36, 1187–1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganichkin, O.M.; Xu, X.M.; Carlson, B.A.; Mix, H.; Hatfield, D.L.; Gladyshev, V.N.; Wahl, M.C. Structure and catalytic mechanism of eukaryotic selenocysteine synthase. J. Biol. Chem. 2008, 283, 5849–5865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forchhammer, K.; Leinfelder, W.; Boesmiller, K.; Veprek, B.; Böck, A. Selenocysteine synthase from Escherichia coli. Nucleotide sequence of the gene (selA) and purification of the protein. J. Biol. Chem. 1991, 266, 6318–6323. [Google Scholar] [CrossRef]
- Itoh, Y.; Sekine, S.; Yokoyama, S. Crystal structure of the full-length bacterial selenocysteine-specific elongation factor SelB. Nucleic Acids Res. 2015, 43, 9028–9038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serrao, V.H.B.; Silva, L.R.; da Silva, M.T.A.; Scortecci, J.F.; Fernandes, A.D.F.; Thiemann, O.H. The unique tRNA(Sec) and its role in selenocysteine biosynthesis. Amino Acids 2018, 50, 1145–1167. [Google Scholar] [CrossRef]
- Wolfe, M.D. Selenophosphate Synthetase. EcoSal Plus 2004, 1, 1–7. [Google Scholar] [CrossRef]
- Itoh, Y.; Sekine, S.; Matsumoto, E.; Akasaka, R.; Takemoto, C.; Shirouzu, M.; Yokoyama, S. Structure of Selenophosphate Synthetase Essential for Selenium Incorporation into Proteins and RNAs. J. Mol. Biol. 2009, 385, 1456–1469. [Google Scholar] [CrossRef]
- Xu, X.M.; Carlson, B.A.; Irons, R.; Mix, H.; Zhong, N.X.; Gladyshev, V.N.; Hatfield, D.L. Selenophosphate synthetase 2 is essential for selenoprotein biosynthesis. Biochem. J. 2007, 404, 115–120. [Google Scholar] [CrossRef] [Green Version]
- Sirko, A.; Hryniewicz, M.; Hulanicka, D.; Böck, A. Sulfate and thiosulfate transport in Escherichia coli K-12: Nucleotide sequence and expression of the cysTWAM gene cluster. J. Bacteriol. 1990, 172, 3351–3357. [Google Scholar] [CrossRef] [Green Version]
- Guzzo, J.; Dubow, M.S. A novel selenite- and tellurite-inducible gene in Escherichia coli. Appl. Environ. Microbiol. 2000, 66, 4972–4978. [Google Scholar] [CrossRef] [Green Version]
- Ledgham, F.; Quest, B.; Vallaeys, T.; Mergeay, M.; Coves, J. A probable link between the DedA protein and resistance to selenite. Res. Microbiol. 2005, 156, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Bebien, M.; Chauvin, J.P.; Adriano, J.M.; Grosse, S. Effect of selenite on growth and protein synthesis in the phototrophic bacterium Rhodobacter sphaeroides. Appl. Environ. Microbiol. 2001, 67, 4440–4447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mapelli, V.; Hillestrom, P.R.; Patil, K.; Larsen, E.H.; Olsson, L. The interplay between sulphur and selenium metabolism influences the intracellular redox balance in Saccharomyces cerevisiae. FEMS Yeast Res. 2012, 12, 20–32. [Google Scholar] [CrossRef] [Green Version]
- Lazard, M.; Blanquet, S.; Fisicaro, P.; Labarraque, G.; Plateau, P. Uptake of selenite by Saccharomyces cerevisiae involves the high and low affinity orthophosphate transporters. J. Biol. Chem. 2010, 285, 32029–32037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDermott, J.R.; Rosen, B.P.; Liu, Z.J. Jen1p: A High Affinity Selenite Transporter in Yeast. Mol. Biol. Cell 2010, 21, 3934–3941. [Google Scholar] [CrossRef] [Green Version]
- Xu, Q.L.; Wu, W.L.; Zhao, G.S.; Zhu, Y.Y.; Guo, Y.B. Selenium metabolism in microorganisms. Microbiol. China 2017, 44, 207–216. [Google Scholar]
- Hoefig, C.S.; Renko, K.; Kohrle, J.; Birringer, M.; Schomburg, L. Comparison of different selenocompounds with respect to nutritional value vs. toxicity using liver cells in culture. J. Nutr. Biochem. 2011, 22, 945–955. [Google Scholar] [CrossRef]
- Mapelli, V.; Hillestrom, P.R.; Kapolna, E.; Larsen, E.H.; Olsson, L. Metabolic and bioprocess engineering for production of selenized yeast with increased content of seleno-methylselenocysteine. Metab. Eng. 2011, 13, 282–293. [Google Scholar] [CrossRef] [Green Version]
- Kitajima, T.; Chiba, Y. Selenomethionine metabolism and its toxicity in yeast. Biomol. Concepts 2013, 4, 611–616. [Google Scholar] [CrossRef]
- Kuroda, M.; Yamashita, M.; Miwa, E.; Imao, K.; Fujimoto, N.; Ono, H.; Nagano, K.; Sei, K.; Ike, M. Molecular Cloning and Characterization of the srdBCA Operon, Encoding the Respiratory Selenate Reductase Complex, from the Selenate-Reducing Bacterium Bacillus selenatarsenatis SF-1. J. Bacteriol. 2011, 193, 2141–2148. [Google Scholar] [CrossRef] [Green Version]
- Losi, M.E.; Frankenberger, W.T. Reduction of Selenium Oxyanions by Enterobacter cloacae SLD1a-1: Isolation and Growth of the Bacterium and Its Expulsion of Selenium Particles. Appl. Environ. Microbiol. 1997, 63, 3079–3084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, C.S.; Debieux, C.M.; Dridge, E.J.; Splatt, P.; Wright, M. Biomineralization of selenium by the selenate-respiring bacterium Thauera selenatis. Biochem. Soc. Trans. 2012, 40, 1239–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viña, J. Glutathione: Metabolism and Physiological Functions; The Chemical Rubber Company Press: Boca Raton, FL, USA, 1990; p. 392. [Google Scholar]
- Kessi, J.; Hanselmann, K.W. Similarities between the abiotic reduction of selenite with glutathione and the dissimilatory reaction mediated by Rhodospirillum rubrum and Escherichia coli. J. Biol. Chem. 2004, 279, 50662–50669. [Google Scholar] [CrossRef]
- Liang, W.X.; Rudd, K.E.; Deutscher, M.P. A Role for REP Sequences in Regulating Translation. Mol. Cell 2015, 58, 431–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tormay, P.; Böck, A. Barriers to heterologous expression of a selenoprotein gene in bacteria. J. Bacteriol. 1997, 179, 576–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, J. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 1989, 53, 273–298. [Google Scholar] [CrossRef]
- Gene-Wei, L.; Burkhardt, D.; Gross, C. Quantifying Absolute Protein Synthesis Rates Reveals Principles Underlying Allocation of Cellular Resources. Cell 2014, 157, 624–635. [Google Scholar]
- Cheng, Q.; Arnér, E.S.J. Selenocysteine Insertion at a Predefined UAG Codon in a Release Factor 1 (RF1)-depleted Escherichia coli Host Strain Bypasses Species Barriers in Recombinant Selenoprotein Translation. J. Biol. Chem. 2017, 292, 5476–5487. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, J.-J.; Yue, S.-Y.; Fang, Y.-H.; Liu, X.-L.; Wang, C.-H. Mechanisms Affecting the Biosynthesis and Incorporation Rate of Selenocysteine. Molecules 2021, 26, 7120. https://doi.org/10.3390/molecules26237120
Peng J-J, Yue S-Y, Fang Y-H, Liu X-L, Wang C-H. Mechanisms Affecting the Biosynthesis and Incorporation Rate of Selenocysteine. Molecules. 2021; 26(23):7120. https://doi.org/10.3390/molecules26237120
Chicago/Turabian StylePeng, Jing-Jing, Shi-Yang Yue, Yu-Hui Fang, Xiao-Ling Liu, and Cheng-Hua Wang. 2021. "Mechanisms Affecting the Biosynthesis and Incorporation Rate of Selenocysteine" Molecules 26, no. 23: 7120. https://doi.org/10.3390/molecules26237120





