Gut Microbiome and Its Cofactors Are Linked to Lipoprotein Distribution Profiles
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Lipoprotein Distribution Profiles
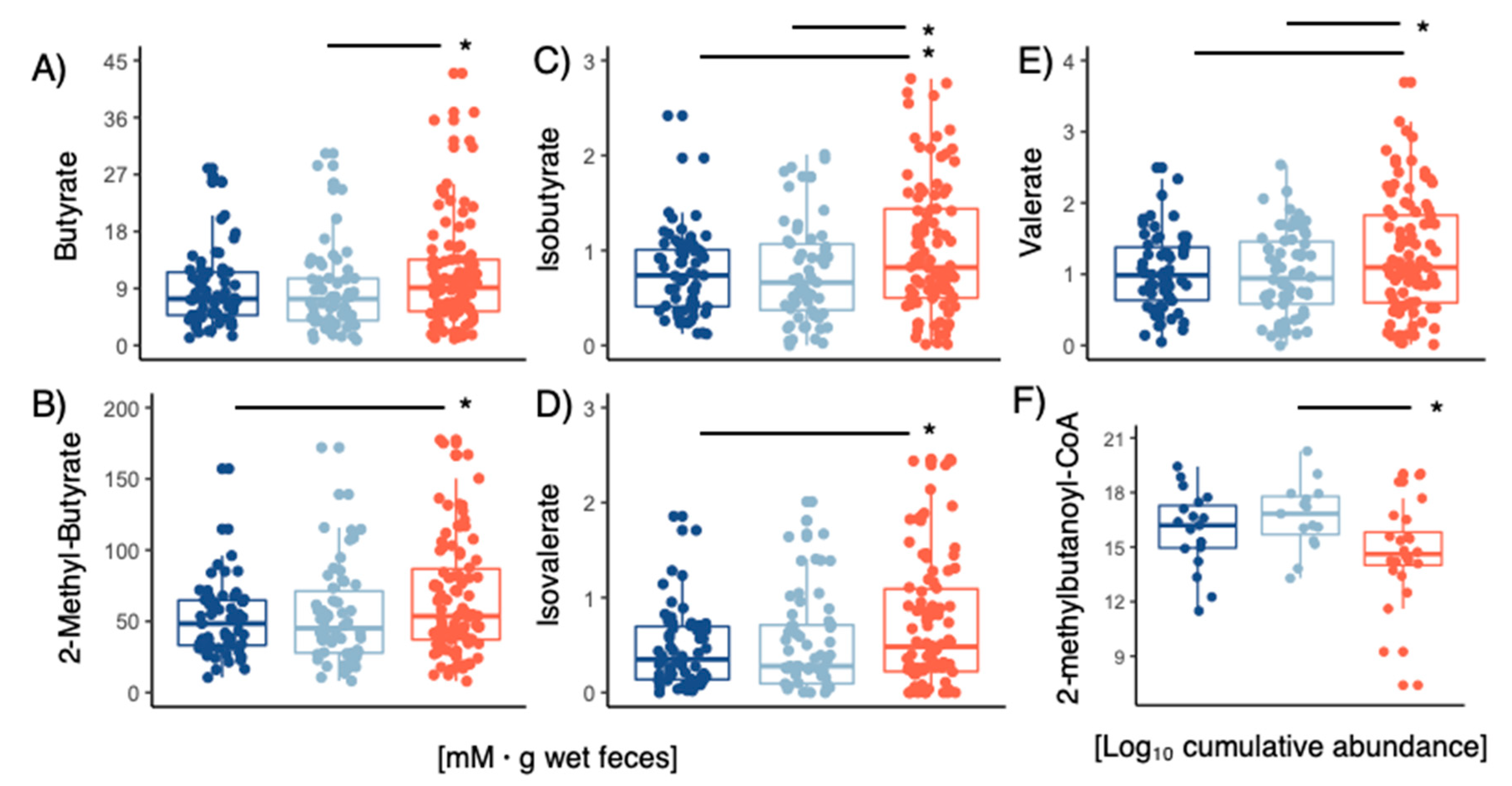
2.3. Short Chain Fatty Acids (SCFAs) Quantification
2.4. Samples Processing, Library Preparation and DNA Sequencing
2.5. Analysis of Sequencing Data
2.6. Statistical Analysis
3. Results
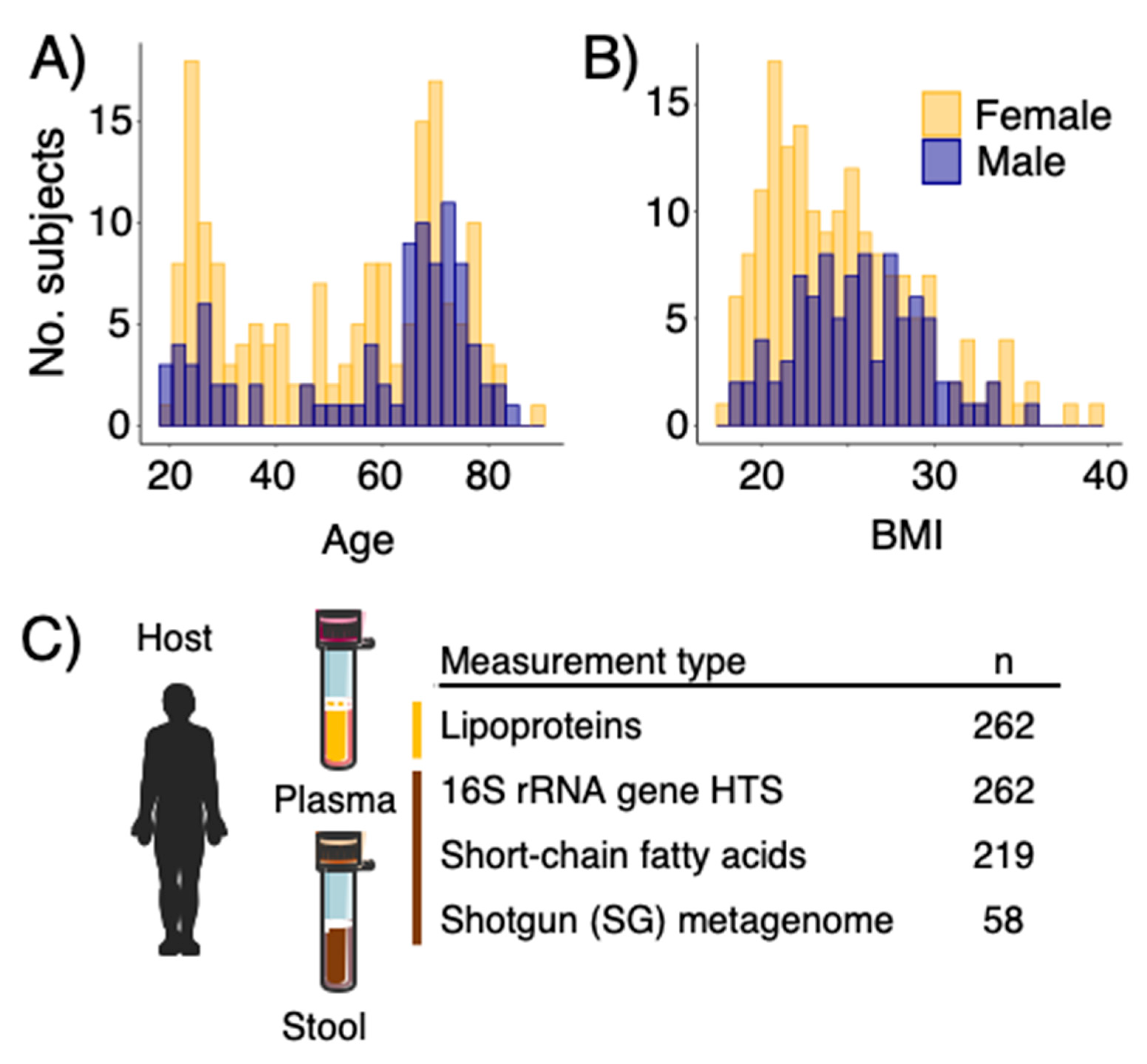
3.1. Participants and Data Collection
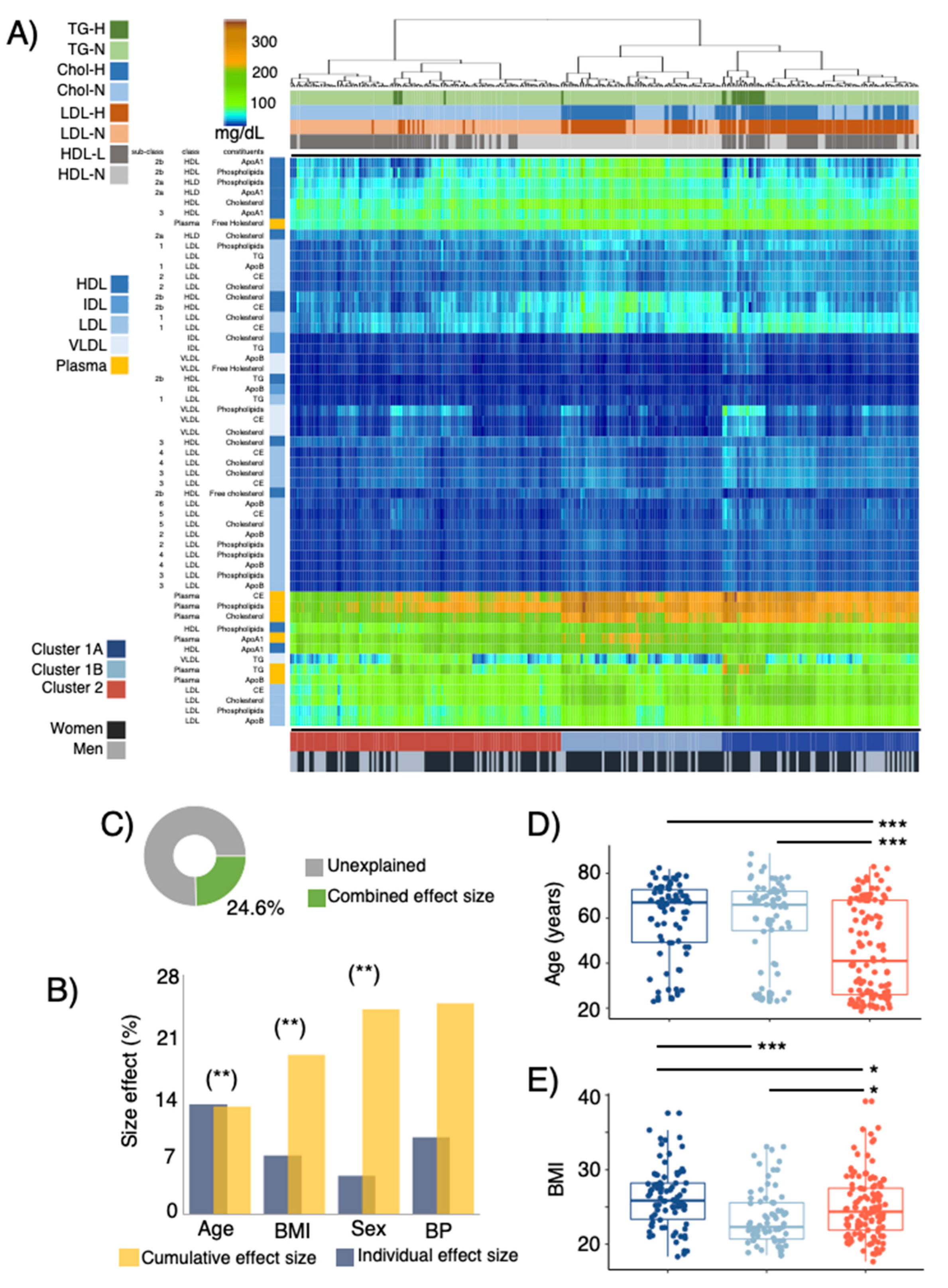
3.2. LPD Profiles, Stratification, and Host Covariates
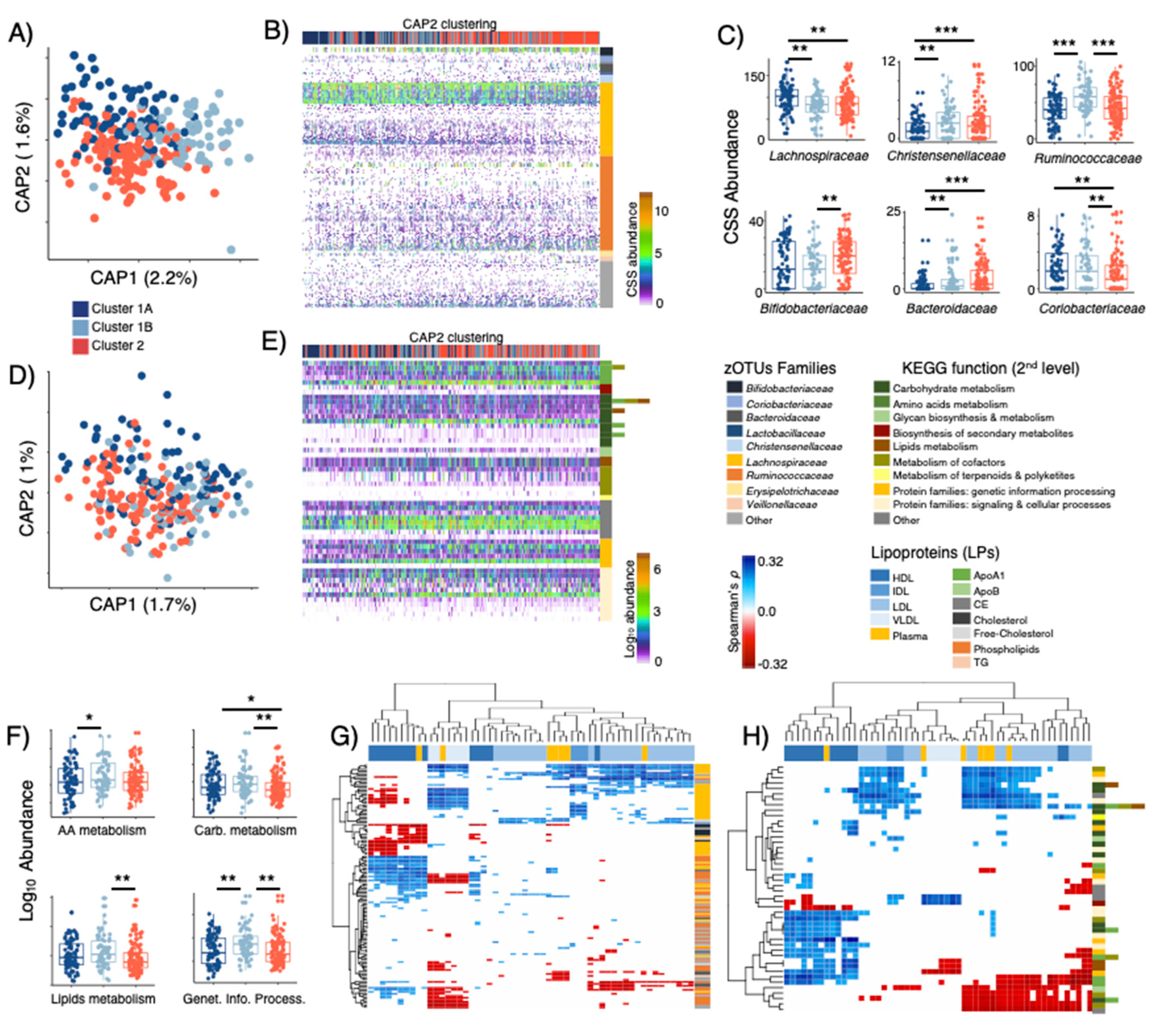
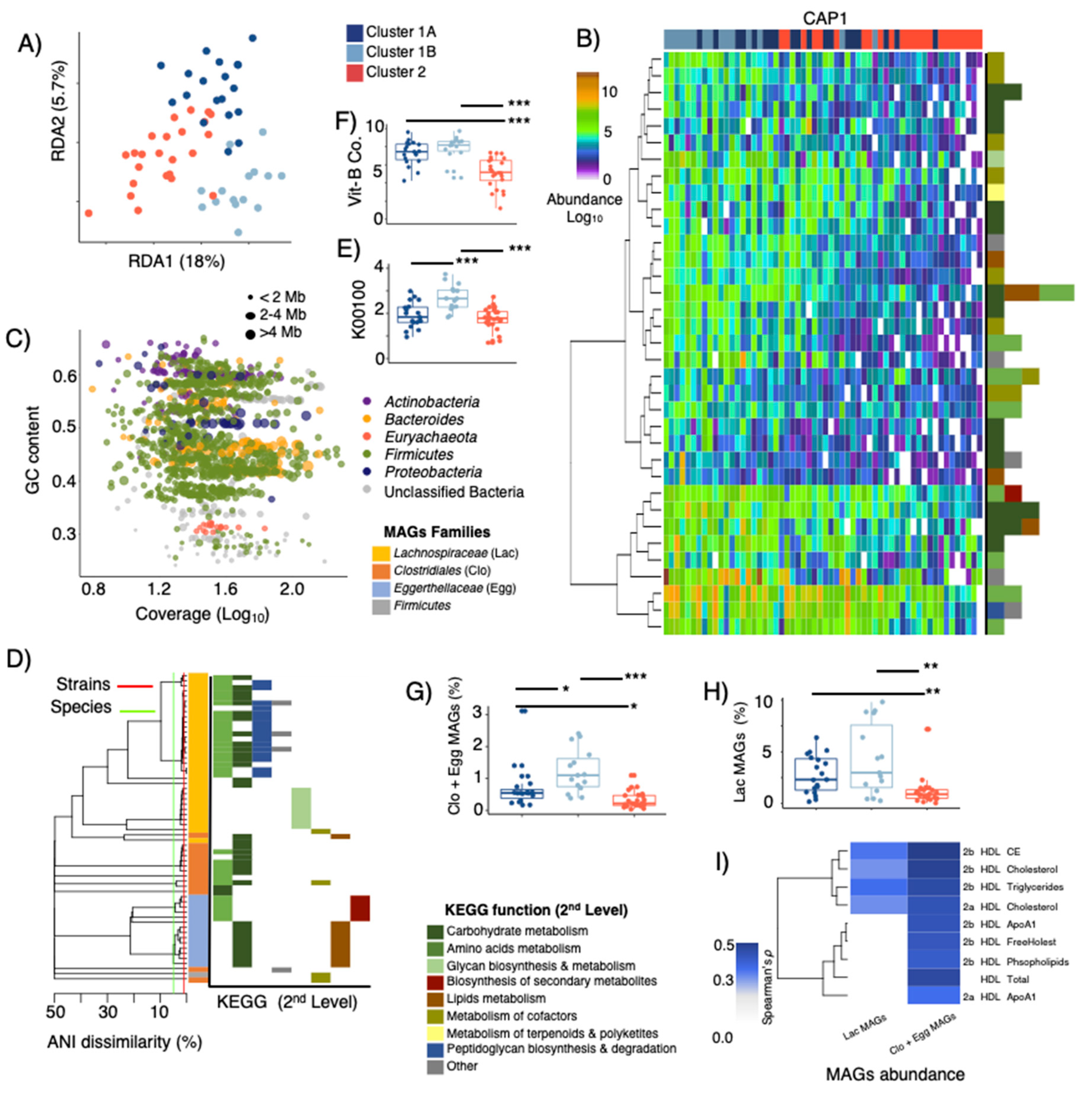
3.3. LPD Clusters Are Linked with GM Profiles
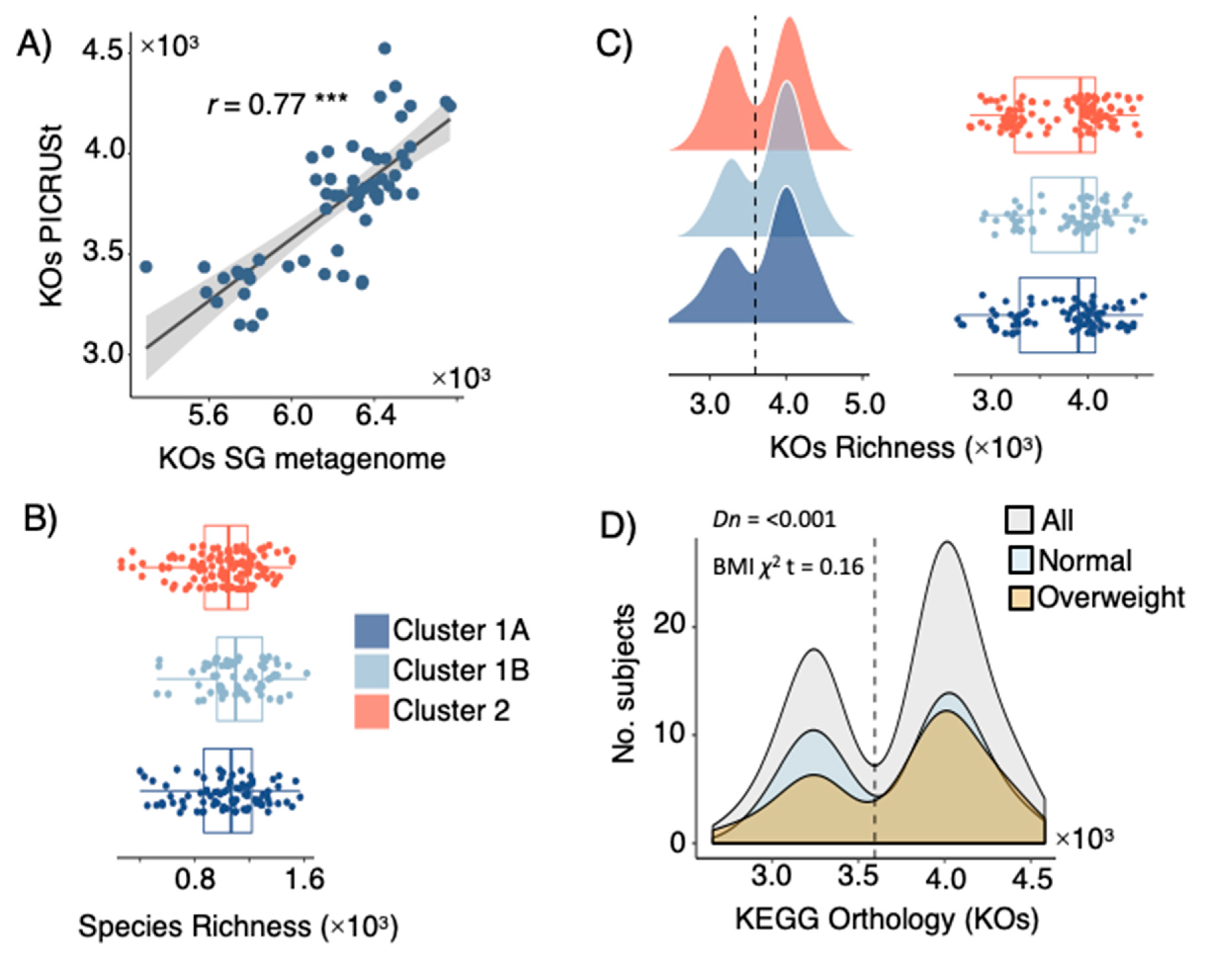
3.4. LPD Clusters Correspond with GM and KOs Features
3.5. Metagenome Bins and Functions Associated with LPD Clusters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, H.; Temel, R.E.; Martel, C. Cholesterol and Lipoprotein Metabolism: Early Career Committee Contribution. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 1791–1794. [Google Scholar] [CrossRef] [PubMed]
- Vallejo-Vaz, A.J.; Robertson, M.; Catapano, A.L.; Watts, G.F.; Kastelein, J.J.; Packard, C.J.; Ford, I.; Ray, K.K. Low-Density Lipoprotein Cholesterol Lowering for the Primary Prevention of Cardiovascular Disease among Men with Primary Elevations of Low-Density Lipoprotein Cholesterol Levels of 190 Mg/DL or above: Analyses from the WOSCOPS. Circulation 2017, 136, 1878–1891. [Google Scholar] [CrossRef] [PubMed]
- Nelson, E.R.; Chang, C.Y.; McDonnell, D.P. Cholesterol and Breast Cancer Pathophysiology. Trends Endocrinol. Metab. 2014, 25, 649–655. [Google Scholar] [CrossRef] [PubMed]
- Aru, V.; Lam, C.; Khakimov, B.; Hoefsloot, H.C.J.; Zwanenburg, G.; Lind, M.V.; Schäfer, H.; van Duynhoven, J.; Jacobs, D.M.; Smilde, A.K.; et al. Quantification of Lipoprotein Profiles by Nuclear Magnetic Resonance Spectroscopy and Multivariate Data Analysis. TrAC-Trends Anal. Chem. 2017, 94, 210–219. [Google Scholar] [CrossRef]
- Vojinovic, D.; Radjabzadeh, D.; Kurilshikov, A.; Amin, N.; Wijmenga, C.; Franke, L.; Ikram, M.A.; Uitterlinden, A.G.; Zhernakova, A.; Fu, J.; et al. Relationship between Gut Microbiota and Circulating Metabolites in Population-Based Cohorts. Nat. Commun. 2019, 10, 5813. [Google Scholar] [CrossRef]
- Khakimov, B.; Hoefsloot, H.C.J.; Mobaraki, N.; Aru, V.; Kristensen, M.; Mads, V.; Holm, L.; Nielsen, D.S.; Jacobs, D.M.; Smilde, A.K.; et al. Human Blood Lipoprotein Predictions from 1 H NMR Spectra: Protocol, Model Performances and Cage of Covariance. Anal. Chem. 2022, 94, 628–636. [Google Scholar] [CrossRef]
- Holmes, M.V.; Millwood, I.Y.; Kartsonaki, C.; Hill, M.R.; Bennett, D.A.; Boxall, R.; Guo, Y.; Xu, X.; Bian, Z.; Hu, R.; et al. Lipids, Lipoproteins, and Metabolites and Risk of Myocardial Infarction and Stroke. J. Am. Coll. Cardiol. 2018, 71, 620–632. [Google Scholar] [CrossRef]
- Wang, J.; Stančáková, A.; Soininen, P.; Kangas, A.J.; Paananen, J.; Kuusisto, J.; Ala-Korpela, M.; Laakso, M. Lipoprotein Subclass Profiles in Individuals with Varying Degrees of Glucose Tolerance: A Population-Based Study of 9399 Finnish Men. J. Intern. Med. 2012, 272, 562–572. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between Microbiota and Immunity in Health and Disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Fu, J.; Bonder, M.J.; Cenit, M.C.; Tigchelaar, E.F.; Maatman, A.; Dekens, J.A.M.; Brandsma, E.; Marczynska, J.; Imhann, F.; Weersma, R.K.; et al. The Gut Microbiome Contributes to a Substantial Proportion of the Variation in Blood Lipids. Circ. Res. 2015, 117, 817–824. [Google Scholar] [CrossRef]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.; et al. Population-Level Analysis of Gut Microbiome Variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment Dominates over Host Genetics in Shaping Human Gut Microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal Microbiota Metabolism of L-Carnitine, a Nutrient in Red Meat, Promotes Atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Le Roy, T.; Lécuyer, E.; Chassaing, B.; Rhimi, M.; Lhomme, M.; Boudebbouze, S.; Ichou, F.; Haro Barceló, J.; Huby, T.; Guerin, M.; et al. The Intestinal Microbiota Regulates Host Cholesterol Homeostasis. BMC Biol. 2019, 17, 94. [Google Scholar] [CrossRef]
- Yu, Y.; Raka, F.; Adeli, K. The Role of the Gut Microbiota in Lipid and Lipoprotein Metabolism. J. Clin. Med. 2019, 8, 2227. [Google Scholar] [CrossRef]
- Liong, M.T.; Shah, N.P. Acid and Bile Tolerance and Cholesterol Removal Ability of Lactobacilli Strains. J. Dairy Sci. 2005, 88, 55–66. [Google Scholar] [CrossRef]
- Kasahara, K.; Krautkramer, K.A.; Org, E.; Romano, K.A.; Kerby, R.L.; Vivas, E.I.; Mehrabian, M.; Denu, J.M.; Bäckhed, F.; Lusis, A.J.; et al. Interactions between Roseburia Intestinalis and Diet Modulate Atherogenesis in a Murine Model. Nat. Microbiol. 2018, 3, 1461–1471. [Google Scholar] [CrossRef]
- Khakimov, B.; Mobaraki, N.; Trimigno, A.; Aru, V.; Balling, S. Analytica Chimica Acta Signature Mapping (SigMa): An Efficient Approach for Processing Complex Human Urine 1 H NMR Metabolomics Data. Anal. Chim. Acta 2020, 1108, 142–151. [Google Scholar] [CrossRef]
- Castro-Mejía, J.L.; Khakimov, B.; Krych, Ł.; Bülow, J.; Bechshøft, R.L.; Højfeldt, G.; Mertz, K.H.; Garne, E.S.; Schacht, S.R.; Ahmad, H.F.; et al. Physical Fitness in Community-dwelling Older Adults Is Linked to Dietary Intake, Gut Microbiota, and Metabolomic Signatures. Aging Cell 2020, 19, e13105. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive Functional Profiling of Microbial Communities Using 16S RRNA Marker Gene Sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R. UNOISE2: Improved Error-Correction for Illumina 16S and ITS Amplicon Sequencing. bioRxiv 2016, 081257. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Uritskiy, G.V.; Diruggiero, J.; Taylor, J. MetaWRAP-A Flexible Pipeline for Genome-Resolved Metagenomic Data Analysis 08 Information and Computing Sciences 0803 Computer Software 08 Information and Computing Sciences 0806 Information Systems. Microbiome 2018, 6, 158. [Google Scholar] [CrossRef]
- Kang, D.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an Efficient Tool for Accurately Reconstructing Single Genomes from Complex Microbial Communities. PeerJ 2015, 3, e1165. [Google Scholar] [CrossRef]
- Kang, D.D.; Li, F.; Kirton, E.; Thomas, A.; Egan, R.; An, H.; Wang, Z. MetaBAT2: An Adaptive Binning Algorithm for Robust and Efficient Genome Reconstruction from Metagenome Assemblies. PeerJ 2019, 27, e7359. [Google Scholar] [CrossRef]
- Wu, Y.-W.; Tang, Y.-H.; Tringe, S.G.; Simmons, B.A.; Singer, S.W. MaxBin: An Automated Binning Method to Recover Individual Genomes from Metagenomes Using. Microbiome 2014, 2, 26. [Google Scholar] [CrossRef]
- Song, W.Z.; Thomas, T. Binning-Refiner: Improving Genome Bins through the Combination of Different Binning Programs. Bioinformatics 2017, 33, 1873–1875. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Carroll, L.M.; Wiedmann, M.; Kovac, J. Proposal of a Taxonomic Nomenclature for the Bacillus Cereus Group Which Reconciles Genomic Definitions of Bacterial Species with Clinical and Industrial Phenotypes. MBio 2020, 11, e00034-20. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. The Subread Aligner: Fast, Accurate and Scalable Read Mapping by Seed-and-Vote. Nucleic Acids Res. 2013, 41, e108. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Kahlke, T.; Ralph, P.J. BASTA–Taxonomic Classification of Sequences and Sequence Bins Using Last Common Ancestor Estimations. Methods Ecol. Evol. 2019, 10, 100–103. [Google Scholar] [CrossRef]
- Li, J.; Wang, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; et al. An Integrated Catalog of Reference Genes in the Human Gut Microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2014, 12, 59–60. [Google Scholar] [CrossRef]
- Warnes, G.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; et al. Gplots. R Package Version 3.1.1. 2020. Available online: https://CRAN.R-project.org/package=gplots (accessed on 25 September 2022).
- Oksanen, A.J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; Hara, R.B.O.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package. R package Version 2.5-7. 2020. Available online: https://CRAN.R-project.org/package=vegan (accessed on 25 September 2022).
- Hothorn, T.; Buehmann, P.; Dudoit, S.; Molinaro, A.; Van der Laan, M. Survival Ensembles. Biostatistics 2006, 7, 355–373. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by RandomForest. R News 2002, 2, 18–22. [Google Scholar]
- Anderson, N.B. National Cholesterol Education Program (Ncep). Encycl. Health Behav. 2004, 1, 530–533. [Google Scholar] [CrossRef]
- Sun, S.; Jones, R.B.; Fodor, A.A. Inference-Based Accuracy of Metagenome Prediction Tools Varies across Sample Types and Functional Categories. Microbiome 2020, 8, 46. [Google Scholar] [CrossRef] [PubMed]
- Sokooti, S.; Flores-Guerrero, J.L.; Kieneker, L.M.; Heerspink, H.J.L.; Connelly, M.A.; Bakker, S.J.L.; Dullaart, R.P.F. HDL Particle Subspecies and Their Association with Incident Type 2 Diabetes: The PREVEND Study. J. Clin. Endocrinol. Metab. 2021, 106, 1761–1772. [Google Scholar] [CrossRef]
- Ala-Korpela, M.; Zhao, S.; Järvelin, M.R.; Mäkinen, V.P.; Ohukainen, P. Apt Interpretation of Comprehensive Lipoprotein Data in Large-Scale Epidemiology: Disclosure of Fundamental Structural and Metabolic Relationships. Int. J. Epidemiol. 2022, 51, 996–1011. [Google Scholar] [CrossRef] [PubMed]
- Behbodikhah, J.; Ahmed, S.; Elyasi, A.; Kasselman, L.J.; De Leon, J.; Glass, A.D.; Reiss, A.B. Apolipoprotein b and Cardiovascular Disease: Biomarker and Potential Therapeutic Target. Metabolites 2021, 11, 690. [Google Scholar] [CrossRef]
- Shen, J.; Tong, X.; Sud, N.; Khound, R.; Song, Y.; Maldonado-Gomez, M.X.; Walter, J.; Su, Q. Low-Density Lipoprotein Receptor Signaling Mediates the Triglyceride-Lowering Action of Akkermansia muciniphila in Genetic-Induced Hyperlipidemia. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 1448–1456. [Google Scholar] [CrossRef]
- Kiouptsi, K.; Jäckel, S.; Pontarollo, G.; Grill, A.; Kuijpers, M.J.E.; Wilms, E.; Weber, C.; Sommer, F.; Nagy, M.; Neideck, C.; et al. The Microbiota Promotes Arterial Thrombosis in Low-Density Lipoprotein Receptor-Deficient Mice. MBio 2019, 10, e02298-19. [Google Scholar] [CrossRef]
- Rune, I.; Rolin, B.; Larsen, C.; Nielsen, D.S.; Kanter, J.E.; Bornfeldt, K.E.; Lykkesfeldt, J.; Buschard, K.; Kirk, R.K.; Christoffersen, B.; et al. Modulating the Gut Microbiota Improves Glucose Tolerance, Lipoprotein Profile and Atherosclerotic Plaque Development in ApoE-Deficient Mice. PLoS ONE 2016, 11, e0146439. [Google Scholar] [CrossRef]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.M.; Kennedy, S.; et al. Richness of Human Gut Microbiome Correlates with Metabolic Markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef]
- Cotillard, A.; Kennedy, S.P.; Kong, L.C.; Prifti, E.; Pons, N.; Le Chatelier, E.; Almeida, M.; Quinquis, B.; Levenez, F.; Galleron, N.; et al. Dietary Intervention Impact on Gut Microbial Gene Richness. Nature 2013, 500, 585–588. [Google Scholar] [CrossRef]
- Kontush, A. HDL Particle Number and Size as Predictors of Cardiovascular Disease. Front. Pharmacol. 2015, 6, 218. [Google Scholar] [CrossRef] [PubMed]
- Camont, L.; Chapman, M.J.; Kontush, A. Biological Activities of HDL Subpopulations and Their Relevance to Cardiovascular Disease. Trends Mol. Med. 2011, 17, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Kim, M.K.; Choi, B.Y. The Long-Term Relationship between Dietary Pantothenic Acid (Vitamin B5) Intake and C-Reactive Protein Concentration in Adults Aged 40 Years and Older. Nutr. Metab. Cardiovasc. Dis. 2017, 27, 806–816. [Google Scholar] [CrossRef] [PubMed]
- Cui, R.; Iso, H.; Date, C.; Kikuchi, S.; Tamakoshi, A. Dietary Folate and Vitamin B6 and B12 Intake in Relation to Mortality from Cardiovascular Diseases: Japan Collaborative Cohort Study. Stroke 2010, 41, 1285–1289. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.K.; Jacobs, L.; Watts, J.L.; Rottiers, V.; Jiang, K.; Finnegan, D.M.; Shioda, T.; Hansen, M.; Yang, F.; Niebergall, L.J.; et al. A Conserved SREBP-1/Phosphatidylcholine Feedback Circuit Regulates Lipogenesis in Metazoans. Cell 2011, 147, 840–850. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.D.; Prykhodko, O.; Fåk Hållenius, F.; Nyman, M. Monovalerin and Trivalerin Increase Brain Acetic Acid, Decrease Liver Succinic Acid, and Alter Gut Microbiota in Rats Fed High-Fat Diets. Eur. J. Nutr. 2019, 58, 1545–1560. [Google Scholar] [CrossRef]
- Jiao, A.R.; Diao, H.; Yu, B.; He, J.; Yu, J.; Zheng, P.; Huang, Z.Q.; Luo, Y.H.; Luo, J.Q.; Mao, X.B.; et al. Oral Administration of Short Chain Fatty Acids Could Attenuate Fat Deposition of Pigs. PLoS ONE 2018, 13, e0196867. [Google Scholar] [CrossRef]
- Du, Y.; Li, X.; Su, C.; Xi, M.; Zhang, X.; Jiang, Z.; Wang, L.; Hong, B. Butyrate Protects against High-Fat Diet-Induced Atherosclerosis via up-Regulating ABCA1 Expression in Apolipoprotein E-Deficiency Mice. Br. J. Pharmacol. 2020, 177, 1754–1772. [Google Scholar] [CrossRef]
- Karlsson, F.H.; Fåk, F.; Nookaew, I.; Tremaroli, V.; Fagerberg, B.; Petranovic, D.; Bäckhed, F.; Nielsen, J. Symptomatic Atherosclerosis Is Associated with an Altered Gut Metagenome. Nat. Commun. 2012, 3, 1245. [Google Scholar] [CrossRef]
- Denou, E.; Lolmède, K.; Garidou, L.; Pomie, C.; Chabo, C.; Lau, T.C.; Fullerton, M.D.; Nigro, G.; Zakaroff-Girard, A.; Luche, E.; et al. Defective NOD2 Peptidoglycan Sensing Promotes Diet-induced Inflammation, Dysbiosis, and Insulin Resistance. EMBO Mol. Med. 2015, 7, 259–274. [Google Scholar] [CrossRef]
- Clarke, T.B.; Davis, K.M.; Lysenko, E.S.; Zhou, A.Y.; Yu, Y.; Weiser, J.N. Recognition of Peptidoglycan from the Microbiota by NOD1 Enhances Systemic Innate Immunity. Nat. Med. 2010, 16, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Tosoni, G.; Conti, M.; Diaz Heijtz, R. Bacterial Peptidoglycans as Novel Signaling Molecules from Microbiota to Brain. Curr. Opin. Pharmacol. 2019, 48, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Baik, J.E.; Jang, Y.O.; Kang, S.S.; Cho, K.; Yun, C.H.; Han, S.H. Differential Profiles of Gastrointestinal Proteins Interacting with Peptidoglycans from Lactobacillus plantarum and Staphylococcus aureus. Mol. Immunol. 2015, 65, 77–85. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castro-Mejía, J.L.; Khakimov, B.; Aru, V.; Lind, M.V.; Garne, E.; Paulová, P.; Tavakkoli, E.; Hansen, L.H.; Smilde, A.K.; Holm, L.; et al. Gut Microbiome and Its Cofactors Are Linked to Lipoprotein Distribution Profiles. Microorganisms 2022, 10, 2156. https://doi.org/10.3390/microorganisms10112156
Castro-Mejía JL, Khakimov B, Aru V, Lind MV, Garne E, Paulová P, Tavakkoli E, Hansen LH, Smilde AK, Holm L, et al. Gut Microbiome and Its Cofactors Are Linked to Lipoprotein Distribution Profiles. Microorganisms. 2022; 10(11):2156. https://doi.org/10.3390/microorganisms10112156
Chicago/Turabian StyleCastro-Mejía, Josué L., Bekzod Khakimov, Violetta Aru, Mads V. Lind, Eva Garne, Petronela Paulová, Elnaz Tavakkoli, Lars H. Hansen, Age K. Smilde, Lars Holm, and et al. 2022. "Gut Microbiome and Its Cofactors Are Linked to Lipoprotein Distribution Profiles" Microorganisms 10, no. 11: 2156. https://doi.org/10.3390/microorganisms10112156
APA StyleCastro-Mejía, J. L., Khakimov, B., Aru, V., Lind, M. V., Garne, E., Paulová, P., Tavakkoli, E., Hansen, L. H., Smilde, A. K., Holm, L., Engelsen, S. B., & Nielsen, D. S. (2022). Gut Microbiome and Its Cofactors Are Linked to Lipoprotein Distribution Profiles. Microorganisms, 10(11), 2156. https://doi.org/10.3390/microorganisms10112156







