Brain Tumor Diagnosis Using Machine Learning, Convolutional Neural Networks, Capsule Neural Networks and Vision Transformers, Applied to MRI: A Survey
Abstract
:1. Introduction
- Most review papers considered one or two types of ML-based techniques, while we included four types of brain tumor segmentation and classification techniques: classical ML algorithms, CNN-based techniques, CapsNet-based techniques, and ViT-based techniques. This survey summarizes the current state-of-the-art in brain tumor segmentation, grade estimation, and classification. It outlines the advances made in ML-based, CNN-based, CapsNet-based, and ViT-based brain tumor diagnosis between 2019 and 2022.
- Most of the previous studies [9,21,22,23] presented an overview of either CNN-based techniques, ML-based techniques, or both ML- and CNN-based approaches. In addition to CNN-based and ML-based techniques, we present a summary of CapsNet-based, and ViT-based brain tumor segmentation and classification techniques. CapsNet is one of the state-of-the-art techniques for brain tumor segmentation and classification. The findings of this survey show that CapsNet outperformed CNN-based brain tumor diagnosis techniques, as they require significantly less training data compared to CNNs. Moreover, CapsNets are very robust to different rotations and image transformations. Furthermore, unlike CNN, ViT-based models can effectively model local and global feature information, which is very critical for accurate brain tumor segmentation and classification.
- Some review papers did not provide a comprehensive discussion and pertinent findings. This survey presents significant findings and a comprehensive discussion of approaches for segmenting and classifying brain tumors. We also identify open problems and significant areas for future research, which should be beneficial to interested scholars and medical practitioners.
2. Background
2.1. Brain Tumors and Magnetic Resonance Imaging
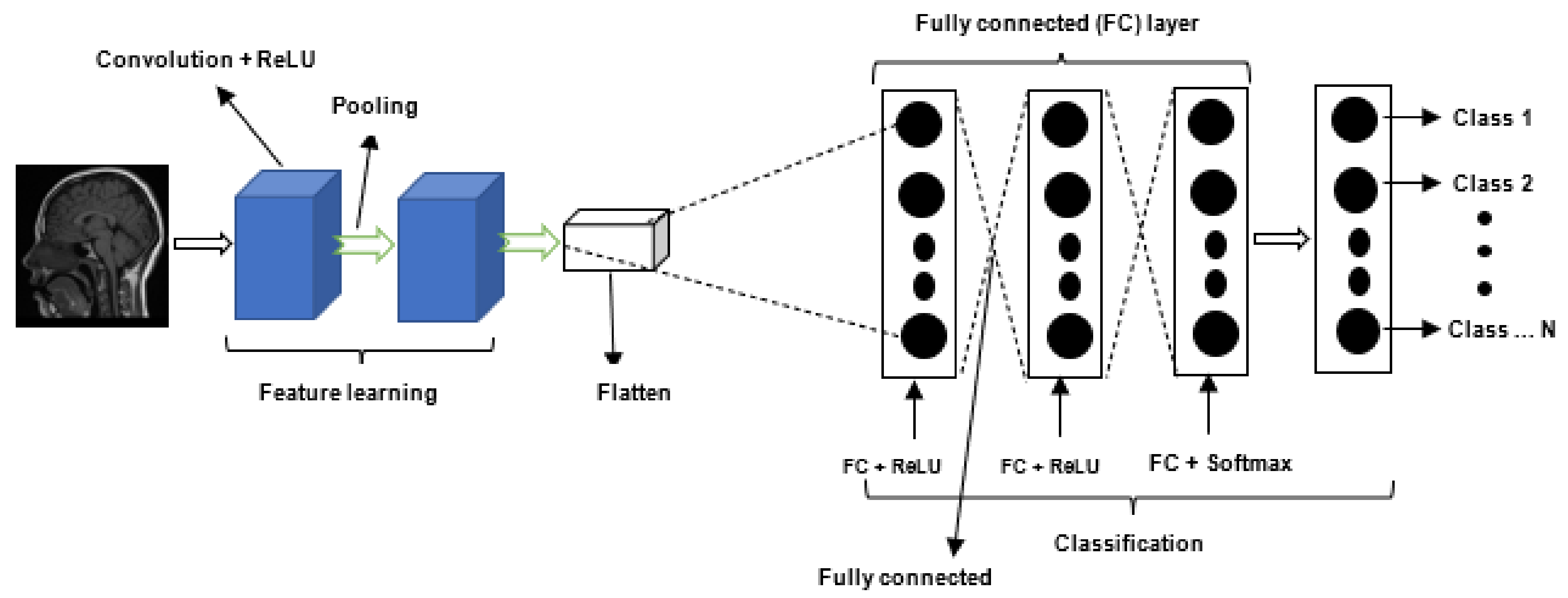
2.2. Deep Learning
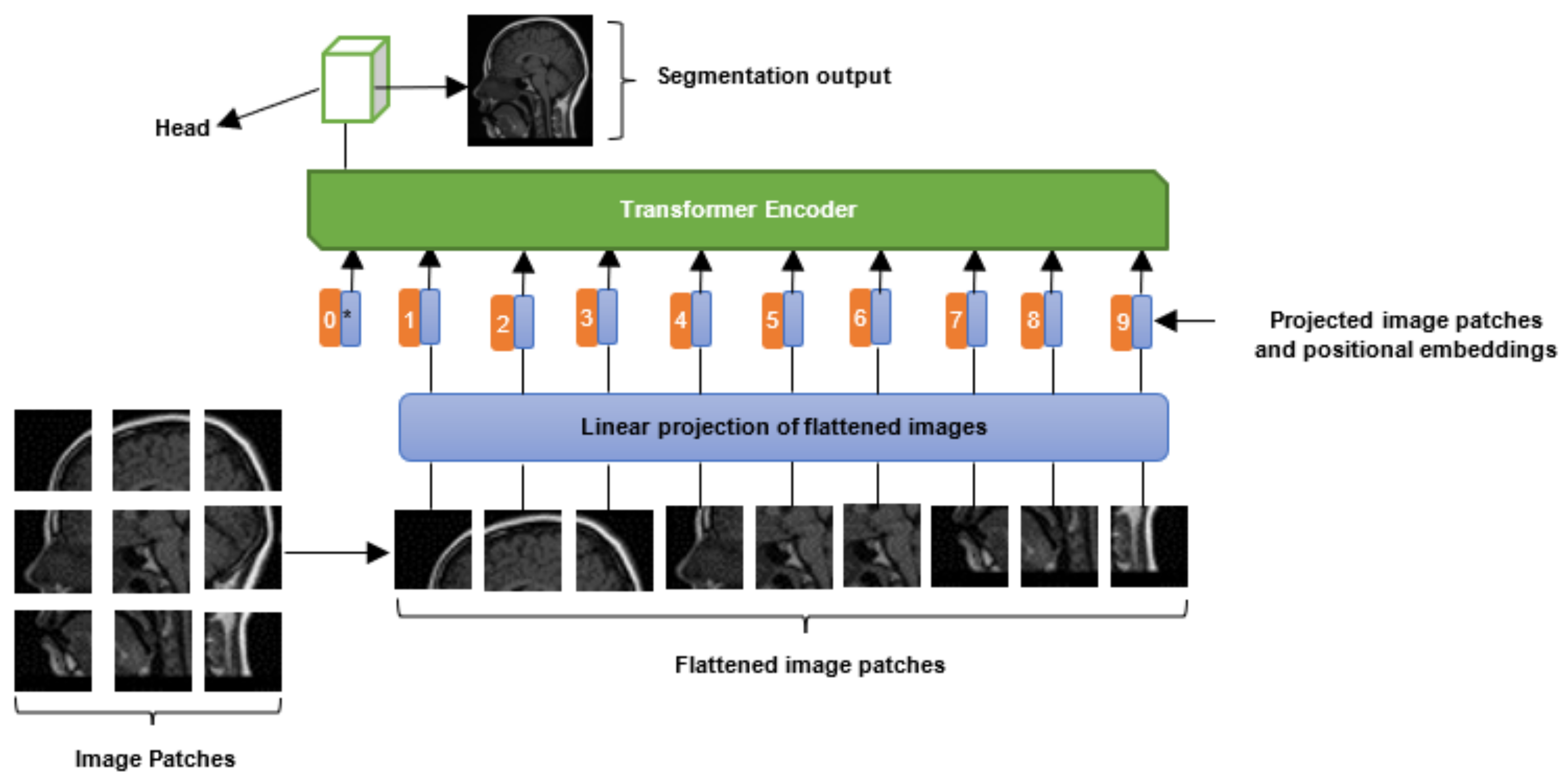
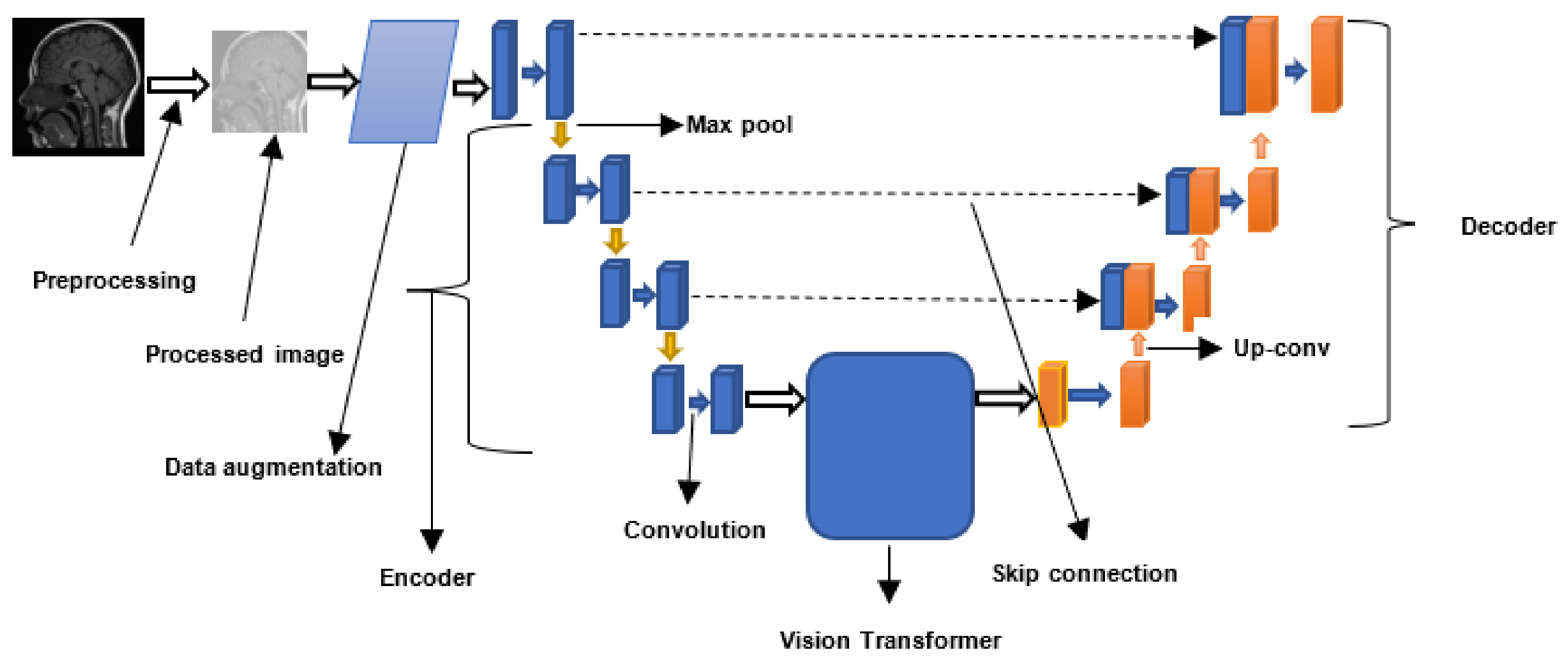
2.3. Vision Transformers
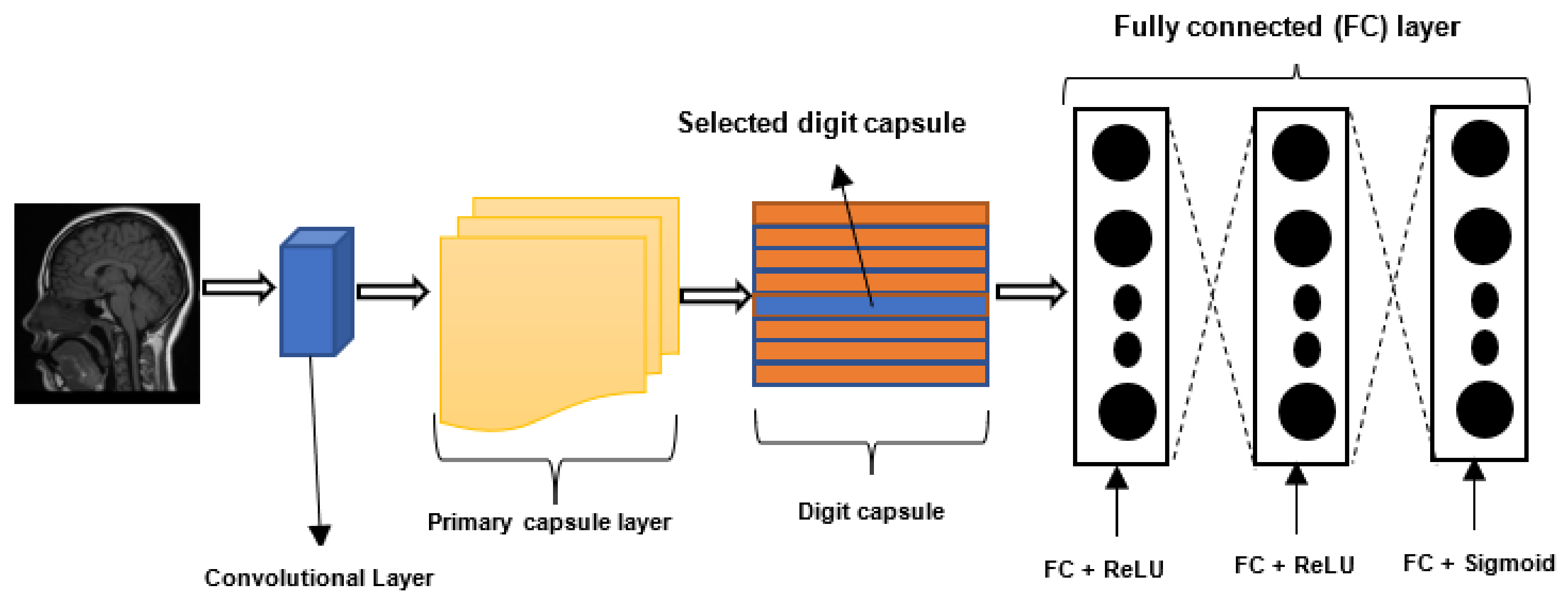
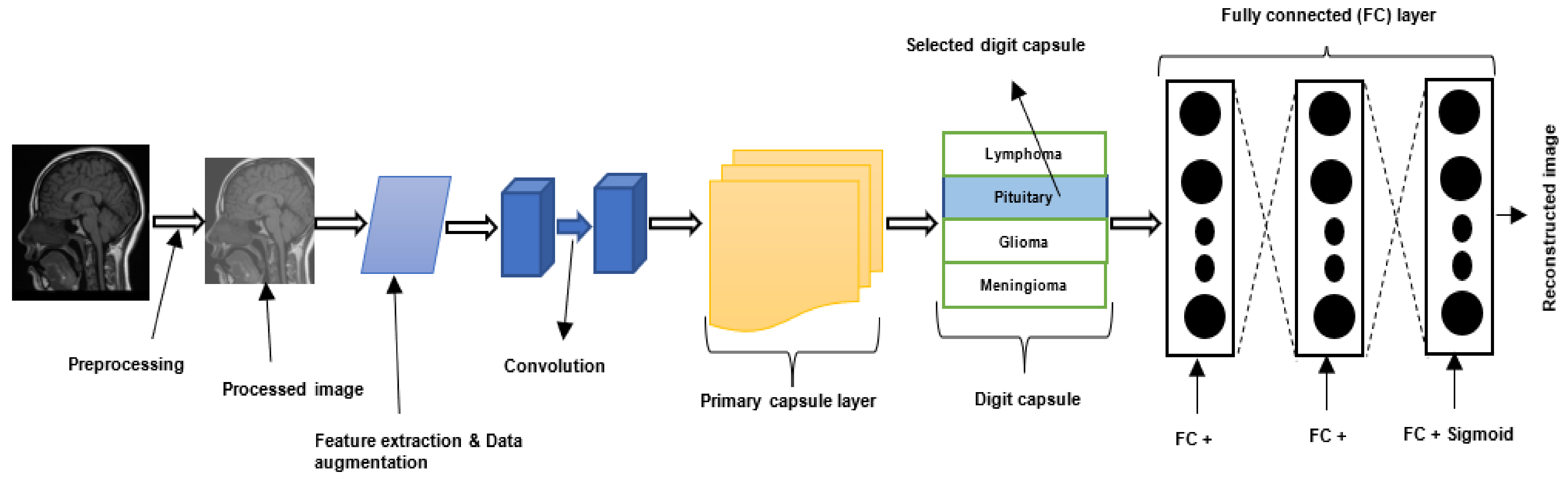
2.4. Capsule Neural Networks
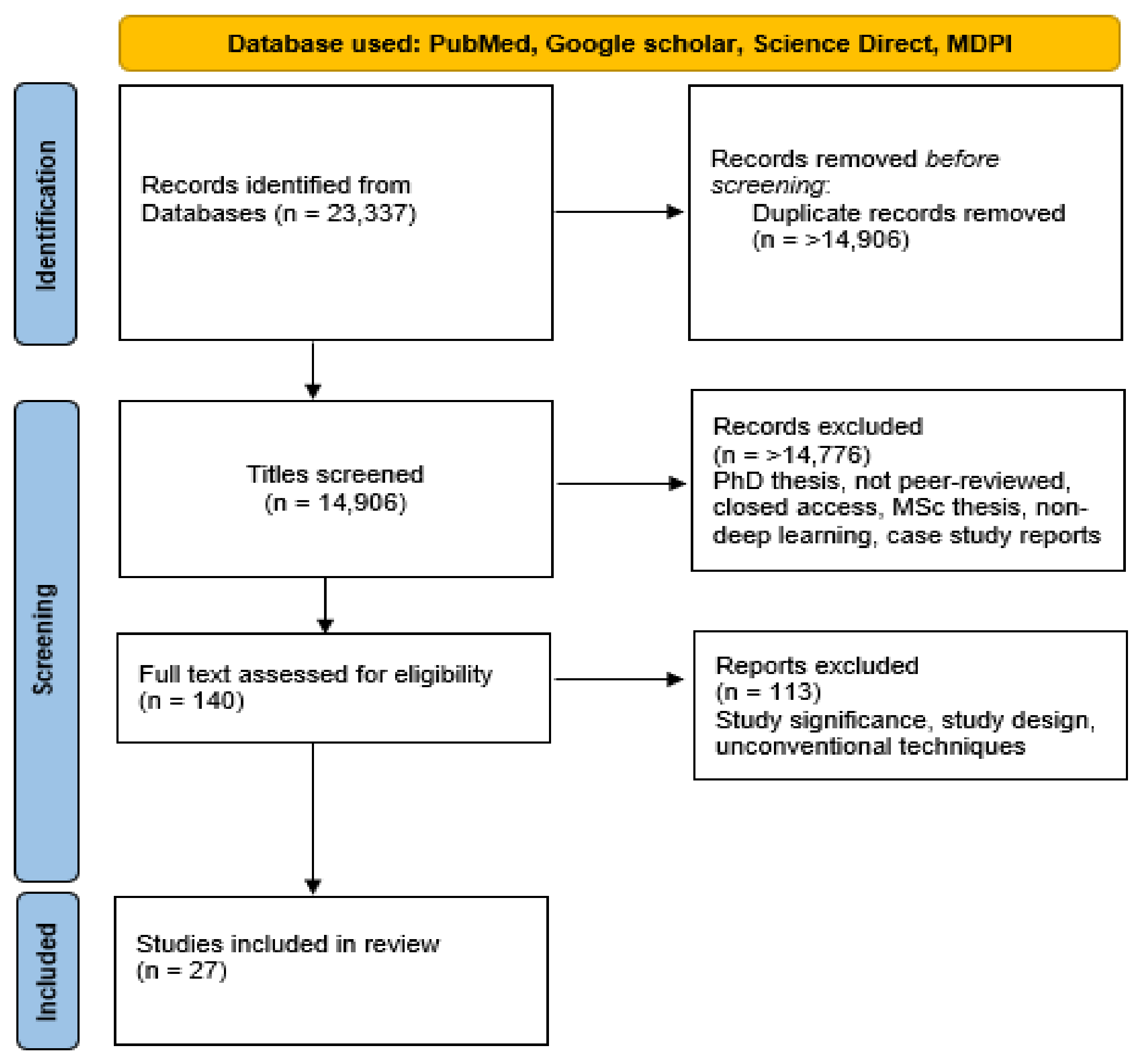
3. Materials and Methods
3.1. Datasets
3.2. Image Pre-Processing Techniques
3.3. Performance Metrics
4. Literature Survey
4.1. Classical Machine Learning Based Techniques
4.1.1. Brain Tumor Classification and Segmentation Using Hybrid Texture-Based Features
4.1.2. Brain Tumor Classification Using GoogleNet Features and ML
4.1.3. Brain Tumor Classification Using Ensemble of Deep Features and ML Classifiers
4.1.4. Brain Tumor Detection Using Metaheuristics and Machine Learning
4.1.5. Categorization of Brain Tumor Using CNN-Based Features and SVM
4.2. Deep Learning-Based Brain Tumor Classification Techniques
4.2.1. Brain Tumor Multi-Grade Classification
4.2.2. MR Brain Image Classification Using Differential Feature Maps
4.2.3. Brain Tumor Classification for Multi-Class Brain Tumor Image Using Block-Wise Fine-Tuning
4.2.4. Multi-Scale 3D CNN for MRI Brain Tumor Grade Classification
4.2.5. Brain Tumor Classification Using Pairwise Generative Adversarial Networks
4.3. Deep Learning-Based Brain Tumor Segmentation Techniques
4.3.1. Enhanced CNN and Bat Algorithm for Brain Tumor Classification
4.3.2. Encoder–Decoder Architecture for Brain Tumor Segmentation
4.3.3. Patching-Based Technique for Tumor Segmentation
4.3.4. Two-Path or Cascade Architecture for Brain Tumor Segmentation
4.3.5. Triple CNN Architecture for Brain Tumor Segmentation
4.3.6. Brain Tumor Classification and Segmentation Using a Combination of YOLOv2 and CNN
4.4. Vision Transformers for Brain Tumor Segmentation and Classification
4.4.1. Brain Tumor Segmentation using Bi Transformer U-Net
4.4.2. Multi-Modal Brain Tumor Segmentation Using Encoder–Transformer–Decoder Structure
4.4.3. Brain Tumor Segmentation Using Transformers Encoders and CNN Decoder
4.4.4. Brain Tumor Segmentation Using Swin Transformers
4.4.5. Convolution-Free 3D Brain Tumor Segmentation Using Vision Transformers
4.5. Capsule Neural Network-Based Brain Tumor Classification and Segmentation
4.5.1. Brain Tumor Classification Using Capsule Neural Network
4.5.2. Brain Tumor Classification Using Bayesian Theory and Capsule Neural Network
4.5.3. CapsNet-Based Brain Tumor Classification Using an Improved Activation Function
4.5.4. Brain Tumor Classification Using a Dilated Capsule Neural Network
4.5.5. CapsNet-Based Image Pre-Processing Framework for Improved Brain Tumor Classification
4.5.6. Brain Tumor Segmentation Using a Modified Version of Capsule Neural Network
5. Discussion
5.1. Machine Learning-Based Brain Tumor Classification and Segmentation Techniques
5.2. Deep Learning-Based Brain Tumor Segmentation and Classification Techniques
5.3. Vision Transformer-Based Tumor Segmentation and Classification
5.4. Capsule Neural Network-Based Brain Tumor Classification and Segmentation Techniques
5.5. Hybrid Brain Tumor Classification and Segmentation Techniques
5.6. Small-Scale and Imbalance Dataset
5.7. Multi-Class and Binary Classification
5.8. Network Architectures and Data Augmentation
5.9. Performance Overview of Brain Tumor Classification and Segmentation Techniques
6. Conclusions and Future Research Directions
- Most of the current research is devoted to brain tumor detection, segmentation, or grade estimation. Most studies did not develop frameworks that can perform these three tasks simultaneously. Moreover, most studies focused on binary-grade classification with less attention paid to multi-grade classification. Designing a framework that can handle brain tumor segmentation, tumor classification (benign versus malignant), and multi-grade estimation would be valuable in improving the decisions and accuracy of medical practitioners when diagnosing brain tumors.
- Generally, feature maps in CNNs are generated using transfer learning or random initialization. Few studies developed techniques that can be used to generate feature maps for CNNs. Isselmou et al. [95] combined user-defined hyperactive values and a differential operator to generate feature maps for CNN. Results show that the differential feature maps improved the accuracy of the CNN model. Future research can develop more techniques that can generate effective feature maps for improved CNN-based brain tumor segmentation and classification.
- Most of the existing DL brain tumor techniques are based on CNNs. However, these architectures require a huge quantity of data for training [9]. They are also incapable of correctly distinguishing between inputs of different rotations [10]. In addition, obtaining and labelling large-scale datasets is a demanding task [9]. Unfortunately, most publicly available brain cancer datasets are small and imbalanced. The accuracy and generalization performance of a CNN model will be affected if it is trained on small-scale or imbalanced datasets. CapsNet [11] is a recently developed network architecture that has been proposed to address the above-mentioned shortcomings of CNNs. CapsNet are particularly appealing because of their robustness to rotation and affine transformation. Additionally, as demonstrated in [2], CapsNets require significantly less training data than CNN, which is the case for medical imaging datasets such as brain MRI images [3]. Moreover, results reported in the literature [2] show that CapsNets have the potential to improve the accuracy of CNN-based brain tumor diagnosis using a very small number of network parameters [12]. Most studies did not explore the use of CapsNet for brain cancer diagnosis.
- While ViT has demonstrated outstanding performance in NLP, its potential has not been fully explored for medical imaging analysis, such as brain tumor segmentation [33]. Future research can focus on improving the effectiveness of ViT-based approaches for classifying and segmenting brain tumors, as this is an active research area. Additionally, future research could further investigate the use of Swin transformers, as they seem to perform better than standard ViTs.
- Most studies focused on 2D network architecture for tumor segmentation and classification. However, few studies explored 3D network architectures. 3D convolutional layers provide a detailed feature map that can explore the volumetric information in MRI scans [62]. The 3D feature map can also learn both local and global features with high classifying power [62]. Future researchers can explore 3D network architectures for improved brain tumor segmentation and classification.
- Most of the techniques developed in the literature do not tackle the problem of model uncertainly in CNN-based and CapsNet-based models. Developing a network that can handle model uncertainty is important because it serves as a medium for returning the uncertain predictions to experts for further verification. Most CNN networks use the softmax activation function. However, the output of softmax does not reflect the degree of uncertainty of a model’s prediction [108]. Afshar et al. [40] developed a CapsNet-based technique that can handle model uncertainty using Bayesian theory. However, experiments performed in the study show that CapsNet (without Bayesian theory) outperformed the Bayesian variant. This confirms that more work is still required. Future research could focus on developing robust techniques that can effectively handle model uncertainty without affecting the performance of the models.
- Most of the CNN-based and CapsNet-based architectures developed in the literature used the ReLU activation function. Few studies explored the use of other activation functions. Results reported by Adu et al. [19] show that PSTanh activation function outperformed the ReLU activation function. Future research could explore more activation functions for improved brain tumor classification and segmentation methods.
- The max-pooling operation used in network architectures normally affects the image quality and resolution of an image, and consequently the accuracy of a network. Very few studies developed techniques that maintain high-resolution feature maps. Further work is therefore required in this area. Future research can develop effective techniques that can preserve the quality and spatial recognizability of an image.
- Most of the currently available CNN-based approaches were developed for a specific form of cancer. A general-purpose DL-based framework that can diagnose different types of cancer will be very useful to medical practitioners
- Most datasets cited in the literature suffer from data imbalance problems. For instance, 98% of samples in one of the benchmark datasets–Brain Tumor Segmentation (BraTS) dataset–belong to a single class, whereas the remaining 2% belong to another class. Clearly, building a model on such an imbalanced dataset will result in overfitting. Furthermore, most studies did not explore the use of advanced data enrichment methods, such as GANs, for improving the performance of brain tumor diagnosis. In addition, most studies did not investigate the performance of different data augmentation techniques for brain cancer diagnosis. Moreover, most studies did not investigate the use of different state-of-the-art pre-trained networks for brain cancer diagnosis. The problem of data imbalance and small-scale dataset in brain tumor diagnosis may be addressed by developing techniques that combine advanced data augmentation techniques and state-of-the-art pre-trained network architectures.
- AlBadawy et al. [73] reported that there was a significant decrease in the performance of CNN models when they were trained for patients from the same institution compared to when they are trained for patients from different institutions. The reason behind the reduced performance requires systematic investigations.
- The fusion of multi-modal data can improve the performance of brain tumor diagnosis models. Utilizing complementary information from multiple imaging modalities has sparked a rise in recent research interest in cross-modality MR image synthesis. Recent studies [119,120] have developed multi-modal brain tumor segmentation systems that can learn high-level features from multi-modal data. Future research can design enhanced multi-modal diagnostic frameworks for brain tumors.
- Correct classification of multi-modal images of brain tumors is a vital step towards accurate diagnosis and successful treatment of brain tumors. However, resolving incomplete multi-modal issues is a challenging task in brain tumor diagnosis. Some techniques [121,122] have been proposed to address this difficulty, but more research is still required.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G. The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- The Brain Tumor Charity. Brain Tumor Basics. Available online: https://www.thebraintumourcharity.org/ (accessed on 1 June 2022).
- Tandel, G.S.; Biswas, M.; Kakde, O.G.; Tiwari, A.; Suri, H.S.; Turk, M.; Laird, J.R.; Asare, C.K.; Ankrah, A.A.; Khanna, N.N. A review on a deep learning perspective in brain cancer classification. Cancers 2019, 11, 111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamimi, A.F.; Juweid, M. Epidemiology and Outcome of Glioblastoma; Exon Publications: Brisbane, Australia, 2017; pp. 143–153. [Google Scholar] [CrossRef]
- Villanueva-Meyer, J.E.; Mabray, M.C.; Cha, S. Current clinical brain tumor imaging. Neurosurgery 2017, 81, 397–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overcast, W.B.; Davis, K.M.; Ho, C.Y.; Hutchins, G.D.; Green, M.A.; Graner, B.D.; Veronesi, M.C. Advanced imaging techniques for neuro-oncologic tumor diagnosis, with an emphasis on PET-MRI imaging of malignant brain tumors. Curr. Oncol. Rep. 2021, 23, 34. [Google Scholar] [CrossRef]
- Zaccagna, F.; Grist, J.T.; Quartuccio, N.; Riemer, F.; Fraioli, F.; Caracò, C.; Halsey, R.; Aldalilah, Y.; Cunningham, C.H.; Massoud, T.F. Imaging and treatment of brain tumors through molecular targeting: Recent clinical advances. Eur. J. Radiol. 2021, 142, 109842. [Google Scholar] [CrossRef]
- Zhang, Z.; Sejdić, E. Radiological images and machine learning: Trends, perspectives, and prospects. Comput. Biol. Med. 2019, 108, 354–370. [Google Scholar] [CrossRef] [Green Version]
- Biratu, E.S.; Schwenker, F.; Ayano, Y.M.; Debelee, T.G. A survey of brain tumor segmentation and classification algorithms. J. Imaging 2021, 7, 179. [Google Scholar] [CrossRef]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.M.; Larochelle, H. Brain tumor segmentation with deep neural networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Işın, A.; Direkoğlu, C.; Şah, M. Review of MRI-based brain tumor image segmentation using deep learning methods. Procedia Comput. Sci. 2016, 102, 317–324. [Google Scholar] [CrossRef] [Green Version]
- Chen, A.; Zhu, L.; Zang, H.; Ding, Z.; Zhan, S. Computer-aided diagnosis and decision-making system for medical data analysis: A case study on prostate MR images. J. Manag. Sci. Eng. 2019, 4, 266–278. [Google Scholar] [CrossRef]
- Deepak, S.; Ameer, P.M. Automated categorization of brain tumor from mri using cnn features and svm. J. Ambient Intell. Humaniz. Comput. 2021, 12, 8357–8369. [Google Scholar] [CrossRef]
- Sekhar, A.; Biswas, S.; Hazra, R.; Sunaniya, A.K.; Mukherjee, A.; Yang, L. Brain tumor classification using fine-tuned GoogLeNet features and machine learning algorithms: IoMT enabled CAD system. IEEE J. Biomed. Health Inform. 2021, 26, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Jena, B.; Nayak, G.K.; Saxena, S. An empirical study of different machine learning techniques for brain tumor classification and subsequent segmentation using hybrid texture feature. Mach. Vis. Appl. 2022, 33, 6. [Google Scholar] [CrossRef]
- Sajjad, M.; Khan, S.; Muhammad, K.; Wu, W.; Ullah, A.; Baik, S.W. Multi-grade brain tumor classification using deep CNN with extensive data augmentation. J. Comput. Sci. 2019, 30, 174–182. [Google Scholar] [CrossRef]
- Thaha, M.M.; Kumar, K.P.M.; Murugan, B.S.; Dhanasekeran, S.; Vijayakarthick, P.; Selvi, A.S. Brain tumor segmentation using convolutional neural networks in MRI images. J. Med. Syst. 2019, 43, 1240–1251. [Google Scholar] [CrossRef]
- Afshar, P.; Mohammadi, A.; Plataniotis, K.N. Brain tumor type classification via capsule networks. In Proceedings of the 2018 25th IEEE International Conference on Image Processing (ICIP), Athens, Greece, 7–10 October 2018; pp. 3129–3133. [Google Scholar]
- Adu, K.; Yu, Y.; Cai, J.; Asare, I.; Quahin, J. The influence of the activation function in a capsule network for brain tumor type classification. Int. J. Imaging Syst. Technol. 2021, 32, 123–143. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Muhammad, K.; Khan, S.; Del Ser, J.; De Albuquerque, V.H.C. Deep learning for multigrade brain tumor classification in smart healthcare systems: A prospective survey. IEEE Trans. Neural Netw. Learn. Syst. 2020, 32, 507–522. [Google Scholar] [CrossRef]
- Shamshad, F.; Khan, S.; Zamir, S.W.; Khan, M.H.; Hayat, M.; Khan, F.S.; Fu, H. Transformers in Medical Imaging: A Survey. arXiv 2022, arXiv:2201.09873. [Google Scholar]
- Magadza, T.; Viriri, S. Deep learning for brain tumor segmentation: A survey of state-of-the-art. J. Imaging 2021, 7, 19. [Google Scholar] [CrossRef]
- Zaccagna, F.; Riemer, F.; Priest, A.N.; McLean, M.A.; Allinson, K.; Grist, J.T.; Dragos, C.; Matys, T.; Gillard, J.H.; Watts, C. Non-invasive assessment of glioma microstructure using VERDICT MRI: Correlation with histology. Eur. Radiol. 2019, 29, 5559–5566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, H.-F.; Chen, Z.-Y.; Lou, X.; Wang, Y.-L.; Gui, Q.-P.; Wang, Y.; Shi, K.-N.; Zhou, Z.-Y.; Zheng, D.-D.; Wang, D.J.J. Astrocytic tumour grading: A comparative study of three-dimensional pseudocontinuous arterial spin labelling, dynamic susceptibility contrast-enhanced perfusion-weighted imaging, and diffusion-weighted imaging. Eur. Radiol. 2015, 25, 3423–3430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dijkstra, M.; van Nieuwenhuizen, D.; Stalpers, L.J.A.; Wumkes, M.; Waagemans, M.; Vandertop, W.P.; Heimans, J.J.; Leenstra, S.; Dirven, C.M.; Reijneveld, J.C. Late neurocognitive sequelae in patients with WHO grade I meningioma. J. Neurol. Neurosurg. Psychiatry 2009, 80, 910–915. [Google Scholar] [CrossRef]
- Waagemans, M.L.; van Nieuwenhuizen, D.; Dijkstra, M.; Wumkes, M.; Dirven, C.M.F.; Leenstra, S.; Reijneveld, J.C.; Klein, M.; Stalpers, L.J.A. Long-term impact of cognitive deficits and epilepsy on quality of life in patients with low-grade meningiomas. Neurosurgery 2011, 69, 72–79. [Google Scholar] [CrossRef]
- van Alkemade, H.; de Leau, M.; Dieleman, E.M.T.; Kardaun, J.W.P.F.; van Os, R.; Vandertop, W.P.; van Furth, W.R.; Stalpers, L.J.A. Impaired survival and long-term neurological problems in benign meningioma. Neuro. Oncol. 2012, 14, 658–666. [Google Scholar] [CrossRef]
- Grist, J.T.; Miller, J.J.; Zaccagna, F.; McLean, M.A.; Riemer, F.; Matys, T.; Tyler, D.J.; Laustsen, C.; Coles, A.J.; Gallagher, F.A. Hyperpolarized 13C MRI: A novel approach for probing cerebral metabolism in health and neurological disease. J. Cereb. Blood Flow Metab. 2020, 40, 1137–1147. [Google Scholar] [CrossRef]
- Fan, J.; Fang, L.; Wu, J.; Guo, Y.; Dai, Q. From brain science to artificial intelligence. Engineering 2020, 6, 248–252. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016; ISBN 0262337371. [Google Scholar]
- Hatamizadeh, A.; Nath, V.; Tang, Y.; Yang, D.; Roth, H.; Xu, D. Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images. arXiv 2022, arXiv:2201.01266. [Google Scholar]
- Dai, Y.; Gao, Y.; Liu, F. Transmed: Transformers advance multi-modal medical image classification. Diagnostics 2021, 11, 1384. [Google Scholar] [CrossRef]
- Raghu, M.; Unterthiner, T.; Kornblith, S.; Zhang, C.; Dosovitskiy, A. Do vision transformers see like convolutional neural networks? Adv. Neural Inf. Process. Syst. 2021, 34, 12116–12128. [Google Scholar]
- Wang, W.; Chen, C.; Ding, M.; Yu, H.; Zha, S.; Li, J. Transbts: Multimodal brain tumor segmentation using transformer. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Virtual Event, 27 September–1 October 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 109–119. [Google Scholar]
- Rodriguez, R.; Dokladalova, E.; Dokládal, P. Rotation invariant CNN using scattering transform for image classification. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; pp. 654–658. [Google Scholar]
- Aziz, M.J.; Zade, A.A.T.; Farnia, P.; Alimohamadi, M.; Makkiabadi, B.; Ahmadian, A.; Alirezaie, J. Accurate Automatic Glioma Segmentation in Brain MRI images Based on CapsNet. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Virtual Conference, 31 October–4 November 2021; pp. 3882–3885. [Google Scholar]
- Sabour, S.; Frosst, N.; Hinton, G.E. Dynamic routing between capsules. arXiv 2017, arXiv:1710.09829. [Google Scholar]
- Patrick, M.K.; Adekoya, A.F.; Mighty, A.A.; Edward, B.Y. Capsule networks–a survey. J. King Saud Univ. Inf. Sci. 2022, 34, 1295–1310. [Google Scholar]
- Afshar, P.; Mohammadi, A.; Plataniotis, K.N. BayesCap: A Bayesian Approach to Brain Tumor Classification Using Capsule Networks. IEEE Signal Process. Lett. 2020, 27, 2024–2028. [Google Scholar] [CrossRef]
- Zeineldin, R.A.; Karar, M.E.; Coburger, J.; Wirtz, C.R.; Burgert, O. DeepSeg: Deep neural network framework for automatic brain tumor segmentation using magnetic resonance FLAIR images. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 909–920. [Google Scholar] [CrossRef]
- LaLonde, R.; Bagci, U. Capsules for object segmentation. arXiv 2018, arXiv:1804.04241. [Google Scholar]
- Nguyen, H.P.; Ribeiro, B. Advanced capsule networks via context awareness. In Proceedings of the International Conference on Artificial Neural Networks, Munich, Germany, 17–19 September 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 166–177. [Google Scholar]
- BraTS. Multimodal Brain Tumor Segmentation. 2012. Available online: https://www2.imm.dtu.dk/projects/BRATS2012/ (accessed on 8 November 2021).
- QTIM. The Quantitative Translational Imaging in Medicine Lab at the Martinos Center. 2013. Available online: https:%0A//qtim-lab.github.io/ (accessed on 8 December 2021).
- BraTS. MICCAI-BRATS 2014. 2014. Available online: https://sites.google.com/site/miccaibrats2014/ (accessed on 8 December 2021).
- BraTS. BraTS 2015. 2015. Available online: https://sites.google.com/site/braintumorsegmentation/home/brats2015 (accessed on 8 December 2021).
- BraTS. BraTS 2016. 2016. Available online: https://sites.google.com/site/braintumorsegmentation/home/brats_2016 (accessed on 1 December 2021).
- MICCAI 2017. 20th International Conference on Medical Image Computing and Computer Assisted Intervention 2017. Available online: https://www.miccai2017.org/ (accessed on 8 December 2021).
- BraTS. Multimodal Brain Tumor Segmentation Challenge 2018. 2018. Available online: https://www.med.upenn.edu/sbia/brats2018.html (accessed on 8 December 2021).
- BraTS. Multimodal Brain Tumor Segmentation Challenge 2019: Data. 2019. Available online: https://www.med.upenn.edu/cbica/brats2019/data.html (accessed on 8 December 2021).
- BraTS. Multimodal Brain Tumor Segmentation Challenge 2020: Data. 2021. Available online: https://www.med.upenn.edu/cbica/brats2020/data.html (accessed on 8 December 2021).
- BraTS. RSNA-ASNR-MICCAI Brain Tumor Segmentation (BraTS) Challenge 2021. 2021. Available online: http://braintumorsegmentation.org/ (accessed on 8 December 2021).
- TCIA. Cancer Imaging Archive. Available online: https://www.cancerimagingarchive.net/ (accessed on 8 December 2021).
- Radiopaedia. Brain Tumor Dataset. Available online: https://radiopaedia.org/ (accessed on 30 May 2022).
- Cheng, J. Brain Tumor Dataset. Figshare. Dataset 2017. Available online: https://scholar.google.com/scholar_lookup?title=Braintumordataset&author=J.Cheng&publication_year=2017 (accessed on 30 May 2022).
- Chakrabarty, N. Brain MRI Images for Brain Tumor Detection Dataset, 2019. Available online: https://www.kaggle.com/navoneel/brain-mri-images-for-brain-tumor-detection (accessed on 28 January 2022).
- Hamada, A. Br35H Brain Tumor Detection 2020 Dataset. 2020. Available online: https://www.kaggle.com/ahmedhamada0/brain-tumor-detection (accessed on 28 January 2022).
- Simpson, A.L.; Antonelli, M.; Bakas, S.; Bilello, M.; Farahani, K.; Van Ginneken, B.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv 2019, arXiv:1902.09063. [Google Scholar]
- Pereira, S.; Pinto, A.; Alves, V.; Silva, C.A. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans. Med. Imaging 2016, 35, 1240–1251. [Google Scholar] [CrossRef]
- Brooks, J.C.W.; Faull, O.K.; Pattinson, K.T.S.; Jenkinson, M. Physiological noise in brainstem FMRI. Front. Hum. Neurosci. 2013, 7, 623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mzoughi, H.; Njeh, I.; Wali, A.; Slima, M.B.; BenHamida, A.; Mhiri, C.; Mahfoudhe, K.B. Deep multi-scale 3D convolutional neural network (CNN) for MRI gliomas brain tumor classification. J. Digit. Imaging 2020, 33, 903–915. [Google Scholar] [CrossRef]
- Mohan, J.; Krishnaveni, V.; Guo, Y. A survey on the magnetic resonance image denoising methods. Biomed. Signal Process. Control 2014, 9, 56–69. [Google Scholar] [CrossRef]
- Chen, H.; Dou, Q.; Yu, L.; Qin, J.; Heng, P.-A. VoxResNet: Deep voxelwise residual networks for brain segmentation from 3D MR images. Neuroimage 2018, 170, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Liu, B.; Peng, S.; Sun, J.; Qiao, X. Computer-aided grading of gliomas combining automatic segmentation and radiomics. Int. J. Biomed. Imaging 2018, 2018, 2512037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mzoughi, H.; Njeh, I.; Slima, M.B.; Hamida, A.B. Histogram equalization-based techniques for contrast enhancement of MRI brain Glioma tumor images: Comparative study. In Proceedings of the 2018 4th International Conference on Advanced Technologies for Signal and Image Processing (ATSIP), Sousse, Tunisia, 21–24 March 2018; pp. 1–6. [Google Scholar]
- Pizer, S.M.; Johnston, E.; Ericksen, J.P.; Yankaskas, B.C.; Muller, K.E.; Medical Image Display Research Group. Contrast-limited adaptive histogram equalization: Speed and effectiveness. In Proceedings of the First Conference on Visualization in Biomedical Computing, Atlanta, GA, USA, 22–25 May 1990; Volume 337. [Google Scholar]
- Ramesh, S.; Sasikala, S.; Paramanandham, N. Segmentation and classification of brain tumors using modified median noise filter and deep learning approaches. Multimed. Tools Appl. 2021, 80, 11789–11813. [Google Scholar] [CrossRef]
- Zeng, Y.; Zhang, B.; Zhao, W.; Xiao, S.; Zhang, G.; Ren, H.; Zhao, W.; Peng, Y.; Xiao, Y.; Lu, Y. Magnetic resonance image denoising algorithm based on cartoon, texture, and residual parts. Comput. Math. Methods Med. 2020, 2020, 1405647. [Google Scholar] [CrossRef] [PubMed]
- Heo, Y.-C.; Kim, K.; Lee, Y. Image Denoising Using Non-Local Means (NLM) Approach in Magnetic Resonance (MR) Imaging: A Systematic Review. Appl. Sci. 2020, 10, 7028. [Google Scholar] [CrossRef]
- Kidoh, M.; Shinoda, K.; Kitajima, M.; Isogawa, K.; Nambu, M.; Uetani, H.; Morita, K.; Nakaura, T.; Tateishi, M.; Yamashita, Y. Deep learning based noise reduction for brain MR imaging: Tests on phantoms and healthy volunteers. Magn. Reson. Med. Sci. 2020, 19, 195. [Google Scholar] [CrossRef] [Green Version]
- Moreno López, M.; Frederick, J.M.; Ventura, J. Evaluation of MRI Denoising Methods Using Unsupervised Learning. Front. Artif. Intell. 2021, 4, 75. [Google Scholar] [CrossRef] [PubMed]
- AlBadawy, E.A.; Saha, A.; Mazurowski, M.A. Deep learning for segmentation of brain tumors: Impact of cross-institutional training and testing. Med. Phys. 2018, 45, 1150–1158. [Google Scholar] [CrossRef]
- Yogananda, C.G.B.; Shah, B.R.; Vejdani-Jahromi, M.; Nalawade, S.S.; Murugesan, G.K.; Yu, F.F.; Pinho, M.C.; Wagner, B.C.; Emblem, K.E.; Bjørnerud, A. A Fully automated deep learning network for brain tumor segmentation. Tomography 2020, 6, 186–193. [Google Scholar] [CrossRef]
- Ge, C.; Gu, I.Y.-H.; Jakola, A.S.; Yang, J. Enlarged training dataset by pairwise gans for molecular-based brain tumor classification. IEEE Access 2020, 8, 22560–22570. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I.H. Textural features for image classification. IEEE Trans. Syst. Man. Cybern. 1973, 6, 610–621. [Google Scholar] [CrossRef] [Green Version]
- Aouat, S.; Ait-hammi, I.; Hamouchene, I. A new approach for texture segmentation based on the Gray Level Co-occurrence Matrix. Multimed. Tools Appl. 2021, 80, 24027–24052. [Google Scholar] [CrossRef]
- Chu, A.; Sehgal, C.M.; Greenleaf, J.F. Use of gray value distribution of run lengths for texture analysis. Pattern Recognit. Lett. 1990, 11, 415–419. [Google Scholar] [CrossRef]
- Tian, S.; Bhattacharya, U.; Lu, S.; Su, B.; Wang, Q.; Wei, X.; Lu, Y.; Tan, C.L. Multilingual scene character recognition with co-occurrence of histogram of oriented gradients. Pattern Recognit. 2016, 51, 125–134. [Google Scholar] [CrossRef]
- Prakasa, E. Texture feature extraction by using local binary pattern. INKOM J. 2016, 9, 45–48. [Google Scholar] [CrossRef] [Green Version]
- Al-Janobi, A. Performance evaluation of cross-diagonal texture matrix method of texture analysis. Pattern Recognit. 2001, 34, 171–180. [Google Scholar] [CrossRef]
- He, D.-C.; Wang, L. Simplified texture spectrum for texture analysis. J. Commun. Comput. 2010, 7, 44–53. [Google Scholar]
- Khan, R.U.; Zhang, X.; Kumar, R. Analysis of ResNet and GoogleNet models for malware detection. J. Comput. Virol. Hacking Tech. 2019, 15, 29–37. [Google Scholar] [CrossRef]
- Kang, J.; Ullah, Z.; Gwak, J. MRI-Based Brain Tumor Classification Using Ensemble of Deep Features and Machine Learning Classifiers. Sensors 2021, 21, 2222. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Krizhevsky, A. One weird trick for parallelizing convolutional neural networks. arXiv 2014, arXiv:1404.5997. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated residual transformations for deep neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1492–1500. [Google Scholar]
- Ma, N.; Zhang, X.; Zheng, H.-T.; Sun, J. Shufflenet v2: Practical guidelines for efficient cnn architecture design. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 116–131. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4510–4520. [Google Scholar]
- Tan, M.; Chen, B.; Pang, R.; Vasudevan, V.; Sandler, M.; Howard, A.; Le, Q. V Mnasnet: Platform-aware neural architecture search for mobile. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 2820–2828. [Google Scholar]
- Kaur, D.; Singh, S.; Mansoor, W.; Kumar, Y.; Verma, S.; Dash, S.; Koul, A. Computational Intelligence and Metaheuristic Techniques for Brain Tumor Detection through IoMT-Enabled MRI Devices. Wirel. Commun. Mob. Comput. 2022, 2022, 1519198. [Google Scholar] [CrossRef]
- Abd El Kader, I.; Xu, G.; Shuai, Z.; Saminu, S.; Javaid, I.; Salim Ahmad, I. Differential deep convolutional neural network model for brain tumor classification. Brain Sci. 2021, 11, 352. [Google Scholar] [CrossRef] [PubMed]
- Lei, B.; Huang, S.; Li, R.; Bian, C.; Li, H.; Chou, Y.-H.; Cheng, J.-Z. Segmentation of breast anatomy for automated whole breast ultrasound images with boundary regularized convolutional encoder–decoder network. Neurocomputing 2018, 321, 178–186. [Google Scholar] [CrossRef]
- Swati, Z.N.K.; Zhao, Q.; Kabir, M.; Ali, F.; Ali, Z.; Ahmed, S.; Lu, J. Brain tumor classification for MR images using transfer learning and fine-tuning. Comput. Med. Imaging Graph. 2019, 75, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Mzoughi, H.; Njeh, I.; Slima, M.B.; Hamida, A.B.; Mhiri, C.; Mahfoudh, K.B. Denoising and contrast-enhancement approach of magnetic resonance imaging glioblastoma brain tumors. J. Med. Imaging 2019, 6, 44002. [Google Scholar] [CrossRef]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.; Freymann, J.; Farahani, K.; Davatzikos, C. Segmentation labels and radiomic features for the pre-operative scans of the TCGA-GBM collection. The Cancer Imaging Archive. Nat. Sci. Data 2017, 4, 170117. [Google Scholar] [CrossRef] [Green Version]
- Tustison, N.J.; Avants, B.B.; Cook, P.A.; Zheng, Y.; Egan, A.; Yushkevich, P.A.; Gee, J.C. N4ITK: Improved N3 bias correction. IEEE Trans. Med. Imaging 2010, 29, 1310–1320. [Google Scholar] [CrossRef] [Green Version]
- Sharif, M.I.; Li, J.P.; Amin, J.; Sharif, A. An improved framework for brain tumor analysis using MRI based on YOLOv2 and convolutional neural network. Complex Intell. Syst. 2021, 7, 2023–2036. [Google Scholar] [CrossRef]
- Conn, A.R.; Gould, N.I.M.; Toint, P. A globally convergent augmented Lagrangian algorithm for optimization with general constraints and simple bounds. SIAM J. Numer. Anal. 1991, 28, 545–572. [Google Scholar] [CrossRef] [Green Version]
- Kapur, J.N.; Sahoo, P.K.; Wong, A.K.C. A new method for gray-level picture thresholding using the entropy of the histogram. Comput. Vis. Graph. Image Process. 1985, 29, 273–285. [Google Scholar] [CrossRef]
- Jia, Q.; Shu, H. BiTr-Unet: A CNN-Transformer Combined Network for MRI Brain Tumor Segmentation. arXiv 2021, arXiv:2109.12271. [Google Scholar]
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.R.; Xu, D. Unetr: Transformers for 3d medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 4–8 January 2022; pp. 574–584. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Peiris, H.; Hayat, M.; Chen, Z.; Egan, G.; Harandi, M. A Volumetric Transformer for Accurate 3D Tumor Segmentation. arXiv 2021, arXiv:2111.13300. [Google Scholar]
- Gal, Y.; Ghahramani, Z. Dropout as a bayesian approximation: Representing model uncertainty in deep learning. In Proceedings of the International Conference on Machine Learning, New York City, NY, USA, 19–24 June 2016; pp. 1050–1059. [Google Scholar]
- Adu, K.; Yu, Y.; Cai, J.; Tashi, N. Dilated Capsule Network for Brain Tumor Type Classification Via MRI Segmented Tumor Region. In Proceedings of the 2019 IEEE International Conference on Robotics and Biomimetics (ROBIO), Dali, China, 6–8 December 2019; pp. 942–947. [Google Scholar]
- Kurup, R.V.; Sowmya, V.; Soman, K.P. Effect of data pre-processing on brain tumor classification using capsulenet. In Proceedings of the International Conference on Intelligent Computing and Communication Technologies, Chongqing, China, 6–8 December 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 110–119. [Google Scholar]
- Verma, R.; Zacharaki, E.I.; Ou, Y.; Cai, H.; Chawla, S.; Lee, S.-K.; Melhem, E.R.; Wolf, R.; Davatzikos, C. Multiparametric tissue characterization of brain neoplasms and their recurrence using pattern classification of MR images. Acad. Radiol. 2008, 15, 966–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zacharaki, E.I.; Wang, S.; Chawla, S.; Yoo, D.S.; Wolf, R.; Melhem, E.R.; Davatzikos, C. MRI-based classification of brain tumor type and grade using SVM-RFE. In Proceedings of the 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Boston, MA, USA, 28 June–1 July 2009; pp. 1035–1038. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef] [Green Version]
- Sasikala, M.; Kumaravel, N. A wavelet-based optimal texture feature set for classification of brain tumours. J. Med. Eng. Technol. 2008, 32, 198–205. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Wang, X.; Yang, L.T.; Feng, J.; Chen, X.; Deen, M.J. A Tensor-Based Big Service Framework for Enhanced Living Environments. IEEE Cloud Comput. 2016, 3, 36–43. [Google Scholar] [CrossRef]
- Naser, M.A.; Deen, M.J. Brain tumor segmentation and grading of lower-grade glioma using deep learning in MRI images. Comput. Biol. Med. 2020, 121, 103758. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Zhou, T.; Fu, H.; Zhang, Y.; Zhang, C.; Lu, X.; Shen, J.; Shao, L. M2Net: Multi-modal Multi-channel Network for Overall Survival Time Prediction of Brain Tumor Patients. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 221–231. [Google Scholar]
- Fang, F.; Yao, Y.; Zhou, T.; Xie, G.; Lu, J. Self-supervised Multi-modal Hybrid Fusion Network for Brain Tumor Segmentation. IEEE J. Biomed. Health Inform. 2021, 1. [Google Scholar] [CrossRef]
- Yurt, M.; Dar, S.U.H.; Erdem, A.; Erdem, E.; Oguz, K.K.; Çukur, T. mustGAN: Multi-stream generative adversarial networks for MR image synthesis. Med. Image Anal. 2021, 70, 101944. [Google Scholar] [CrossRef]
- Yu, B.; Zhou, L.; Wang, L.; Shi, Y.; Fripp, J.; Bourgeat, P. Ea-GANs: Edge-aware generative adversarial networks for cross-modality MR image synthesis. IEEE Trans. Med. Imaging 2019, 38, 1750–1762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rundo, L.; Militello, C.; Vitabile, S.; Russo, G.; Sala, E.; Gilardi, M.C. A survey on nature-inspired medical image analysis: A step further in biomedical data integration. Fundam. Inform. 2020, 171, 345–365. [Google Scholar] [CrossRef]
- Ressler, K.J.; Williams, L.M. Big data in psychiatry: Multiomics, neuroimaging, computational modeling, and digital phenotyping. Neuropsychopharmacology 2021, 46, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Biswas, N.; Chakrabarti, S. Artificial intelligence (AI)-based systems biology approaches in multi-omics data analysis of cancer. Front. Oncol. 2020, 10, 588221. [Google Scholar] [CrossRef]
- Simidjievski, N.; Bodnar, C.; Tariq, I.; Scherer, P.; Andres Terre, H.; Shams, Z.; Jamnik, M.; Liò, P. Variational autoencoders for cancer data integration: Design principles and computational practice. Front. Genet. 2019, 10, 1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
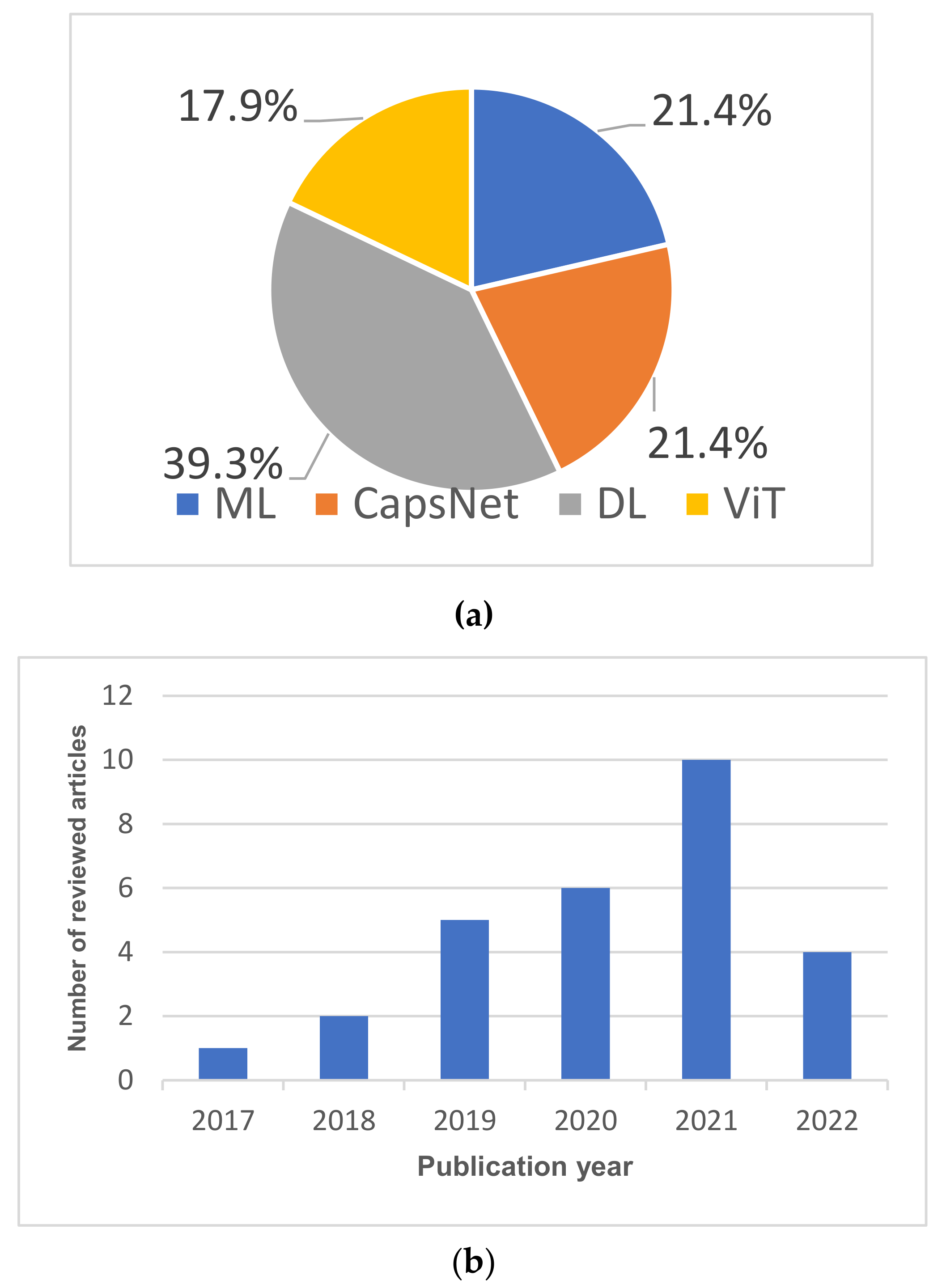
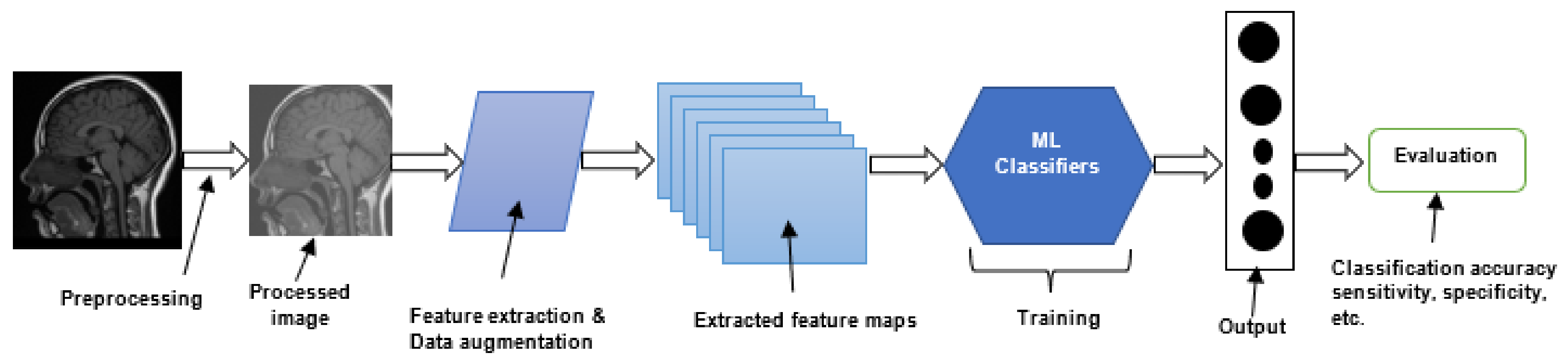
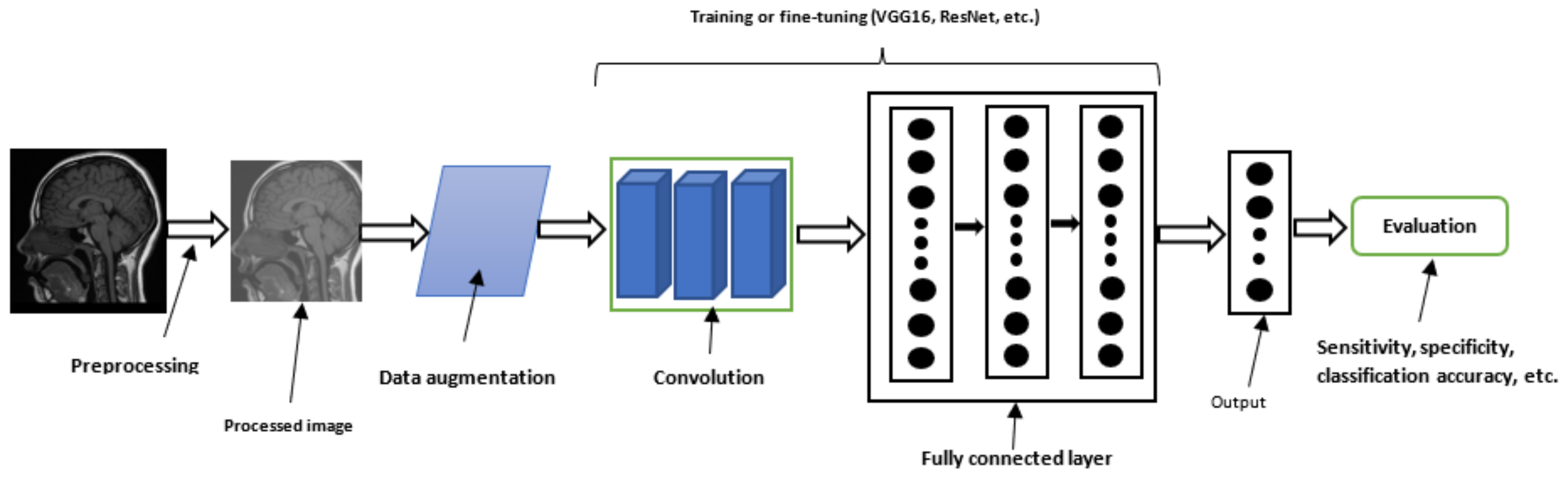
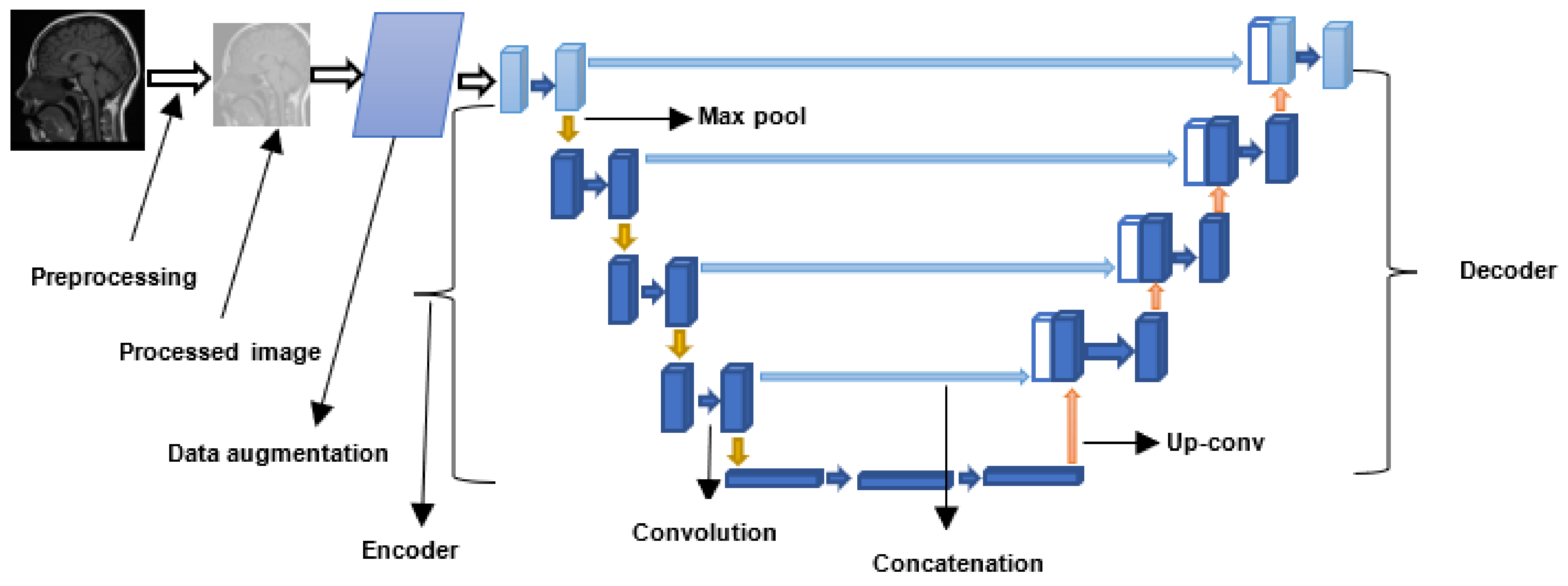
| Dataset Name | Dataset Details | Reference |
|---|---|---|
| BraTS 2012 | 30 MRI, 50 simulated images (25 Low Grade Glioma (LGG) and 25 High Grade Glioma (HGG)) | [44] |
| BraTS 2013 | 30 MRI (20 HGG and 10 LGG), 50 simulated images (25 LGG and 25 HGG) | [45] |
| BraTS 2014 | 190 HGG and 26 LGG MRI | [46] |
| BraTS 2015 | 220 HGG and 54 LGG MRI | [47] |
| BraTS 2016 | 220 HGG and 54 LGG; Testing: 191 images with unknown grades | [48] |
| BraTS 2017 | 285 MRI scan. Contains full masks for brain tumors. | [49] |
| BraTS 2018 | Training dataset: 210 HGG and 75 LGG MRI scans. The validation dataset includes 66 different MRI scans | [50] |
| BraTS 2019 | 259 HGG and 76 LGG MRI scans from 19 institution. | [51] |
| BraTS 2020 | 2640 MRI scans from 369 patients with ground truth in four sequences (T1-weighted (T1w), T2-weighted (T2w), post-gadolinium-based contrast agent (GBCA) T1w (T1w post GBCA), Fluid Attenuated Inversion Recovery (FLAIR)) | [52] |
| BraTS 2021 | 8000 MRI scans from 2000 cases | [53] |
| TCIA | 3929 MRI scans from 110 patients. 1373 tumor images and 2556 normal images. | [54] |
| Radiopedia | 121 MRI | [55] |
| Contrast Enhanced Magnetic Resonance Images (CE-MRI) dataset | 3064 MRI T1w post GBCA images from 233 patients | [56] |
| Brain MRI Images | 253 MRI images, 155 tumor images, 98 non-tumor images | [57] |
| Br35H dataset | 3000 MRI images, 1500 tumor images, and 1500 non-tumor images | [58] |
| MSD dataset | 484 multi-modal multi-site MRI data (FLAIR, T1W, T1w post GBCA, T2W) | [59] |
| Ref. | Year | Method | Classes Considered | Main Highlight | Dataset | Performance |
|---|---|---|---|---|---|---|
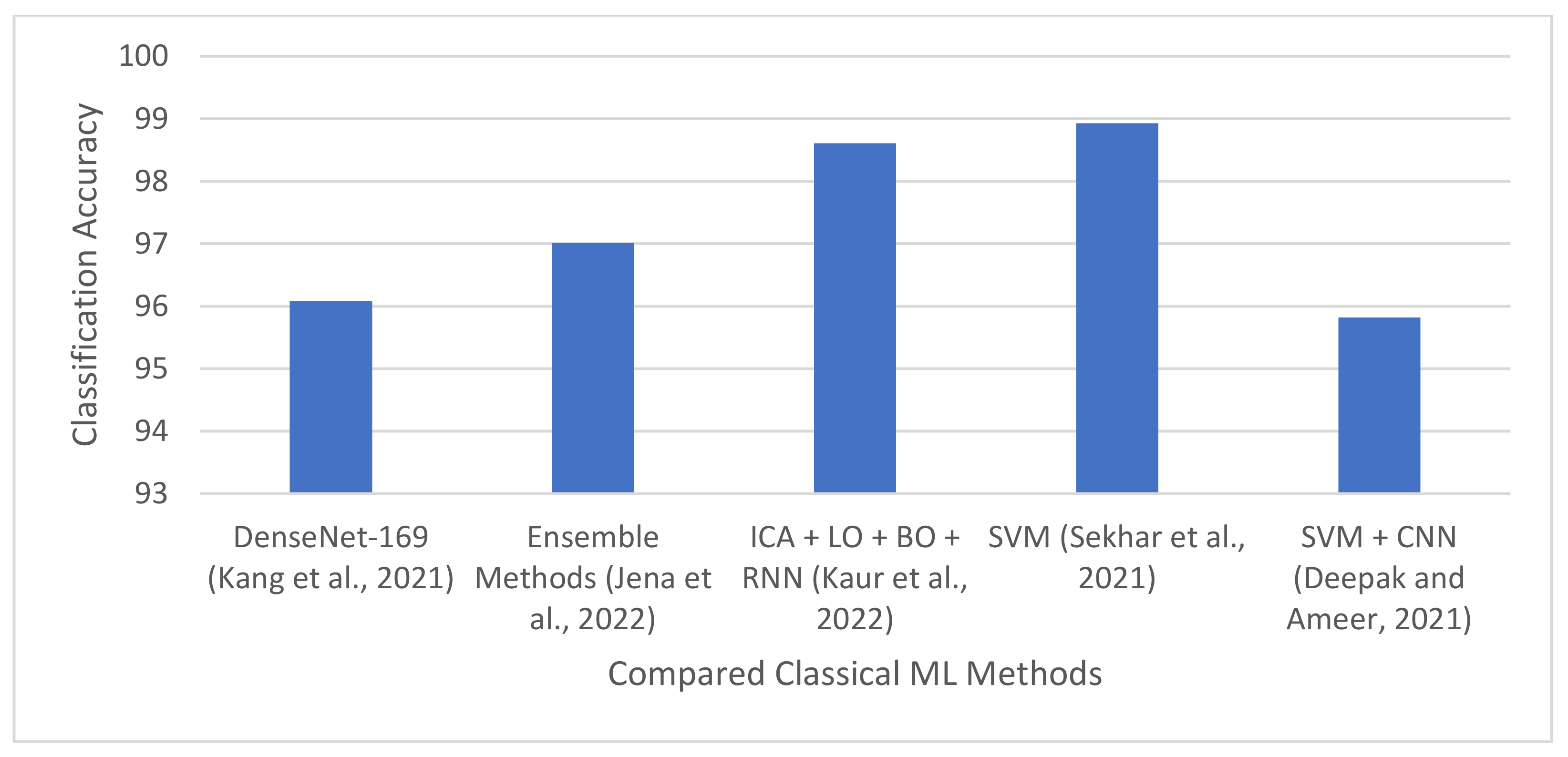
| [84] | 2021 | 13 pre-trained CNN models and nine ML classifiers | Normal and tumor images | Concatenated three deep features from pre-trained CNN models and trained nine ML classifiers. | Three brain MRI datasets [56,57,58] | DenseNet-169, Inception-v3, and ResNeXt-50 produced the best deep features, achieving an accuracy of 96.08%, 92.16% and 94.12%, respectively. |
| [14] | 2021 | SVM, k-NN, and modified GoogleNet pre-trained architecture | Glioma, meningioma, and pituitary. | Extracted features from modified GoogleNet pre-trained architecture and used it to train SVM and k-NN | CE-MRI dataset | SVM and k-NN produced a specificity of 98.93% and 98.63%, respectively. |
| [15] | 2022 | SVM, k-NN, binary decision tree, RF, ensemble methods. | FLAIR, T1w, T1w post GBCA, and T2w | Developed multiple ML models on six texture-based features. Applied a hybrid of k-NN and C-means clustering for tumor segmentation. | BraTS 2017 and BraTS 2019 | Classification accuracy of 96.98% and 97.01% for BraTS 2017 and 2019, respectively. DSC and accuracy of 90.16% and 98.4%, respectively. |
| [94] | 2022 | NB, RNN, bat and lion optimization, PCA, ICA, and cuckoo search. | Tumor and normal images | Designed hybrid techniques model using the combination of metaheuristics and ML algorithms. | TCIA | Classification accuracy of 98.61%. |
| [13] | 2021 | SVM and CNN | Glioma, meningioma, and pituitary tumor | Proposed a hybrid technique using CNN-based features and SVM. | FigShare dataset | 95.82% accuracy |
| Ref. | Year | Classes Considered | Method | Main Highlight | Dataset | Performance |
|---|---|---|---|---|---|---|
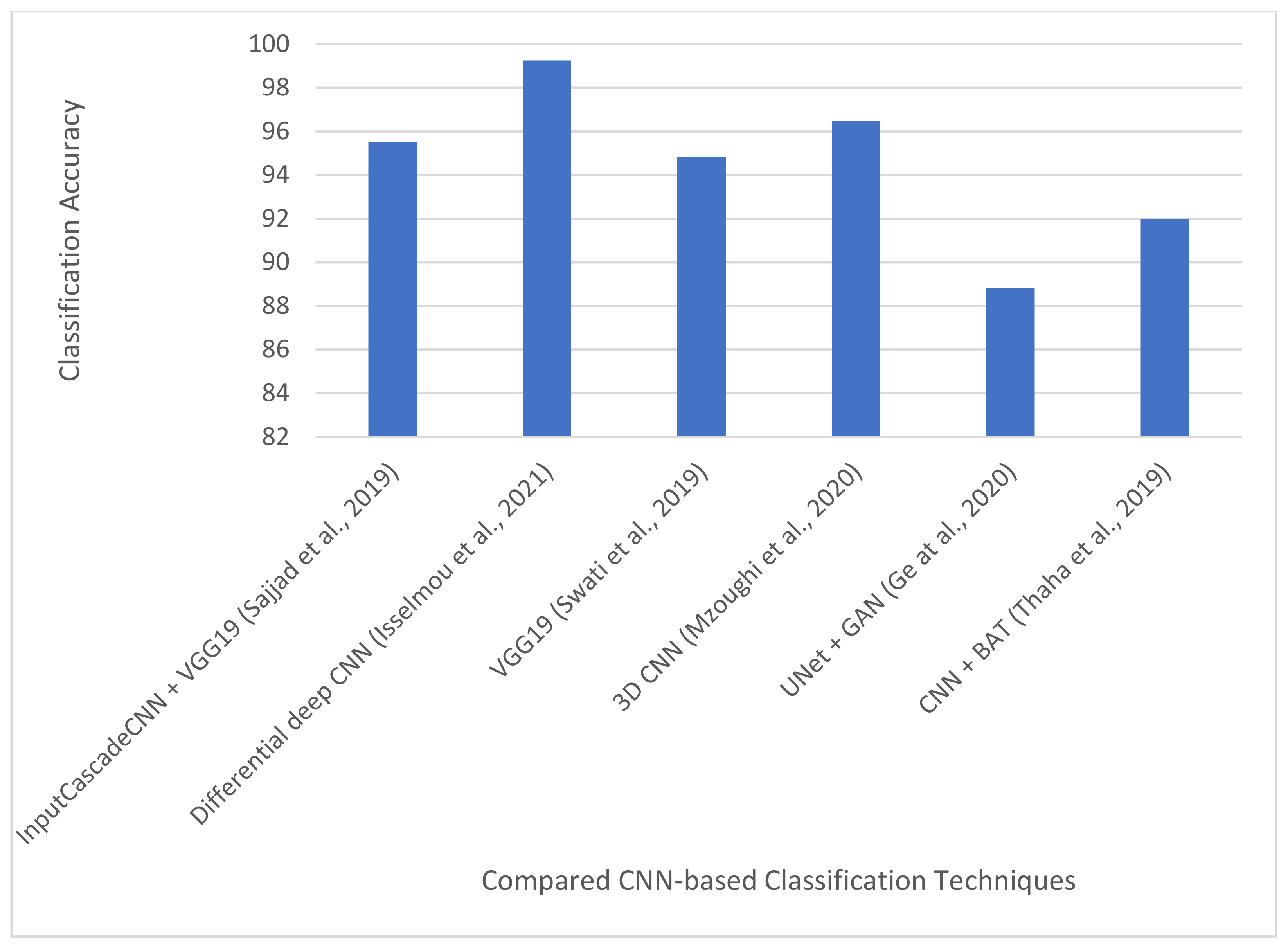
| [16] | 2019 | Grades I–IV | InputCascadeCNN + data augmentation + VGG-19 | Adopted four data augmentation techniques. Also used InputCascadeNN architecture for data augmentation and VGG-19 for fine-tuning. | Radiopedia and Brain tumor dataset | Classification accuracy of 95.5%, 92.66%, 87.77%, and 86.71%. for Grades I–IV, respectively on radiopedia dataset. Sensitivity and specificity 88.41% and 96.12%, respectively, on the brain tumor dataset. |
| [95] | 2021 | T1w, T2w and FLAIR images | Differential deep CNN + Data augmentation | Applied user-defined hyperactive values and a differential operator to generate feature maps for CNN. Proposed several data augmentation techniques. | TUCMD (17,600 MR brain images) | Classification accuracy, sensitivity, and specificity of 99.25%, 95.89%, and 93.75%, respectively |
| [97] | 2019 | Glioma, meningioma, and pituitary tumor | VGG-19 | Introduced a block-wise fine-tuning technique for multi-class brain tumor MRI image. | CE-MRI [56] 3064 images from 233 patients. | Classification accuracy: 94.82% |
| [62] | 2020 | LGG and HGG | 3D CNN | Proposed a multi-scale 3D CNN architecture for grade classification capable of learning both local and global brain tumor features. Applied two image pre-processing techniques for reducing thermal noise and scanner-related artifacts in brain MRI. Used data augmentation. | BraTS2018: Training-209 HGG and 75 LGG from 284 patients. Validation: 67 mixed grades. | Classification accuracy: 96.49% |
| [75] | 2020 | T1w, T1w post GBCA, T2w, FLAIR | U-Net architecture, GANs | GANs was used to generate synthetic images for four modalities of MRI: T1, T1e, T2, FLAIR. | TCGA-GBM [99] and TCGA-LGG [99]. | Average classification accuracy, sensitivity, and specificity of 88.82%, 81.81%, and 92.17%, respectively |
| [17] | 2019 | Complete, core, and enhancing tumors | Custom CNN architecture, Bat algorithm | Used BAT algorithm to optimize the loss function of CNN. In addition, used skull stripping and image enhancement techniques for image pre-processing. | BraTS2015 | Accuracy, recall (or sensitivity), and precision of 92%, 87%, and 90%, respectively |
| Ref. | Year | Classes Considered | Method | Main Highlight | Dataset | Performance |
|---|---|---|---|---|---|---|
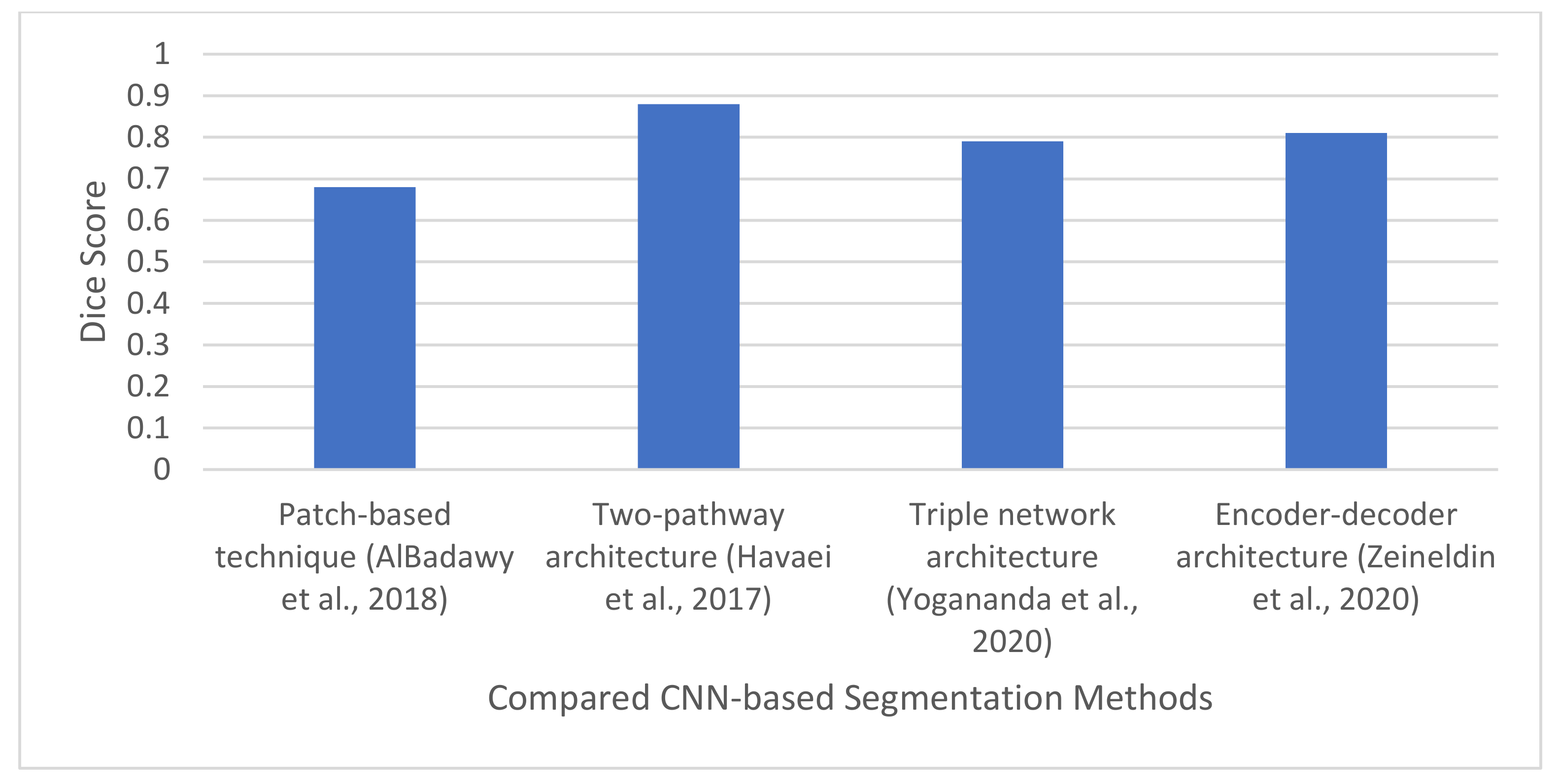
| [73] | 2018 | Metastasis, meningiomas gliomas | CNN | Designed a patching-based technique for brain tumor segmentation. Evaluated the impact of inter-institutional dataset. | TCIA | DSC-Same institution: 0.72 ± 0.17 and 0.76 ± 0.12. Different Institution: 0.68 ± 0.19 and 0.59 ± 0.19 |
| [10] | 2017 | Necrosis, edema, non-ET, ET. | CNN | Designed a two-pathway architecture for capturing global and local features. Also designed three cascade architectures. | BraTS2013 | DSC: 0.88 |
| [74] | 2020 | WT, TC, and ET | Triple CNN architecture for multi-class segmentation. | Developed a triple network architecture to simplify the multiclass segmentation problem to a single binary segmentation problem. | BraT2018 dataset | DSC: 0.90, 0.82, and 0.79 for WT, TC, and ET, respectively |
| [41] | 2020 | T1w, T1w post GBCA, T2w, and FLAIR | Modified U-Net architecture, Data augmentation, batch normalization using the N3 bias correction tool [100]. | Developed an encoder-decoder architecture for brain tumor segmentation. | BraTS 2019 | DSC, sensitivity, and specificity of 0.814, 0.783, 0.999, respectively. |
| [101] | 2021 | HGG and LGG | Inception-v3, NSGA, LDA, SVM, k-NN, softmax, CART, YOLOv2, and McCulloch’s Kapur entropy | Designed a CNN-based hybrid framework for tumor enhancement, feature extraction and selection, localization, and tumor segmentation | BraTS 2018, BraTS2019, and BraTS2020 | Classification accuracy of 98%, 99%, and 99% for BraTS2018, BraTS2019, and BraTS2020, respectively. |
| [17] | 2019 | Complete, core, and enhancing tumors | Custom CNN architecture, Bat algorithm | Used BAT algorithm to optimize the loss function of CNN. In addition, used skull stripping and image enhancement techniques for image pre-processing. | BraTS2015 | Accuracy, recall (or sensitivity), and precision of 92%, 87%, and 90%, respectively |
| Ref. | Year | Classes Considered | Method | Dataset | Main Highlight | Performance |
|---|---|---|---|---|---|---|
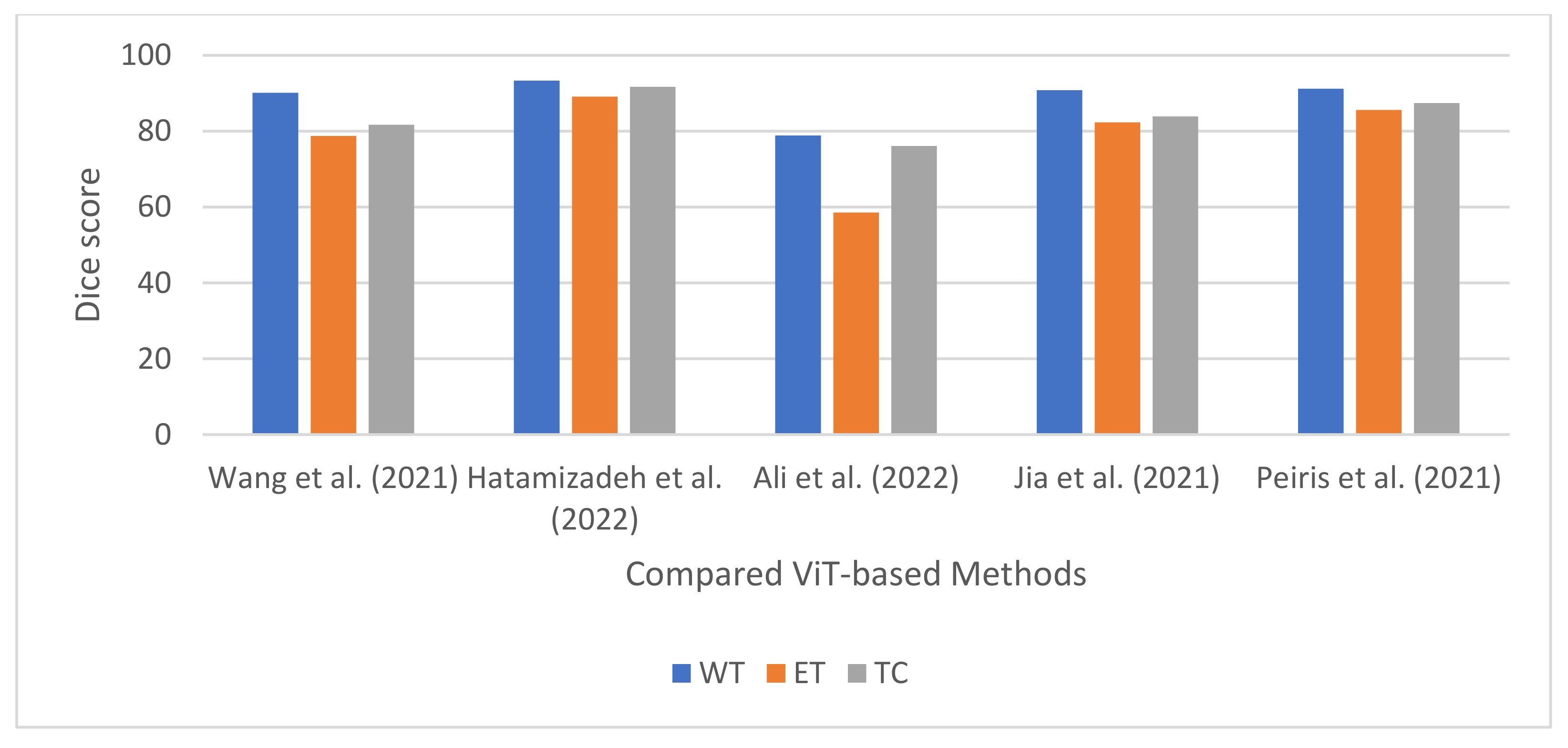
| [35] | 2021 | ET, WT, EC | Transformers and 3D CNN | BraTS2019 and BraTS2020 | Developed a transformer-based network for 3D brain tumor segmentation. | BraTS2020-DSC of 90.09%, 78.73%, and 81.73% for WT, ET, and TC, respectively. BraTS2019–DSC of 90%, 78.93%, and 81.84% for WT, ET, and TC, respectively DSC of 90%, 78.93%, and 81.84% for WT, ET, and TC, respectively |
| [32] | 2022 | ET, WT, and TC | Swin transformers and CNN | BraTS2021 | Developed a technique for multi-modal brain tumor images using Swin transformers and CNN. | DSC of 0.891, 0.933, and 0.917 for ET, WT, and TC, respectively. |
| [105] | 2022 | WT, ET, and TC | Transformers and CNN | MSD dataset | Developed a segmentation technique for multi-modal brain tumor image using transformers and CNN. | DSC of 0.789, 0.585,, and 0.761 for WT, ET, and TC, respectively. |
| [104] | 2021 | WT, ET, and TC | Transformers and 3D CNN | BraTS2021 | Designed a CNN-transformer technique for multi-modal brain MRI scan segmentation. | DSC of 0.823, 0.908, and 0.839 for ET, WT, and TC, respectively. |
| [107] | 2021 | WT, ET, and TC | Transformers and 3D CNN | BraTS2021 | Developed a U-Net shaped encoder-decoder technique using only transformers. The transformer encoder can capture local and global information. The decoder block allows parallel computation of cross- and self-attention. | DSC of 85.59%, 87.41%, and 91.20% for ET, TC, and WT, respectively |
| Ref. | Year | Classes Considered | Method | Dataset | Main Highlight | Performance |
|---|---|---|---|---|---|---|
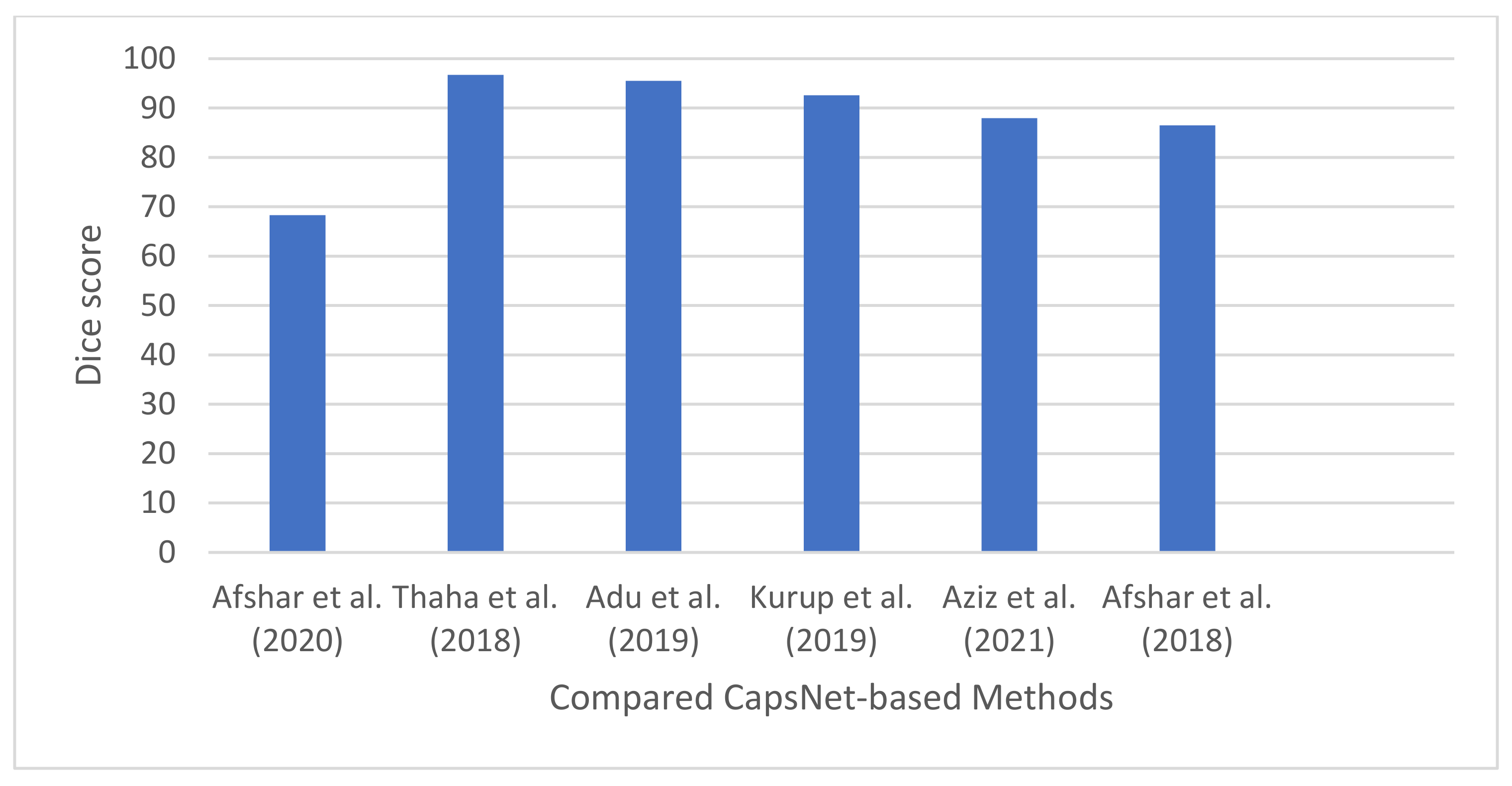
| [40] | 2020 | Meningioma, glioma, and pituitary. | CapsNet and Bayesian theory. | Cancer dataset [56] | Designed a DL technique that can model uncertainty associated with predictions of CapsNet models. | Classification accuracy: 68.3% |
| [19] | 2021 | Meningioma, Glioma, Pituitary, normal | CapsNet | Brain tumor dataset. Meningioma (937 images), Glioma (926 images), Pituitary (901 images), normal (500) | Introduced a new activation function for CapsNet, called PSTanh activation function. | Classification accuracy of 96.70%. |
| [109] | 2019 | Meningioma, Glioma, Pituitary, normal | CapsNet, dilation convolution | Brain tumor dataset [56]. 3064 images from 233 patients. Meningioma (708 slices), Glioma (1426 slices), Pituitary (930 slices), normal | Developed a CapsNet-based technique using dilation convolution with the objective of maintaining the high resolution of the images for accurate classification. | Classification accuracy: 95.54%. |
| [110] | 2019 | Meningioma, Glioma, Pituitary | CapsNet; classification; Data pre-processing | Brain tumor dataset: 3064 [56]. | Presented a performance analysis on the effect of image pre-processing on CapsNet for brain tumor segmentation. | Classification accuracy: 92.6% |
| [37] | 2021 | T1w, T2w, T1 w post GBCA and FLAIR | SegCaps–Capsule network; brain tumor segmentation | BraTS 2020 | Designed a modified version of CapsNet using SegCaps network. | DSC of 87.96%. |
| [18] | 2018 | Meningioma, Pituitary, and Glioma | Capsule network | Brain tumor dataset proposed by [56] | Developed different CapsNet for brain tumor segmentation. Investigated the performance of input data on capsule network. Developed a visualization paradigm for the output of capsule network. | 86.5% for segmented tumor, and 78% for whole brain image |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akinyelu, A.A.; Zaccagna, F.; Grist, J.T.; Castelli, M.; Rundo, L. Brain Tumor Diagnosis Using Machine Learning, Convolutional Neural Networks, Capsule Neural Networks and Vision Transformers, Applied to MRI: A Survey. J. Imaging 2022, 8, 205. https://doi.org/10.3390/jimaging8080205
Akinyelu AA, Zaccagna F, Grist JT, Castelli M, Rundo L. Brain Tumor Diagnosis Using Machine Learning, Convolutional Neural Networks, Capsule Neural Networks and Vision Transformers, Applied to MRI: A Survey. Journal of Imaging. 2022; 8(8):205. https://doi.org/10.3390/jimaging8080205
Chicago/Turabian StyleAkinyelu, Andronicus A., Fulvio Zaccagna, James T. Grist, Mauro Castelli, and Leonardo Rundo. 2022. "Brain Tumor Diagnosis Using Machine Learning, Convolutional Neural Networks, Capsule Neural Networks and Vision Transformers, Applied to MRI: A Survey" Journal of Imaging 8, no. 8: 205. https://doi.org/10.3390/jimaging8080205
APA StyleAkinyelu, A. A., Zaccagna, F., Grist, J. T., Castelli, M., & Rundo, L. (2022). Brain Tumor Diagnosis Using Machine Learning, Convolutional Neural Networks, Capsule Neural Networks and Vision Transformers, Applied to MRI: A Survey. Journal of Imaging, 8(8), 205. https://doi.org/10.3390/jimaging8080205










