Comparative Genomics, Transcriptome, and Prokaryotic Expression Analysis of alkB1_1 in Acinetobacter vivianii KJ-1: Revealing the Mechanism of Petroleum Hydrocarbon Degradation
Abstract
:1. Introduction
2. Results
2.1. Degradation of Various Alkanes with A. vivianii KJ-1
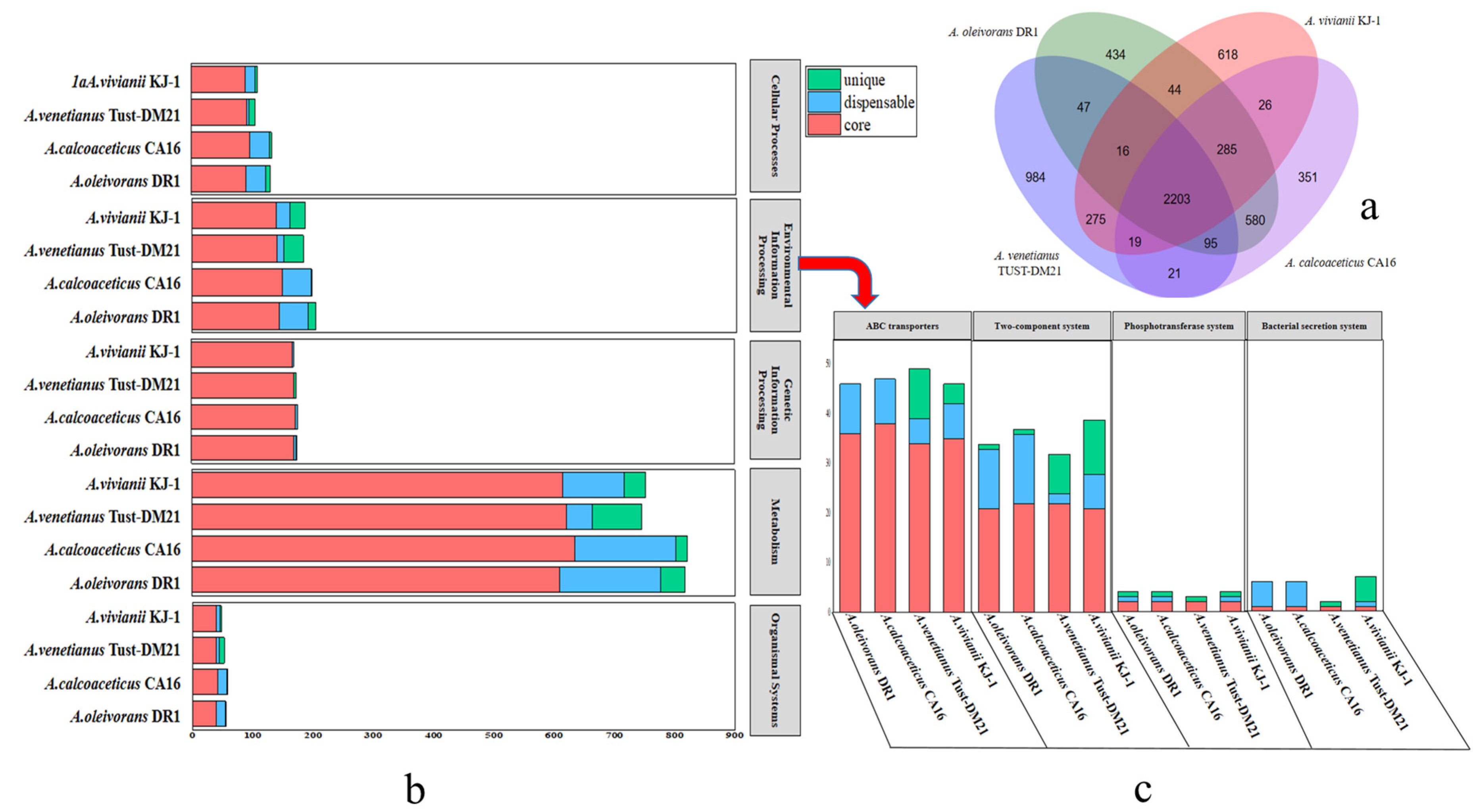
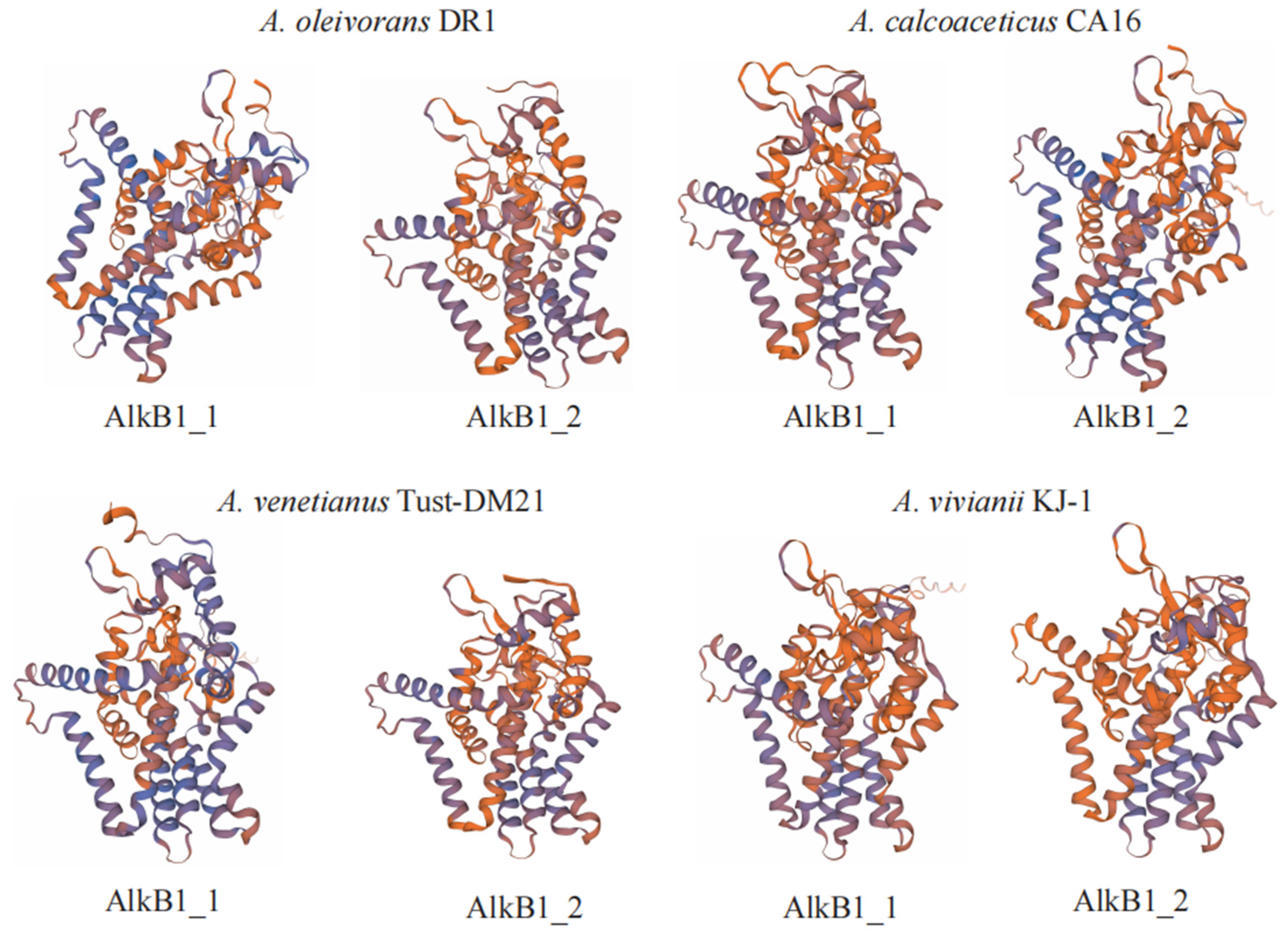
2.2. Genomic Comparison
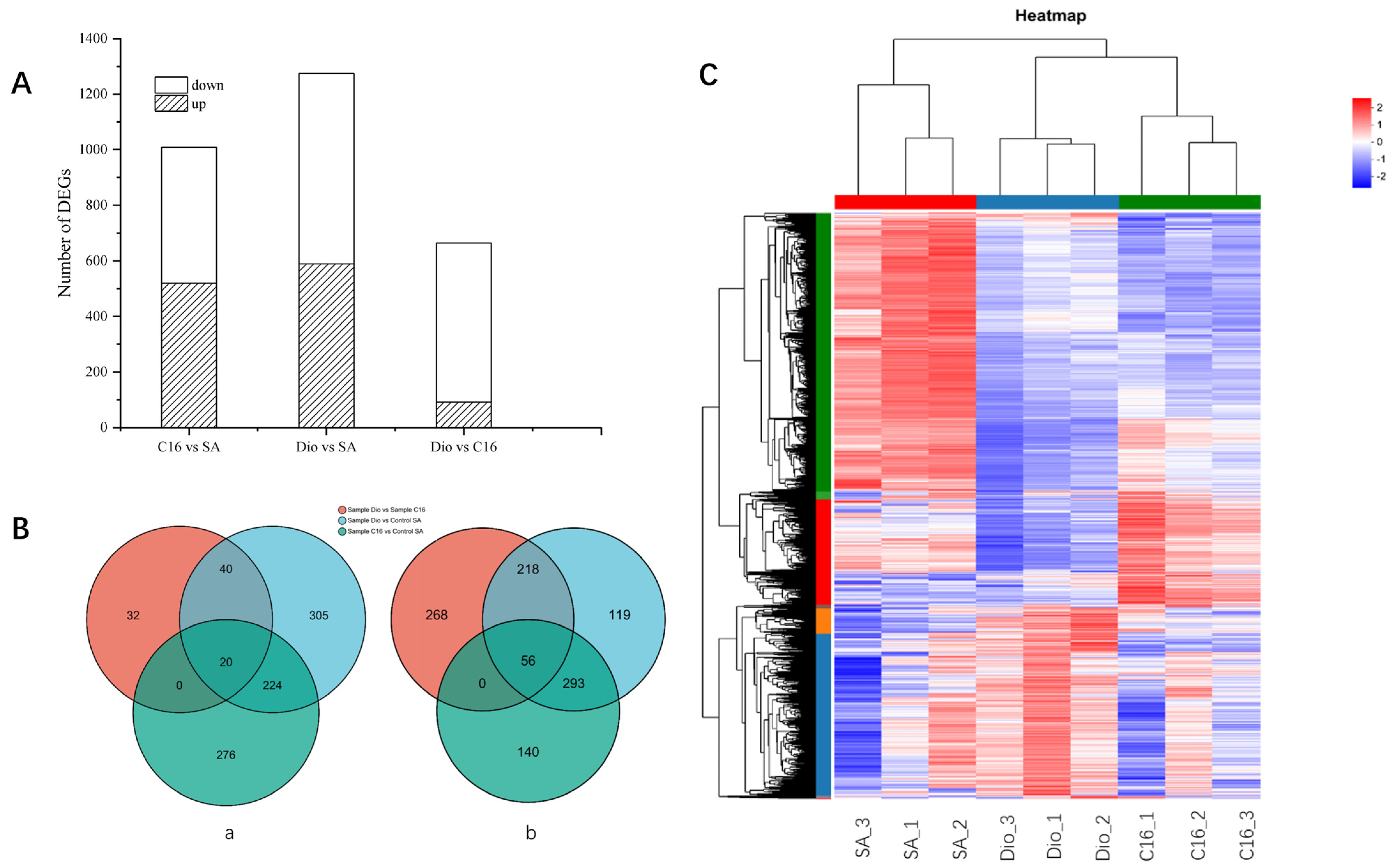
2.3. General Features of Transcriptome in Different Carbon Sources
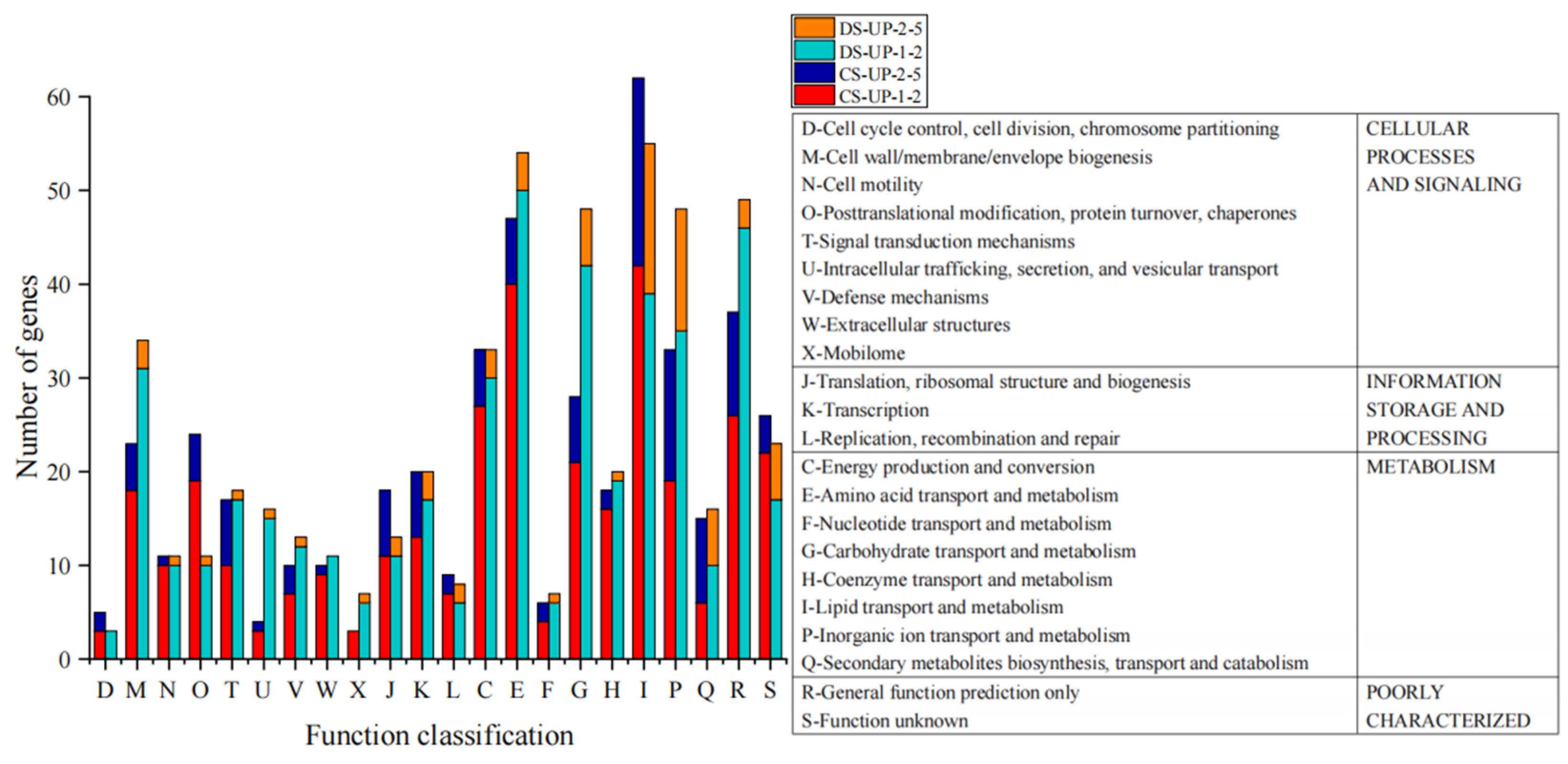
2.4. Analysis of Upregulated DEGs
2.5. Differential Expression of Alkane Hydrolases
2.6. Expression of Alkane Monooxygenase Gene
2.7. Enzyme Activity Assay
3. Discussion
4. Materials and Methods
4.1. Strains, Plasmids and Growth Conditions
4.2. Comparative Genomic Analysis Based on the Whole Genome
4.3. Transcriptome Sample Preparation and Analysis
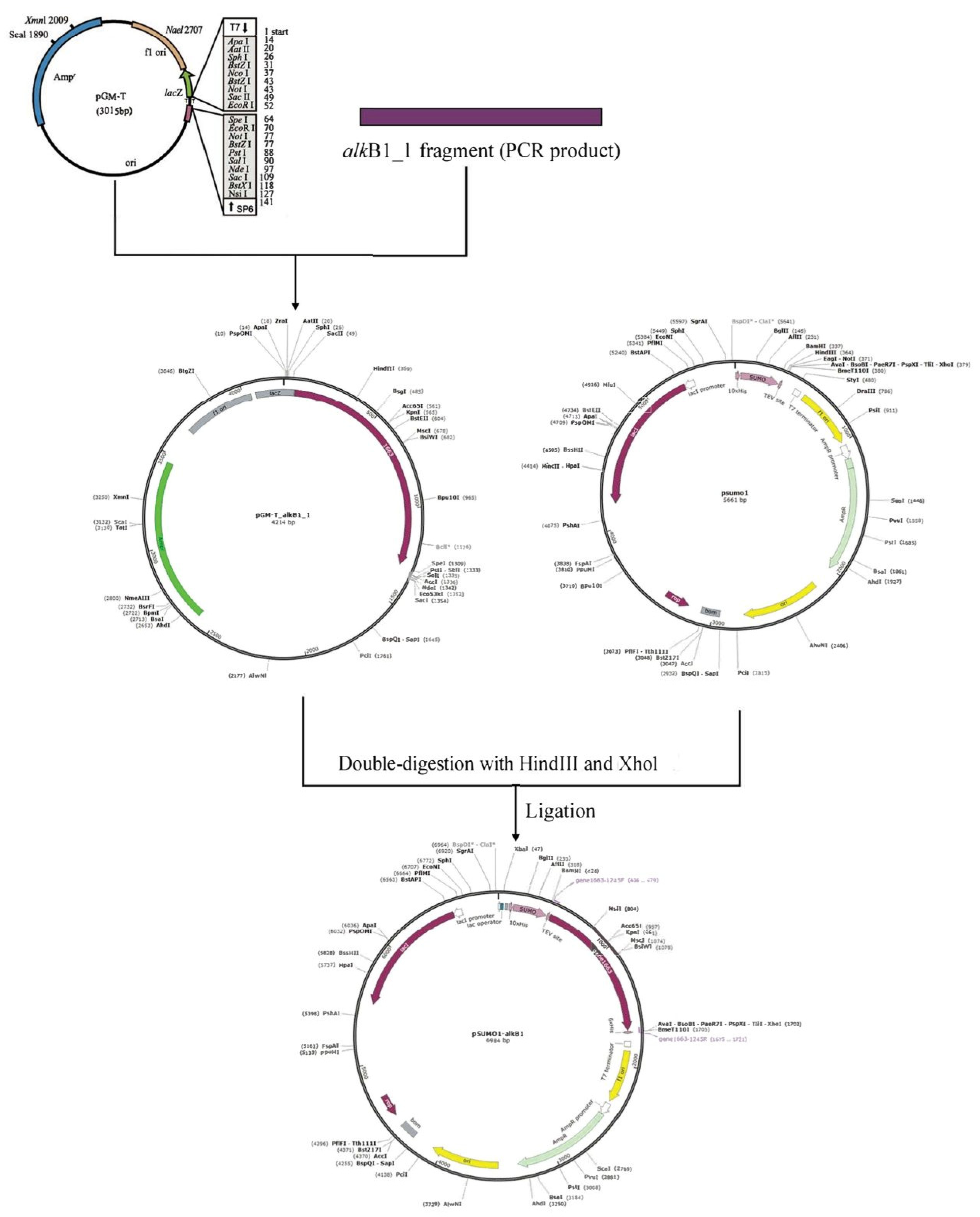
4.4. Expression of the Alkane Monooxygenase Gene alkB1_1
- alkB1_1-F: 5′-CGAAGCTTCACCATGAATGCGCCAGTAAATGTAGAGC-3′
- alkB1_1-R: 5′-CGGAGCTCTGCAGTTGCTACTTGTTCTG-3′
4.5. Enzyme Activity Assay and Its Degradation of n-Hexadecane
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoang, S.A.; Sarkar, B.; Seshadri, B.; Lamb, D.; Wijesekara, H.; Vithanage, M.; Liyanage, C.; Kolivabandara, P.A.; Rinklebe, J.; Lam, S.S.; et al. Mitigation of Petroleum-Hydrocarbon-Contaminated Hazardous Soils Using Organic Amendments: A Review. J. Hazard. Mater. 2021, 416, 125702. [Google Scholar] [CrossRef] [PubMed]
- Hoang, S.A.; Lamb, D.; Seshadri, B.; Sarkar, B.; Choppala, G.; Kirkham, M.B.; Bolan, N.S. Rhizoremediation as a Green Technology for the Remediation of Petroleum Hydrocarbon-Contaminated Soils. J. Hazard. Mater. 2021, 401, 123282. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, A.; Siddiqui, S.; Bano, A.; Sattar, S.; Hashmi, M.Z.; Qin, M.; Shakoor, A. Hydrocarbon Degradation in Oily Sludge by Bacterial Consortium Assisted with Alfalfa (Medicago sativa L.) and Maize (Zea mays L.). Arab. J. Geosci. 2020, 13, 879. [Google Scholar] [CrossRef]
- Hou, J.; Wang, Q.; Liu, W.; Zhong, D.; Ge, Y.; Christie, P.; Luo, Y. Soil Microbial Community and Association Network Shift Induced by Several Tall Fescue Cultivars during the Phytoremediation of a Petroleum Hydrocarbon-Contaminated Soil. Sci. Total Environ. 2021, 792, 148411. [Google Scholar] [CrossRef]
- Manzoor, M.; Khan, A.H.A.; Ullah, R.; Khan, M.Z.; Ahmad, I. Environmental Epidemiology of Cancer in South Asian Population: Risk Assessment Against Exposure to Polycyclic Aromatic Hydrocarbons and Volatile Organic Compounds. Arab. J. Sci. Eng. 2016, 41, 2031–2043. [Google Scholar] [CrossRef]
- Sun, W.-J.; Li, Q.; Luo, B.-Y.; Sun, R.; Ke, C.-Y.; Wang, S.-C.; Zhang, Q.-Z.; Zhang, X.-L. Pilot-Scale Field Studies on Activated Microbial Remediation of Petroleum-Contaminated Soil. Environ. Geochem. Health 2024, 46, 243. [Google Scholar] [CrossRef]
- Farzadkia, M.; Esrafili, A.; Gholami, M.; Koolivand, A. Effect of Immature and Mature Compost Addition on Petroleum Contaminated Soils Composting: Kinetics. J. Environ. Health Sci. Eng. 2019, 17, 839–846. [Google Scholar] [CrossRef]
- Mekonnen, B.A.; Aragaw, T.A.; Genet, M.B. Bioremediation of Petroleum Hydrocarbon Contaminated Soil: A Review on Principles, Degradation Mechanisms, and Advancements. Front. Environ. Sci. 2024, 12, 1354422. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, X.; Huang, Y.; Zhou, F.; Zhao, Q.; Zhang, W.; Chen, G.; Gao, Y. Isolation of Petroleum Degraders and Petroleum-Degradation Characteristics of Crude Enzymes from Providencia rettgeri L1. Pol. J. Environ. Stud. 2022, 31, 3859–3866. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, Z.; Zhang, Y.; Zhang, X.; Wang, L.; Zhang, W.; Zhao, Q.; Zheng, L.; Song, F.; Wang, J. Characterization of a Diesel-Degrading Rhodococcus Qingshengii RHZ01and Its Genome Analysis. Pol. J. Environ. Stud. 2022, 32, 99–112. [Google Scholar] [CrossRef]
- Zhang, Y.; Song, F.; Wang, J.; Zhao, Q.; Zheng, L.; Wang, Z.; Zhang, X.; Gao, Y.; Chen, G.; Huang, Y. Complete Genome Sequence Analysis of a Novel Alkane-Degrading Bacterial Strain, Acinetobacter vivianii KJ-1, and Its Diesel Degradation Ability. Front. Environ. Sci. 2022, 10, 1044754. [Google Scholar] [CrossRef]
- Fu, X.; Lai, Q.; Dong, C.; Wang, W.; Shao, Z. Complete Genome Sequence of Alcanivorax xenomutans P40, an Alkane-Degrading Bacterium Isolated from Deep Seawater. Mar. Genom. 2018, 38, 1–4. [Google Scholar] [CrossRef]
- Hossain, M.F.; Akter, M.A.; Sohan, M.S.R.; Sultana, D.N.; Reza, M.A.; Hoque, K.M.F. Bioremediation Potential of Hydrocarbon Degrading Bacteria: Isolation, Characterization, and Assessment. Saudi J. Biol. Sci. 2022, 29, 211–216. [Google Scholar] [CrossRef]
- Xiang, W.; Liang, Y.; Hong, S.; Wang, G.; You, J.; Xue, Y.; Ma, Y. Degradation of Long-Chain n-Alkanes by a Novel Thermal-Tolerant Rhodococcus Strain. Arch. Microbiol. 2022, 204, 259. [Google Scholar] [CrossRef]
- Labbé, D.; Margesin, R.; Schinner, F.; Whyte, L.G.; Greer, C.W. Comparative Phylogenetic Analysis of Microbial Communities in Pristine and Hydrocarbon-Contaminated Alpine Soils: Microbial Community Phylogeny in Alpine Soil. FEMS Microbiol. Ecol. 2007, 59, 466–475. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Q.; Liu, L. Crude Oil Degrading Fingerprint and the Overexpression of Oxidase and Invasive Genes for N-Hexadecane and Crude Oil Degradation in the Acinetobacter pittii H9-3 Strain. Int. J. Environ. Res. Public Health 2019, 16, 188. [Google Scholar] [CrossRef]
- Acer, Ö.; Güven, K.; Poli, A.; Di Donato, P.; Leone, L.; Buono, L.; Güven, R.G.; Nicolaus, B.; Finore, I. Acinetobacter mesopotamicus Sp. Nov., Petroleum-Degrading Bacterium, Isolated from Petroleum-Contaminated Soil in Diyarbakir, in the Southeast of Turkey. Curr. Microbiol. 2020, 77, 3192–3200. [Google Scholar] [CrossRef]
- Park, C.; Shin, B.; Park, W. Protective Role of Bacterial Alkanesulfonate Monooxygenase under Oxidative Stress. Appl. Environ. Microbiol. 2020, 86, e00692-20. [Google Scholar] [CrossRef]
- Ho, M.T.; Li, M.S.M.; McDowell, T.; MacDonald, J.; Yuan, Z.-C. Characterization and Genomic Analysis of a Diesel-Degrading Bacterium, Acinetobacter calcoaceticus CA16, Isolated from Canadian Soil. BMC Biotechnol. 2020, 20, 39. [Google Scholar] [CrossRef]
- Xu, L.; Zhao, Y.; Li, Y.; Sun, J.-Q. Genomic and Transcriptomic Analyses Provide New Insights into the Allelochemical Degradation Preference of a Novel Acinetobacter Strain. Environ. Res. 2024, 246, 118145. [Google Scholar] [CrossRef]
- Zhan, Y.; Yan, Y.; Zhang, W.; Chen, M.; Lu, W.; Ping, S.; Lin, M. Comparative Analysis of the Complete Genome of an Acinetobacter calcoaceticus Strain Adapted to a Phenol-Polluted Environment. Res. Microbiol. 2012, 163, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Liu, M.; Feng, L.; Wang, Z. Comparative Genomic Analysis Reveals the Evolution and Environmental Adaptation of Acinetobacter johnsonii. Gene 2022, 808, 145985. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wei, H.-M.; Yuan, J.-L.; Xu, L.; Sun, J.-Q. A Comprehensive Genomic Analysis Provides Insights on the High Environmental Adaptability of Acinetobacter Strains. Front. Microbiol. 2023, 14, 1177951. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Zhao, C.; Gao, X.; Wang, L.; Tian, Q.; Liu, Y.; Xue, S.; Han, Z.; Chen, F.; Wang, S. Characterization and Transcriptome Analysis of a Long-Chain n-Alkane-Degrading Strain Acinetobacter pittii SW-1. Int. J. Environ. Res. Public Health 2021, 18, 6365. [Google Scholar] [CrossRef]
- Lin, H.; Guan, W.; Xiao, Y.; Qu, Y.; Ren, Y. Degradation Mechanisms of 2-Chlorophenol by Acinetobacter Sp.: Exploring Cellular Membrane Structure, Intra- and Extracellular Components, and Transcriptomic Analysis. J. Environ. Chem. Eng. 2023, 11, 109865. [Google Scholar] [CrossRef]
- Xiang, W.; Hong, S.; Xue, Y.; Ma, Y. Functional Analysis of Novel alkB Genes Encoding Long-Chain n-Alkane Hydroxylases in Rhodococcus sp. Strain CH91. Microorganisms 2023, 11, 1537. [Google Scholar] [CrossRef]
- Van Beilen, J.B.; Funhoff, E.G. Alkane Hydroxylases Involved in Microbial Alkane Degradation. Appl. Microbiol. Biotechnol. 2007, 74, 13–21. [Google Scholar] [CrossRef]
- Wang, S.; Li, G.; Liao, Z.; Liu, T.; Ma, T. A Novel Alkane Monooxygenase (alkB) Clade Revealed by Massive Genomic Survey and Its Dissemination Association with IS Elements. PeerJ 2022, 10, e14147. [Google Scholar] [CrossRef]
- Jeon, H.J.; Kim, M.N. Functional Analysis of Alkane Hydroxylase System Derived from Pseudomonas aeruginosa E7 for Low Molecular Weight Polyethylene Biodegradation. Int. Biodeterior. Biodegrad. 2015, 103, 141–146. [Google Scholar] [CrossRef]
- Dombrowski, N.; Donaho, J.A.; Gutierrez, T.; Seitz, K.W.; Teske, A.P.; Baker, B.J. Reconstructing Metabolic Pathways of Hydrocarbon-Degrading Bacteria from the Deepwater Horizon Oil Spill. Nat. Microbiol. 2016, 1, 16057. [Google Scholar] [CrossRef]
- Liu, Q.; Peng, Y.; Liao, J.; Liu, X.; Peng, J.; Wang, J.-H.; Shao, Z. Broad-Spectrum Hydrocarbon-Degrading Microbes in the Global Ocean Metagenomes. Sci. Total Environ. 2024, 926, 171746. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, H.L.; Dias, G.M.; Neves, B.C. Genome Sequence of Pseudomonas Aeruginosa PA1-Petro—A Role Model of Environmental Adaptation and a Potential Biotechnological Tool. Heliyon 2022, 8, e11566. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Liu, W.; Tian, S.; Wang, W.; Qi, Q.; Jiang, P.; Gao, X.; Li, F.; Li, H.; Yu, H. Petroleum Hydrocarbon-Degrading Bacteria for the Remediation of Oil Pollution Under Aerobic Conditions: A Perspective Analysis. Front. Microbiol. 2018, 9, 2885. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Duo, H.; Li, S.; Qin, D.; Xie, L.; Xiao, Y.; Sun, J.; Tao, J.; Zhang, X.; Li, Y.; et al. Unlocking Biological Insights from Differentially Expressed Genes: Concepts, Methods, and Future Perspectives. J. Adv. Res. 2024, S2090123224005605. [Google Scholar] [CrossRef]
- Neilands, J.B. Siderophores: Structure and Function of Microbial Iron Transport Compounds. J. Biol. Chem. 1995, 270, 26723–26726. [Google Scholar] [CrossRef]
- Bacosa, H.P.; Erdner, D.L.; Rosenheim, B.E.; Shetty, P.; Seitz, K.W.; Baker, B.J.; Liu, Z. Hydrocarbon Degradation and Response of Seafloor Sediment Bacterial Community in the Northern Gulf of Mexico to Light Louisiana Sweet Crude Oil. ISME J. 2018, 12, 2532–2543. [Google Scholar] [CrossRef]
- Medić, A.; Lješević, M.; Inui, H.; Beškoski, V.; Kojić, I.; Stojanović, K.; Karadžić, I. Efficient Biodegradation of Petroleum N-Alkanes and Polycyclic Aromatic Hydrocarbons by Polyextremophilic Pseudomonas Aeruginosa San Ai with Multidegradative Capacity. RSC Adv. 2020, 10, 14060–14070. [Google Scholar] [CrossRef]
- Throne-Holst, M.; Wentzel, A.; Ellingsen, T.E.; Kotlar, H.-K.; Zotchev, S.B. Identification of Novel Genes Involved in Long-Chain n -Alkane Degradation by Acinetobacter sp. Strain DSM 17874. Appl. Environ. Microbiol. 2007, 73, 3327–3332. [Google Scholar] [CrossRef]
- Li, L.; Liu, X.; Yang, W.; Xu, F.; Wang, W.; Feng, L.; Bartlam, M.; Wang, L.; Rao, Z. Crystal Structure of Long-Chain Alkane Monooxygenase (LadA) in Complex with Coenzyme FMN: Unveiling the Long-Chain Alkane Hydroxylase. J. Mol. Biol. 2008, 376, 453–465. [Google Scholar] [CrossRef]
- Wang, W.; Shao, Z. Diversity of Flavin-Binding Monooxygenase Genes (almA) in Marine Bacteria Capable of Degradation Long-Chain Alkanes. FEMS Microbiol. Ecol. 2012, 80, 523–533. [Google Scholar] [CrossRef]
- Yin, C.-F.; Nie, Y.; Li, T.; Zhou, N.-Y. AlmA Involved in the Long-Chain n -Alkane Degradation Pathway in Acinetobacter Baylyi ADP1 Is a Baeyer–Villiger Monooxygenase. Appl. Environ. Microbiol. 2024, 90, e01625-23. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, Z.; Kong, D.; Zhang, X.; Wang, C.; Wang, Y. Synergistic Action of Acinetobacter Baumannii and Talaromyces Sp.: Function of Enzymes in Crude Oil Degradation. Biochem. Eng. J. 2024, 201, 109144. [Google Scholar] [CrossRef]
- Baburam, C.; Feto, N.A. Mining of Two Novel Aldehyde Dehydrogenases (DHY-SC-VUT5 and DHY-G-VUT7) from Metagenome of Hydrocarbon Contaminated Soils. BMC Biotechnol. 2021, 21, 18. [Google Scholar] [CrossRef]
- Chen, S.; Liu, J.; Gao, G.; Li, M.; Cao, L.; Liu, T.; Li, G.; Ma, T. An NAD+-Dependent Group III Alcohol Dehydrogenase Involved in Long-Chain Alkane Degradation in Acinetobacter venetianus RAG-1. Enzym. Microb. Technol. 2024, 172, 110343. [Google Scholar] [CrossRef]
- Ji, L.; Fu, X.; Wang, M.; Xu, C.; Chen, G.; Song, F.; Guo, S.; Zhang, Q. Enzyme Cocktail Containing NADH Regeneration System for Efficient Bioremediation of Oil Sludge Contamination. Chemosphere 2019, 233, 132–139. [Google Scholar] [CrossRef]
- Meng, L.; Li, W.; Bao, M.; Sun, P. Promoting the Treatment of Crude Oil Alkane Pollution through the Study of Enzyme Activity. Int. J. Biol. Macromol. 2018, 119, 708–716. [Google Scholar] [CrossRef]



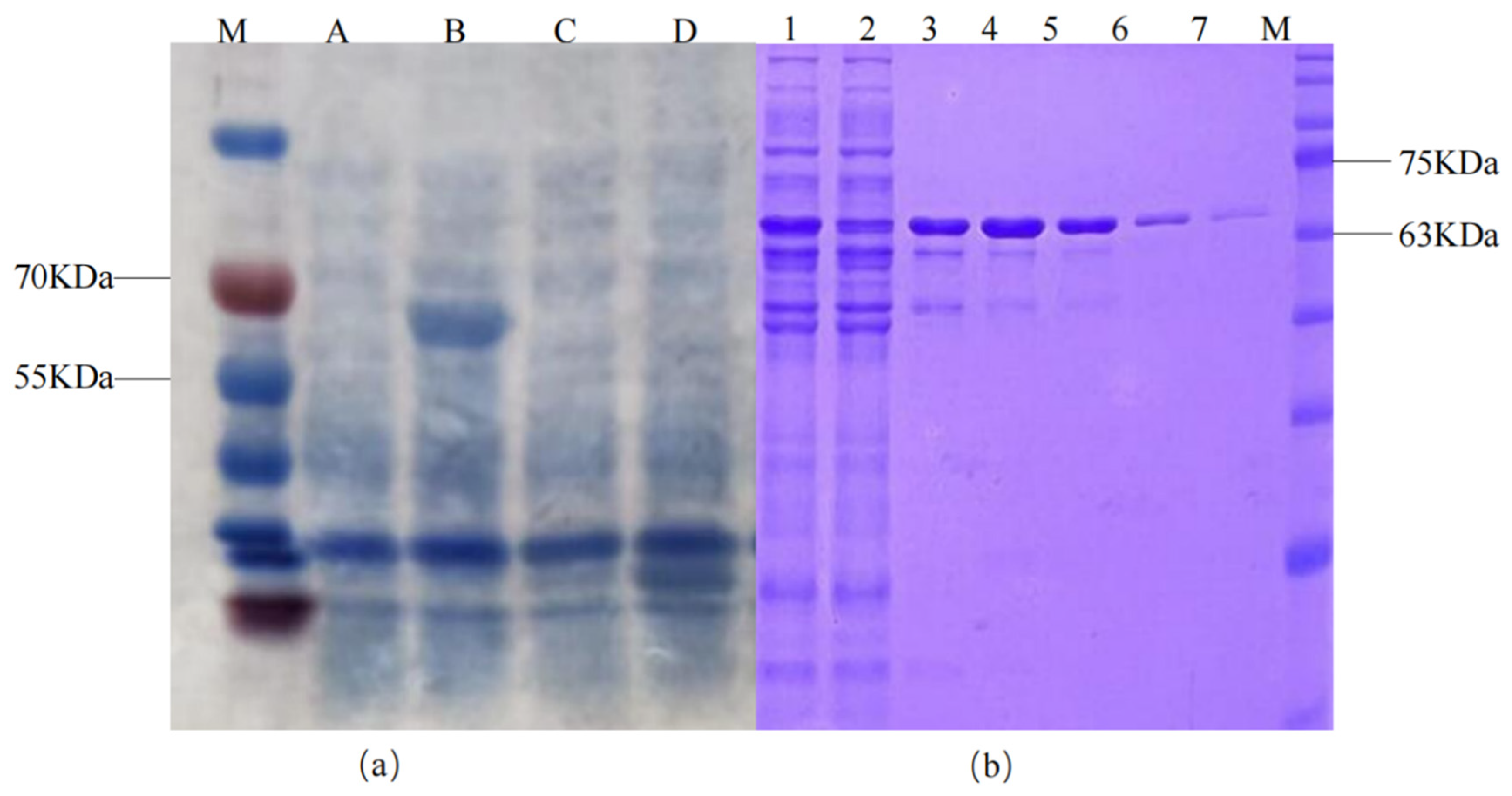
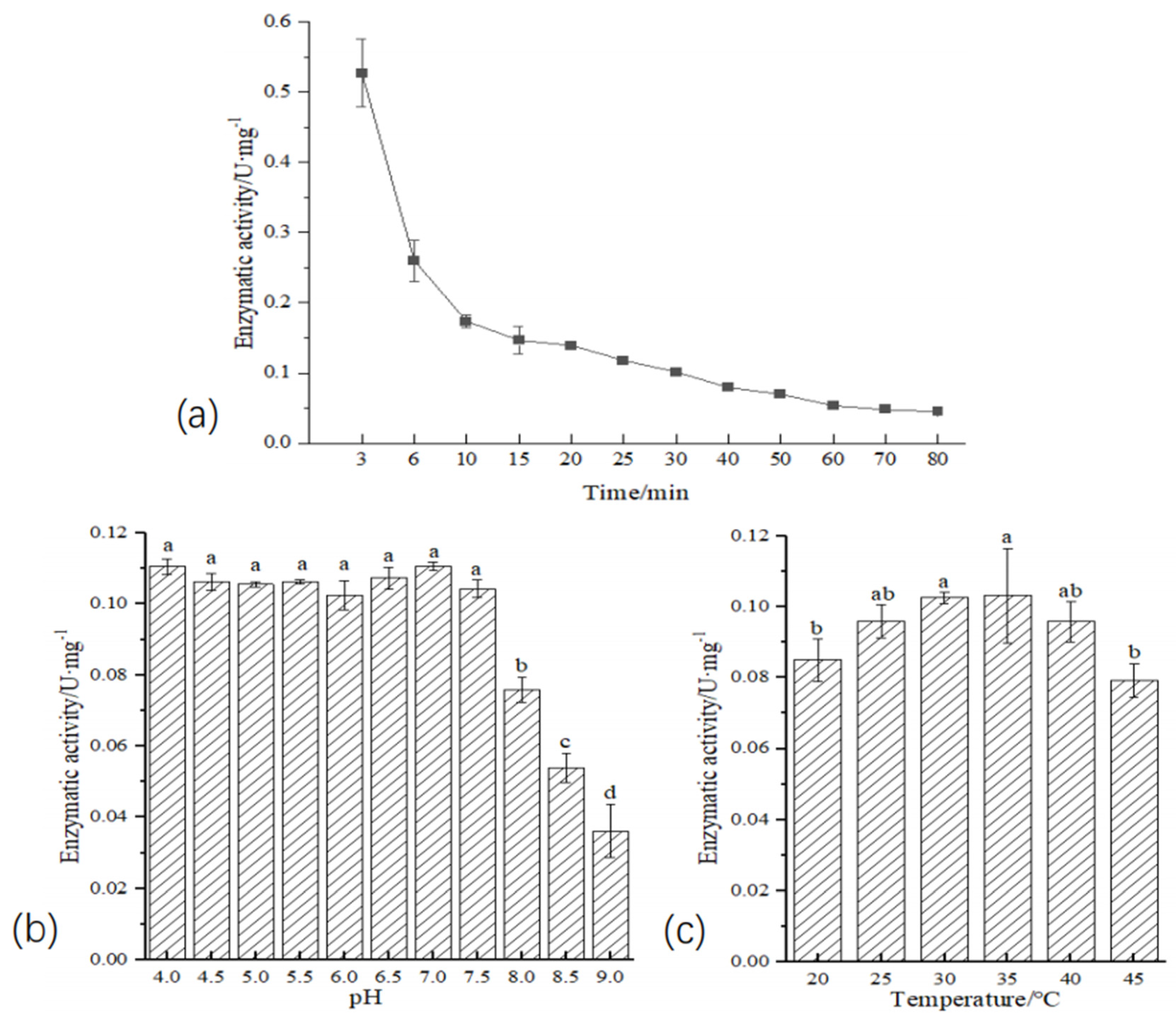

| Sample Name | A. calcoaceticus CA16 | A. oleivorans DR1 | A. venetianus Tust-DM21 | A. vivianii KJ-1 |
|---|---|---|---|---|
| Genome Size (Mb) | 4.12 | 4.15 | 4.12 | 3.93 |
| Chromosome No. | 1 | 1 | 1 | 1 |
| Plasmid No. | 1 | 0 | 3 | 0 |
| G + C (%) | 38.72 | 38.73 | 41.37 | 41.50 |
| CDS | 3879 | 3874 | 3846 | 3648 |
| rRNA | 18 | 18 | 18 | 18 |
| tRNA | 65 | 71 | 75 | 74 |
| House-keeping gene No. | 31 | 31 | 31 | 31 |
| Genbank Number | GCA_002055515.1 | GCA_000196795.1 | GCA_014250455.1 | GCA_020673335.1 |
| Degradation substrate | n-alkane (where n = 12–18) | diesel oil | crude petroleum | diesel oil |
| Degradation rate | 82–92% | 60–80% | 77.63–97.34% | >60% |
| Strains or Plasmids | Characteristics | Source |
|---|---|---|
| Strains | ||
| E. coli Top10 | Suitable for efficient DNA cloning and plasmid amplification, ensuring stable inheritance of high copy plasmids | Lab Store |
| E. coli BL21(DE3) | Used to efficient expression of genes cloned into an expression vector containing a bacteriophage T7 promoter | TianGen-China |
| A. vivianii KJ-1 | Wild-type, with diesel degradation capability | Lab Store |
| E. coli TO-alkB1_1 | Ampr, TOP10 containing pGM-T-alkB1 | This study |
| E. coli BL-alkB1_1 | Ampr, BL21(DE3) containing pSUMO1-alkB1 | This study |
| E. coli BL-C1 | Ampr, BL21(DE3) containing pSUMO1 | This study |
| plasmids | ||
| pGM-T vector | Ampr, contains T7 and SP6 RNA polymerase promoters, and the polyclonal site region is located in β- Galactosidase α within the peptide coding region | TianGen-China |
| pGM-T-alkB1_1 | Ampr, pGM-T vector containing gene alkB1_1 | This study |
| pSUMO1 vector | Ampr, contains His-sumo tag | Abiotech-Jinan |
| pSUMO1-alkB1_1 | Ampr, pSUMO1 vector containing gene alkB1_1 | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, Q.; Zhang, Y.; Wang, J.; Wang, J.; Zhao, Q.; Song, F.; Wang, L.; Zhang, W.; Huang, Y. Comparative Genomics, Transcriptome, and Prokaryotic Expression Analysis of alkB1_1 in Acinetobacter vivianii KJ-1: Revealing the Mechanism of Petroleum Hydrocarbon Degradation. Int. J. Mol. Sci. 2025, 26, 4083. https://doi.org/10.3390/ijms26094083
Cui Q, Zhang Y, Wang J, Wang J, Zhao Q, Song F, Wang L, Zhang W, Huang Y. Comparative Genomics, Transcriptome, and Prokaryotic Expression Analysis of alkB1_1 in Acinetobacter vivianii KJ-1: Revealing the Mechanism of Petroleum Hydrocarbon Degradation. International Journal of Molecular Sciences. 2025; 26(9):4083. https://doi.org/10.3390/ijms26094083
Chicago/Turabian StyleCui, Qiannan, Yali Zhang, Jie Wang, Jianing Wang, Qingqing Zhao, Fanyong Song, Leilei Wang, Wen Zhang, and Yujie Huang. 2025. "Comparative Genomics, Transcriptome, and Prokaryotic Expression Analysis of alkB1_1 in Acinetobacter vivianii KJ-1: Revealing the Mechanism of Petroleum Hydrocarbon Degradation" International Journal of Molecular Sciences 26, no. 9: 4083. https://doi.org/10.3390/ijms26094083
APA StyleCui, Q., Zhang, Y., Wang, J., Wang, J., Zhao, Q., Song, F., Wang, L., Zhang, W., & Huang, Y. (2025). Comparative Genomics, Transcriptome, and Prokaryotic Expression Analysis of alkB1_1 in Acinetobacter vivianii KJ-1: Revealing the Mechanism of Petroleum Hydrocarbon Degradation. International Journal of Molecular Sciences, 26(9), 4083. https://doi.org/10.3390/ijms26094083






