Genome-Wide Analysis of the JAZ Gene Family in Potato and Functional Verification of StJAZ23 Under Drought Stress
Abstract
:1. Introduction
2. Results
2.1. Identification of StJAZ Genes
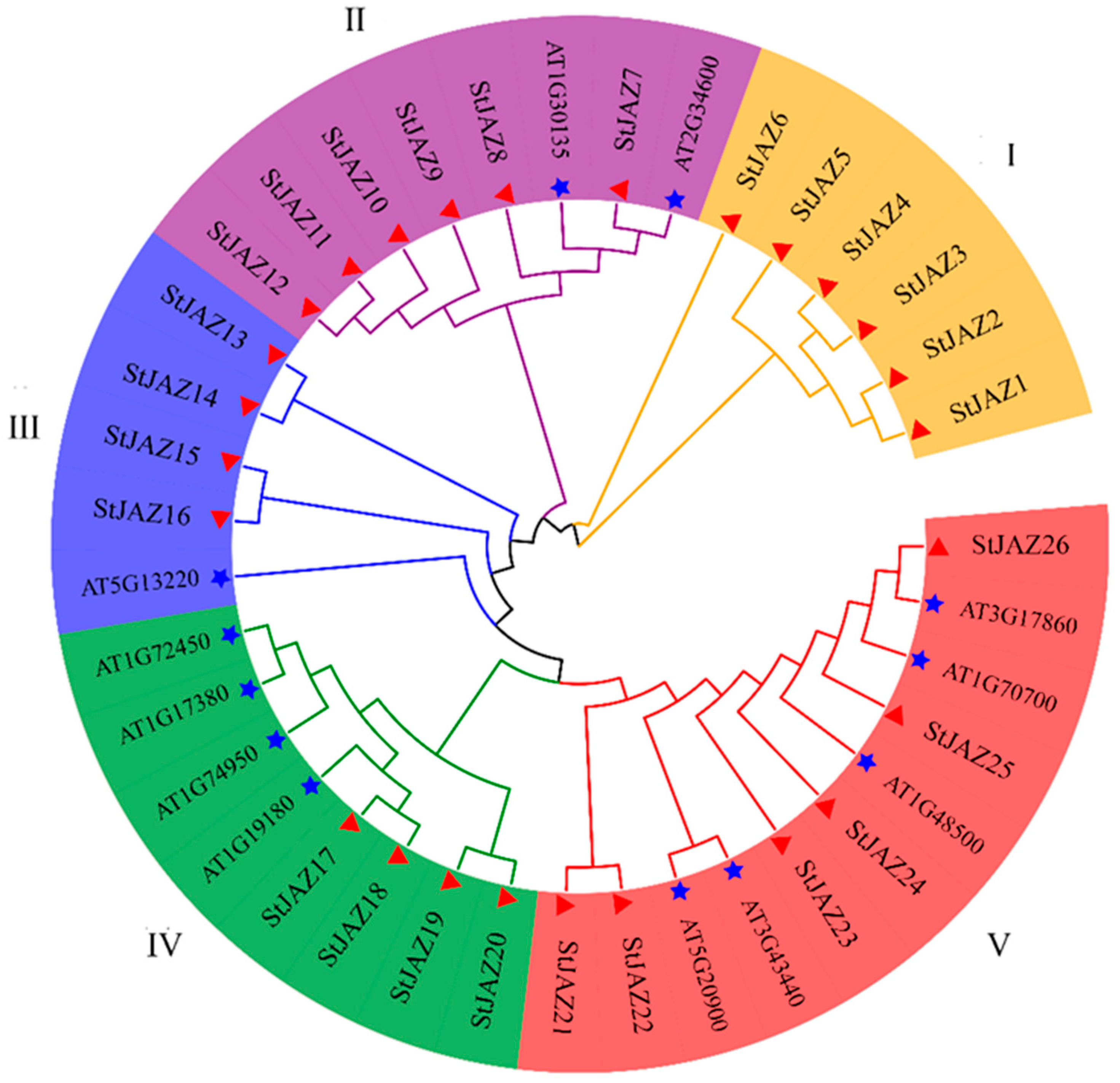
2.2. Multiple Sequence Alignment and Phylogenetic Analysis
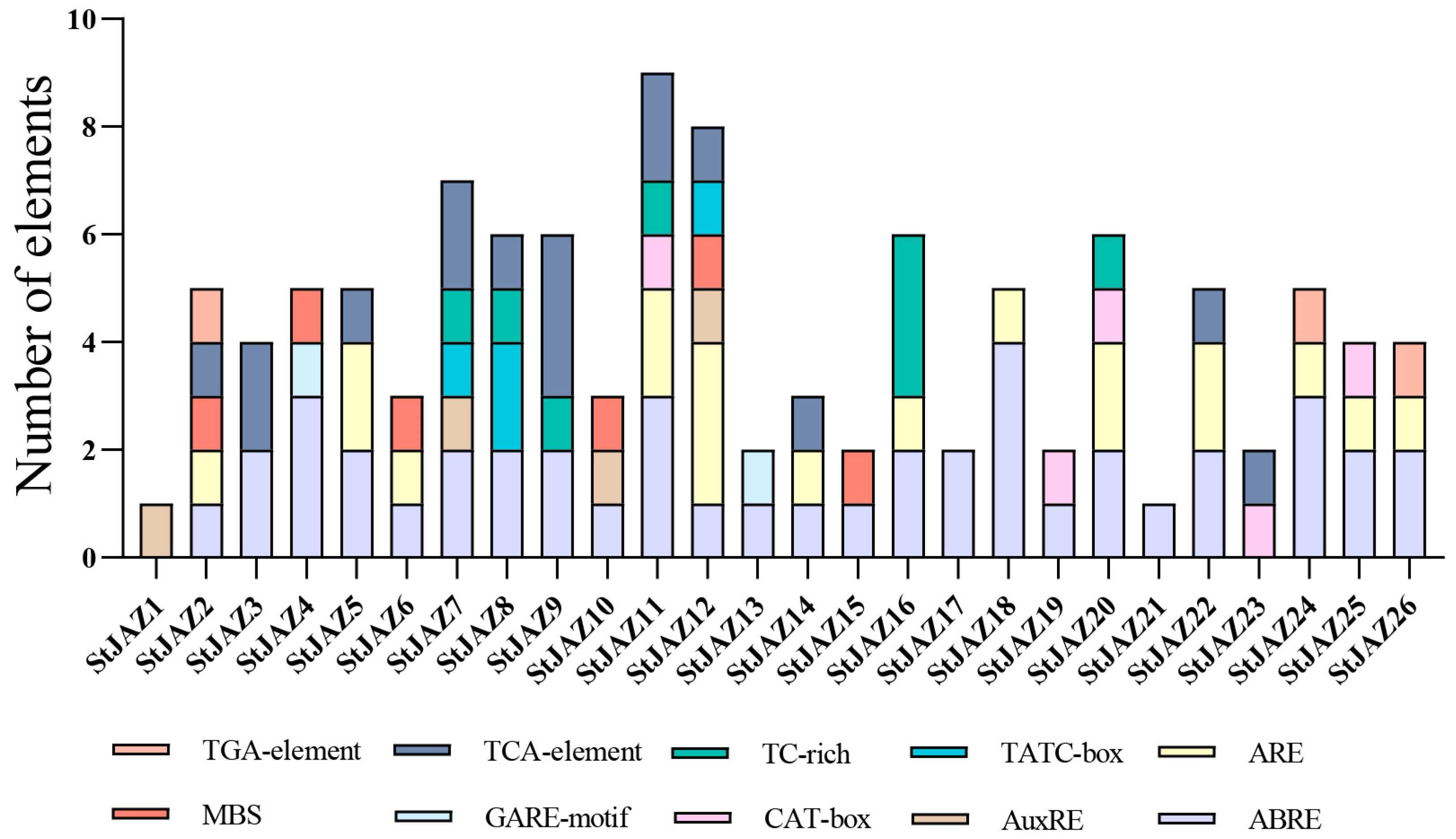
2.3. Promoter Cis-Acting Analysis of StJAZ Genes
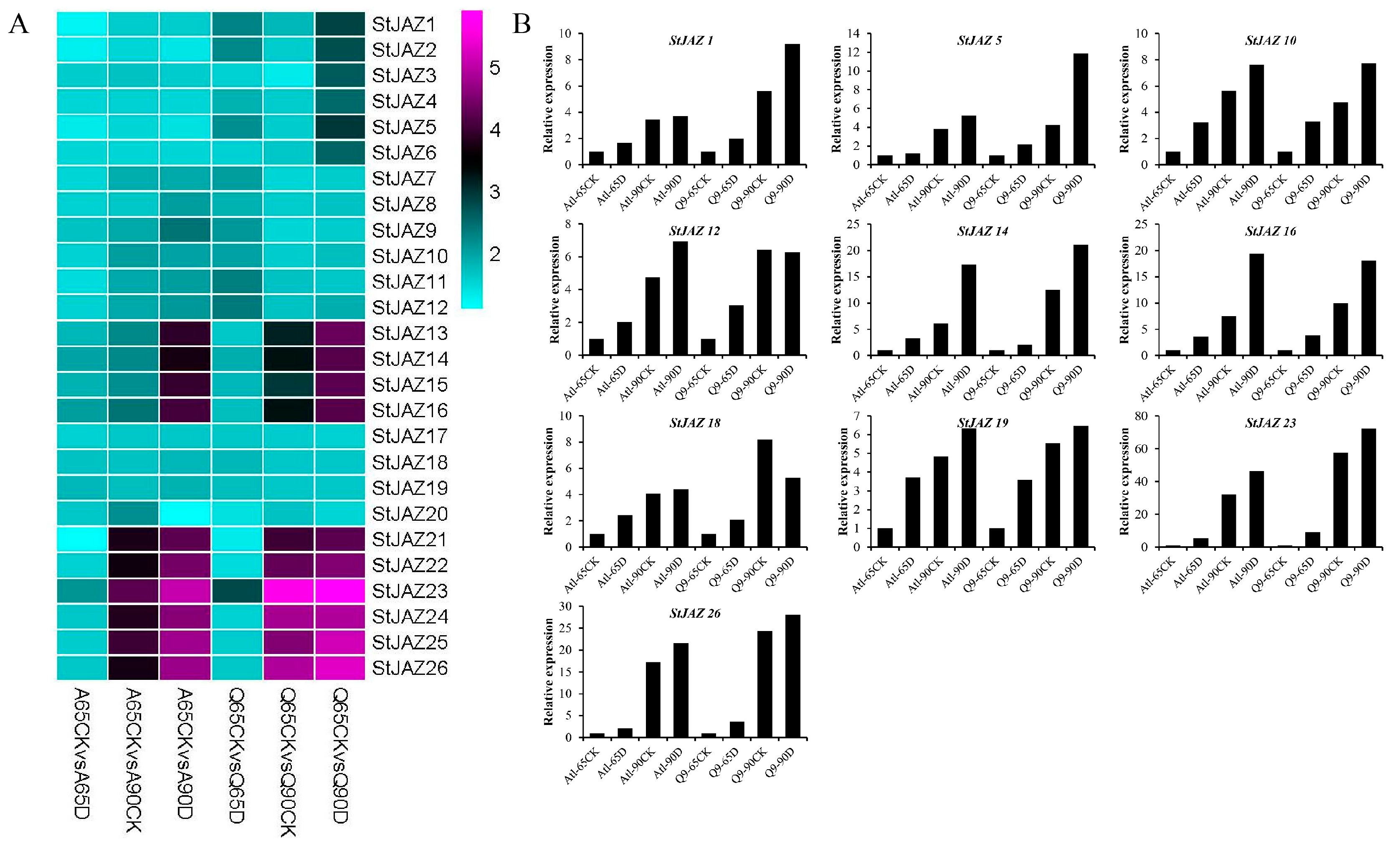
2.4. Expression Analysis of StJAZ Genes in Response to Abiotic Stress
2.5. Subcellular Localization of StJAZ23
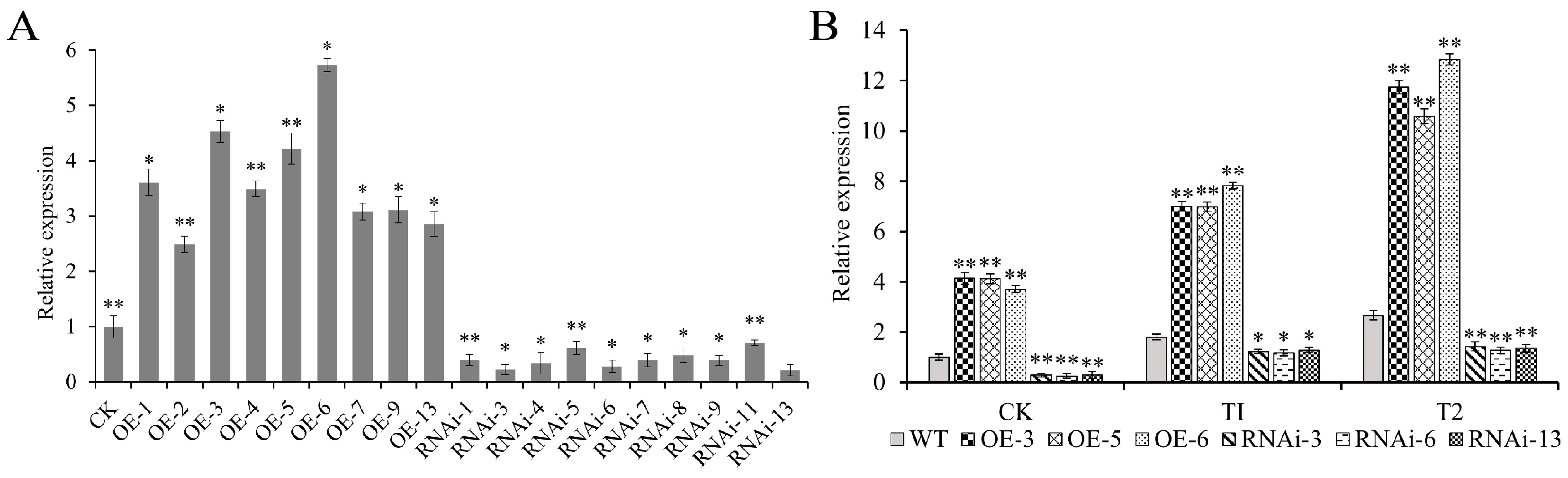
2.6. Generation of Stable Transgenic Potatoes
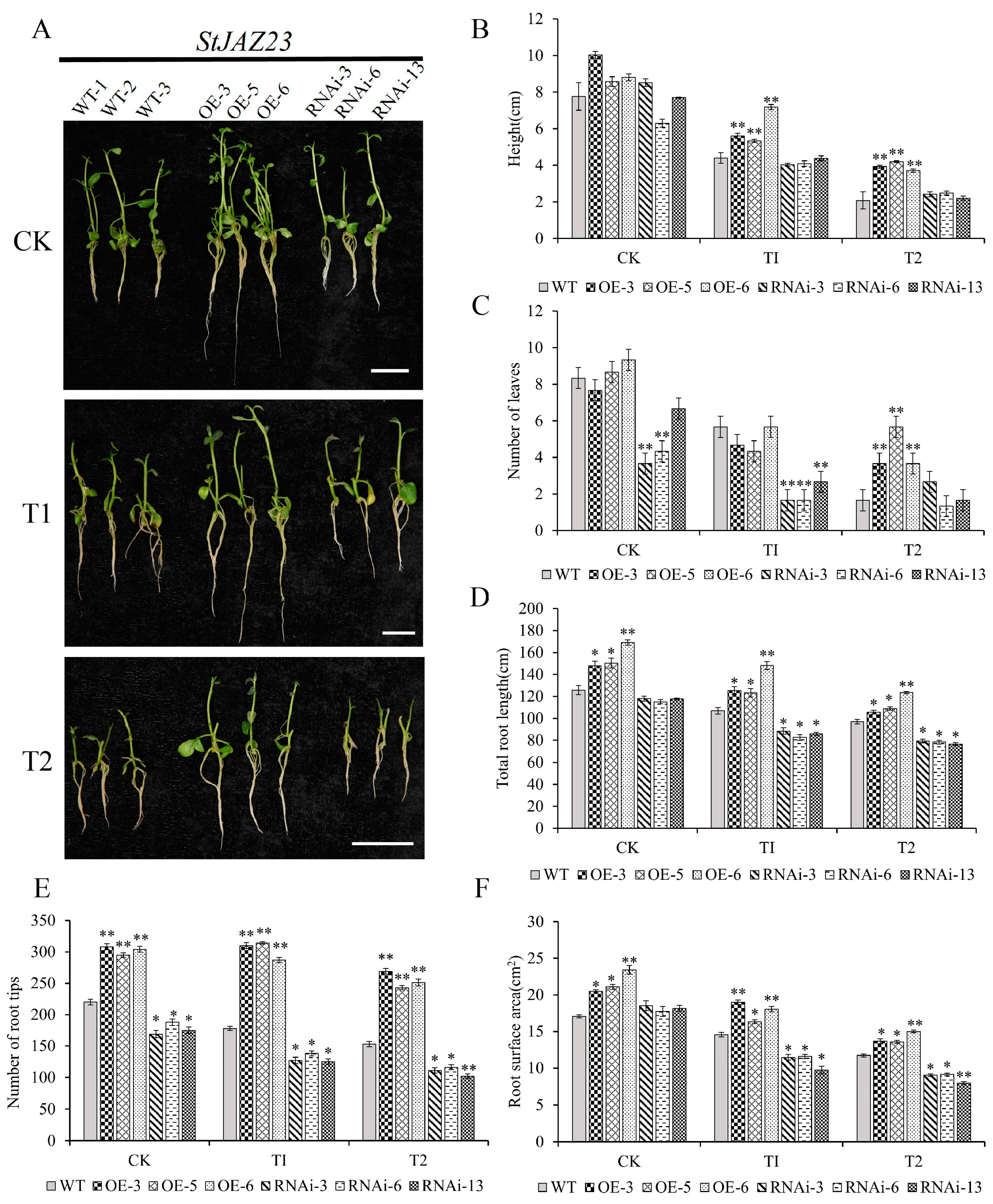
2.7. The Overexpression of StJAZ23 Increases Potato Drought Tolerance
2.8. Overexpression of StJAZ23 Enhances Antioxidant Enzyme Activities and Root Vigor
2.9. Increased Hormone Content in StJAZ23 Overexpression Lines
3. Discussion
4. Materials and Methods
4.1. Identification of Potato StJAZ Genes
4.2. Phylogenetic Analysis
4.3. Analysis of StJAZs Gene Promoter Cis-Acting Element
4.4. Expression Pattern Analysis of Potato StJAZ Gene Family Members
4.5. Plant Material and Growth Conditions
4.6. Cloning and Generation of Transgenic Potato Plants of StJAZ23
4.7. Subcellular Localization of StJAZ in Tobacco
4.8. Gene Expression Analysis Under Drought Stress
4.9. Phenotypic, Physiological and Hormonal Analysis of Potato Plants Under Drought Stress
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zheng, Y.; Chen, X.; Wang, P.; Sun, Y.; Yue, C.; Ye, N. Genome-wide and expression pattern analysis of JAZ family involved in stress responses and postharvest processing treatments in Camellia sinensis. Sci. Rep. 2020, 10, 2792. [Google Scholar] [CrossRef] [PubMed]
- Balbi, V.; Devoto, A. Jasmonate signalling network in Arabidopsis thaliana: Crucial regulatory nodes and new physiological scenarios. New Phytol. 2008, 177, 301–318. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Seong, K.; Pang, S.; Song, J.; Gao, H.; Wang, C.; Zhai, J.; Zhang, Y.; Gao, S.; Li, X.; et al. Functional specificity, diversity, and redundancy of Arabidopsis JAZ family repressors in Jasmonate and COI1-regulated growth, development, and defense. New Phytol. 2021, 231, 1525–1545. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Shang, L.; Li, Y.; Zhang, Q.; Chu, Z.; He, S.; Yang, W.; Ding, X. Ectopic Expression of OsJAZs Alters Plant Defense and Development. Int. J. Mol. Sci. 2022, 23, 4581. [Google Scholar] [CrossRef]
- Han, Y.; Luthe, D. Identification and evolution analysis of the JAZ gene family in maize. BMC Genom. 2021, 22, 256. [Google Scholar] [CrossRef]
- Zhai, Z.; Che, Y.; Geng, S.; Liu, S.; Zhang, S.; Cui, D.; Deng, Z.; Fu, M.; Li, Y.; Zou, X.; et al. Comprehensive Comparative Analysis of the JAZ Gene Family in Common Wheat (Triticum aestivum) and Its D-Subgenome Donor Aegilops tauschii. Plants 2024, 13, 1259. [Google Scholar] [CrossRef]
- Chini, A.; Ben-Romdhane, W.; Hassairi, A.; Aboul-Soud, M.A.M. Identification of TIFY/JAZ family genes in Solanum lycopersicum and their regulation in response to abiotic stresses. PLoS ONE 2017, 12, e0177381. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qiao, L.; Bai, J.; Wang, P.; Duan, W.; Yuan, S.; Yuan, G.; Zhang, F.; Zhang, L.; Zhao, C. Genome-wide characterization of Jasmonate—Zim domain transcription repressors in wheat (Triticum aestivum L.). BMC Genom. 2017, 18, 152. [Google Scholar] [CrossRef]
- Song, M.; Wang, H.; Ma, H.; Zheng, C. Genome-wide analysis of JAZ family genes expression patterns during fig (Ficus carica L.) fruit development and in response to hormone treatment. BMC Genom. 2022, 23, 170. [Google Scholar] [CrossRef]
- Bisht, N.; Anshu, A.; Singh, P.C.; Chauhan, P.S. Comprehensive analysis of OsJAZ gene family deciphers rhizobacteria-mediated nutrient stress modulation in rice. Int. J. Biol. Macromol. 2023, 253, 126832. [Google Scholar] [CrossRef]
- Zhang, B.; Zheng, H.; Wu, H.; Wang, C.; Liang, Z. Recent genome-wide replication promoted expansion and functional differentiation of the JAZs in soybeans. Int. J. Biol. Macromol. 2023, 238, 124064. [Google Scholar] [CrossRef] [PubMed]
- An, X.-H.; Hao, Y.-J.; Li, E.-M.; Xu, K.; Cheng, C.-G. Functional identification of apple MdJAZ2 in Arabidopsis with reduced JA-sensitivity and increased stress tolerance. Plant Cell Rep. 2017, 36, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Peethambaran, P.K.; Glenz, R.; Hoeninger, S.; Islam, S.M.S.; Hummel, S.; Harter, K.; Kolukisaoglu, U.; Meynard, D.; Guiderdoni, E.; Nick, P.; et al. Salt-inducible expression of OsJAZ8 improves resilience against salt-stress. BMC Plant Biol. 2018, 18, 311. [Google Scholar] [CrossRef]
- Thirumalaikumar, V.P.; Devkar, V.; Mehterov, N.; Ali, S.; Ozgur, R.; Turkan, I.; Mueller-Roeber, B.; Balazadeh, S. NAC transcription factor Jungbrunnen1 enhances drought tolerance in tomato. Plant Biotechnol. J. 2018, 16, 354–366. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, M.; Wang, Z.; Li, J.; Liu, K.; Huang, D. The role of arbuscular mycorrhizal symbiosis in plant abiotic stress. Front. Microbiol. 2024, 14, 1323881. [Google Scholar] [CrossRef]
- Guo, W.; Chen, L.; Herrera-Estrella, L.; Cao, D.; Lam-Son Phan, T. Altering Plant Architecture to Improve Performance and Resistance. Trends Plant Sci. 2020, 25, 1154–1170. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, P.; Augstein, F.; Nguyen, V.; Carlsbecker, A. Coping with Water Limitation: Hormones That Modify Plant Root Xylem Development. Front. Plant Sci. 2020, 11, 570. [Google Scholar] [CrossRef]
- Steffens, B.; Wang, J.; Sauter, M. Interactions between ethylene, gibberellin and abscisic acid regulate emergence and growth rate of adventitious roots in deepwater rice. Planta 2006, 223, 604–612. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, W.; Yan, M.; He, S.; Lu, Y.; Zhang, G.; Hao, B.; Fan, W.; Hu, Y.; Yang, S. Ectopic Expression of Erigeron breviscapus JAZ1 Affects JA-Induced Development Processes in Transgenic Arabidopsis. Plant Mol. Biol. Report. 2022, 40, 530–538. [Google Scholar] [CrossRef]
- DeMott, L.; Oblessuc, P.R.; Pierce, A.; Student, J.; Melotto, M. Spatiotemporal regulation of JAZ4 expression and splicing contribute to ethylene- and auxin-mediated responses in Arabidopsis roots. Plant J. 2021, 108, 1266–1282. [Google Scholar] [CrossRef]
- Pandey, B.K.; Verma, L.; Prusty, A.; Singh, A.P.; Bennett, M.J.; Tyagi, A.K.; Giri, J.; Mehra, P. OsJAZ11 regulates phosphate starvation responses in rice. Planta 2021, 254, 8. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.-C.; Hu, Q.-Q.; Li, W.; Chen, Y.; Han, L.-H.; Tao, M.; Wu, W.-Y.; Li, X.-B.; Huang, G.-Q. Cotton (Gossypium hirsutum) JAZ3 and SLR1 function in jasmonate and gibberellin mediated epidermal cell differentiation and elongation. Plant Cell Tissue Organ Cult. 2018, 133, 249–262. [Google Scholar] [CrossRef]
- Wang, X.-Y.; Zhu, N.-N.; Yang, J.-S.; Zhou, D.; Yuan, S.-T.; Pan, X.-J.; Jiang, C.-X.; Wu, Z.-G. CwJAZ4/9 negatively regulates jasmonate-mediated biosynthesis of terpenoids through interacting with CwMYC2 and confers salt tolerance in Curcuma wenyujin. Plant Cell Environ. 2024, 47, 3090–3110. [Google Scholar] [CrossRef]
- Fu, J.; Wu, H.; Ma, S.; Xiang, D.; Liu, R.; Xiong, L. OsJAZ1 Attenuates Drought Resistance by Regulating JA and ABA Signaling in Rice. Front. Plant Sci. 2017, 8, 2108. [Google Scholar] [CrossRef]
- Shewry, P.R.; Jones, H.D.; Halford, N.G. Plant biotechnology: Transgenic crops. Adv. Biochem. Eng. Biotechnol. 2008, 111, 149–186. [Google Scholar] [PubMed]
- Hajlaoui, H.; Akrimi, R.; Guesmi, A.; Djebali, N. Limiting Chemical Fertilization in Drought Stressed Potatoes (Solanum tuberosum L.) by Using Compost: Influence in Tuber Quality and Storability. J. Soil Sci. Plant Nutr. 2024, 24, 3026–3041. [Google Scholar] [CrossRef]
- Zhang, X.C.; Guo, J.; Ma, Y.F.; Yu, X.F.; Hou, H.Z.; Wang, H.L.; Fang, Y.J.; Tang, Y.F. Effects of vertical rotary subsoiling with plastic mulching on soil water availability and potato yield on a semiarid Loess plateau, China. Soil Tillage Res. 2020, 199, 104591. [Google Scholar] [CrossRef]
- Vain, P. Global trends in plant transgenic science and technology (1973–2003). Trends Biotechnol. 2006, 24, 206–211. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, F.; Li, S.; Feng, Y.; Yang, J.; Zhang, N.; Si, H. Calcium-Dependent Protein Kinase 28 Maintains Potato Photosynthesis and Its Tolerance under Water Deficiency and Osmotic Stress. Int. J. Mol. Sci. 2022, 23, 8795. [Google Scholar] [CrossRef]
- Lovas, A.; Bimbo, A.; Szabo, L.; Banfalvi, Z. Antisense repression of StubGAL83 affects root and tuber development in potato. Plant J. Cell Mol. Biol. 2003, 33, 139–147. [Google Scholar] [CrossRef]
- Bi, Z.; Dekomah, S.D.; Wang, Y.; Pu, Z.; Wang, X.; Dormatey, R.; Sun, C.; Liu, Y.; Liu, Z.; Bai, J.; et al. Overexpression of StCDPK13 in Potato Enhances Tolerance to Drought Stress. Int. J. Mol. Sci. 2024, 25, 12620. [Google Scholar] [CrossRef]
- Luo, H.; Yang, J.; Liu, S.; Li, S.; Si, H.; Zhang, N. Control of Plant Height and Lateral Root Development via Stu-miR156 Regulation of SPL9 Transcription Factor in Potato. Plants 2024, 13, 723. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Tang, X.; Zhang, N.; Si, H. StUBC13, a Ubiquitin-Conjugating Enzyme, Positively Regulates Salt and Osmotic Stresses in Potato. Int. J. Mol. Sci. 2024, 25, 13197. [Google Scholar] [CrossRef]
- Qin, T.; Wang, Y.; Pu, Z.; Shi, N.; Dormatey, R.; Wang, H.; Sun, C. Comprehensive Transcriptome and Proteome Analyses Reveal the Drought Responsive Gene Network in Potato Roots. Plants 2024, 13, 1530. [Google Scholar] [CrossRef]
- Loh, H.-S.; Green, B.J.; Yusibov, V. Using transgenic plants and modified plant viruses for the development of treatments for human diseases. Curr. Opin. Virol. 2017, 26, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Jungbauer, A. Transgenic crops and plant biotechnology. Biotechnol. J. 2010, 5, 135. [Google Scholar] [CrossRef] [PubMed]
- Jia, K.; Yan, C.; Zhang, J.; Cheng, Y.; Li, W.; Yan, H.; Gao, J. Genome-wide identification and expression analysis of the JAZ gene family in turnip. Sci. Rep. 2021, 11, 21330. [Google Scholar] [CrossRef]
- Shen, J.; Zou, Z.; Xing, H.; Duan, Y.; Zhu, X.; Ma, Y.; Wang, Y.; Fang, W. Genome-Wide Analysis Reveals Stress and Hormone Responsive Patterns of JAZ Family Genes in Camellia Sinensis. Int. J. Mol. Sci. 2020, 21, 2433. [Google Scholar] [CrossRef]
- Li, W.; Xia, X.-C.; Han, L.-H.; Ni, P.; Yan, J.-Q.; Tao, M.; Huang, G.-Q.; Li, X.-B. Genome-wide identification and characterization of JAZ gene family in upland cotton (Gossypium hirsutum). Sci. Rep. 2017, 7, 2788. [Google Scholar] [CrossRef]
- Zhou, S.-L.; Zhang, J.-X.; Jiang, S.; Lu, Y.; Huang, Y.-S.; Dong, X.-M.; Hu, Q.; Yao, W.; Zhang, M.-Q.; Xiao, S.-H. Genome-wide identification of JAZ gene family in sugarcane and function analysis of ScJAZ1/2 in drought stress response and flowering regulation. Plant Physiol. Biochem. 2024, 210, 108577. [Google Scholar] [CrossRef]
- Song, H.; Duan, Z.; Wang, Z.; Li, Y.; Wang, Y.; Li, C.; Mao, W.; Que, Q.; Chen, X.; Li, P. Genome-wide identification, expression pattern and subcellular localization analysis of the JAZ gene family in Toona ciliata. Ind. Crops Prod. 2022, 178, 114582. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, C.; Liu, Z.; Zhao, T.; Jiang, J.; Li, J.; Xu, X.; Yang, H. Genome-Wide Identification, Characterization and Expression Analysis of the JAZ Gene Family in Resistance to Gray Leaf Spots in Tomato. Int. J. Mol. Sci. 2021, 22, 9974. [Google Scholar] [CrossRef] [PubMed]
- Alabd, A.; Cheng, H.; Ahmad, M.; Wu, X.; Peng, L.; Wang, L.; Yang, S.; Bai, S.; Ni, J.; Teng, Y. Abre-Binding Factor3-Wrky Dna-Binding Protein44 module promotes salinity-induced malate accumulation in pear. Plant Physiol. 2023, 192, 1982–1996. [Google Scholar] [CrossRef]
- Huang, X.; Lu, Z.; Li, F.; Deng, Y.; Wan, F.; Wang, Q.; Folega, F.; Wang, J.; Guo, Z. Evolution history dominantly regulates fine root lifespan in tree species across the world. For. Ecosyst. 2024, 11, 100211. [Google Scholar] [CrossRef]
- Yamauchi, T.; Pedersen, O.; Nakazono, M.; Tsutsumi, N. Key root traits of Poaceae for adaptation to soil water gradients. New Phytol. 2021, 229, 3133–3140. [Google Scholar] [CrossRef]
- Zhang, T.; Zheng, D.; Zhang, C.; Wu, Z.; Yu, R.; Zhang, Z. Heterologous expression of ZmNF-YA12 confers tolerance to drought and salt stress in Arabidopsis. Plant Biotechnol. Rep. 2022, 16, 437–448. [Google Scholar] [CrossRef]
- Feng, Y.; Xu, P.; Li, B.; Li, P.; Wen, X.; An, F.; Gong, Y.; Xin, Y.; Zhu, Z.; Wang, Y.; et al. Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2017, 114, 13834–13839. [Google Scholar] [CrossRef]
- Pucciariello, C.; Banti, V.; Perata, P. ROS signaling as common element in low oxygen and heat stresses. Plant Physiol. Biochem. 2012, 59, 3–10. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, L.; Geng, B.; Feng, J.; Zhu, S. Interactive effects of abscisic acid and nitric oxide on chilling resistance and active oxygen metabolism in peach fruit during cold storage. J. Sci. Food Agric. 2019, 99, 3367–3380. [Google Scholar] [CrossRef]
- Rao, X.; Zhang, Y.; Gao, Y.; Zhao, L.; Wang, P. Influence of Exogenous Abscisic Acid on Germination and Physiological Traits of Sophora viciifolia Seedlings under Drought Conditions. Appl. Sci. 2024, 14, 4359. [Google Scholar] [CrossRef]
- Luo, J.; Li, M.; Ju, J.; Hai, H.; Wei, W.; Ling, P.; Li, D.; Su, J.; Zhang, X.; Wang, C. Genome-Wide Identification of the GhANN Gene Family and Functional Validation of GhANN11 and GhANN4 under Abiotic Stress. Int. J. Mol. Sci. 2024, 25, 1877. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.-L.; Min, Z.; Yue, X.-F.; Zhang, Y.-L.; Zhang, J.-X.; Zhang, Z.-Q.; Fang, Y.-L. Overexpression of grapevine VvNAC08 enhances drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2020, 151, 214–222. [Google Scholar] [CrossRef]
- Zhang, T.; Mo, J.; Zhou, K.; Chang, Y.; Liu, Z. Overexpression of Brassica campestris BcICE1 gene increases abiotic stress tolerance in tobacco. Plant Physiol. Biochem. 2018, 132, 515–523. [Google Scholar] [CrossRef] [PubMed]
- Che, B.; Cheng, C.; Fang, J.; Liu, Y.; Jiang, L.; Yu, B. The Recretohalophyte Tamarix TrSOS1 Gene Confers Enhanced Salt Tolerance to Transgenic Hairy Root Composite Cotton Seedlings Exhibiting Virus-Induced Gene Silencing of GhSOS1. Int. J. Mol. Sci. 2019, 20, 2930. [Google Scholar] [CrossRef]
- Xing, X.; Cao, C.; Xu, Z.; Qi, Y.; Fei, T.; Jiang, H.; Wang, X. Reduced Soybean Water Stress Tolerance by miR393a-Mediated Repression of GmTIR1 and Abscisic Acid Accumulation. J. Plant Growth Regul. 2023, 42, 1067–1083. [Google Scholar] [CrossRef]
- Li, X.; Huang, D.; Lin, X. Interlinked regulator loops of ABA and JA respond to salt and drought stress in Caragana korshinskii. Environ. Exp. Bot. 2024, 225, 105829. [Google Scholar] [CrossRef]
- Lim, C.W.; Baek, W.; Jung, J.; Kim, J.-H.; Lee, S.C. Function of ABA in Stomatal Defense against Biotic and Drought Stresses. Int. J. Mol. Sci. 2015, 16, 15251–15270. [Google Scholar] [CrossRef]
- Lin, Q.; Wang, J.; Gong, J.; Zhang, Z.; Wang, S.; Sun, J.; Li, Q.; Gu, X.; Jiang, J.; Qi, S. The Arabidopsis thaliana trehalose-6-phosphate phosphatase gene AtTPPI improve chilling tolerance through accumulating soluble sugar and JA. Environ. Exp. Bot. 2023, 205, 105117. [Google Scholar] [CrossRef]
- Zhu, H.; Zhai, H.; He, S.; Zhang, H.; Gao, S.; Liu, Q. A novel sweetpotato GATA transcription factor, IbGATA24, interacting with IbCOP9-5a positively regulates drought and salt tolerance. Environ. Exp. Bot. 2022, 194, 104735. [Google Scholar] [CrossRef]
- Liu, R.; Shu, B.; Wang, Y.; Feng, J.; Yu, B.; Gan, Y.; Liang, Y.; Qiu, Z.; Yan, S.; Cao, B. The Jasmonic Acid Biosynthetic Genes SmLOX4 and SmLOX5 Are Involved in Heat Tolerance in Eggplant. Plant Cell Physiol. 2024, 65, 1705–1716. [Google Scholar] [CrossRef]
- Xu, B.-Q.; Wang, J.-J.; Peng, Y.; Huang, H.; Sun, L.-L.; Yang, R.; Suo, L.-N.; Wang, S.-H.; Zhao, W.-C. SlMYC2 mediates stomatal movement in response to drought stress by repressing SlCHS1 expression. Front. Plant Sci. 2022, 13, 952758. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Chen, X.; Chen, Z.; Wang, H.; Shah, S.H.A.; Bai, A.; Liu, T.; Xiao, D.; Hou, X.; Li, Y. BcSRC2 interacts with BcAPX4 to increase ascorbic acid content for responding ABA signaling and drought stress in pak choi. Hortic. Res. 2024, 11, uhae165. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Tang, C.; Zhang, Y.; Sha, X.; Tian, S.; Luo, Z.; Wei, G.; Zhu, L.; Li, Y.; Fu, J.; et al. The SnRK2.2-ZmHsf28-JAZ14/17 module regulates drought tolerance in maize. New Phytol. 2024, 245, 1985–2003. [Google Scholar] [CrossRef]
- Kim, T.; Joo, H. ClustalXeed: A GUI-based grid computation version for high performance and terabyte size multiple sequence alignment. BMC Bioinform. 2010, 11, 467. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Chico, J.M.; Raices, M.; Tellez-Inon, M.T.; Ulloa, R.M. A calcium-dependent protein kinase is systemically induced upon wounding in tomato plants. Plant Physiol. 2002, 128, 256–270. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pu, Z.; Qin, T.; Wang, Y.; Wang, X.; Shi, N.; Yao, P.; Liu, Y.; Bai, J.; Bi, Z.; Sun, C. Genome-Wide Analysis of the JAZ Gene Family in Potato and Functional Verification of StJAZ23 Under Drought Stress. Int. J. Mol. Sci. 2025, 26, 2360. https://doi.org/10.3390/ijms26052360
Pu Z, Qin T, Wang Y, Wang X, Shi N, Yao P, Liu Y, Bai J, Bi Z, Sun C. Genome-Wide Analysis of the JAZ Gene Family in Potato and Functional Verification of StJAZ23 Under Drought Stress. International Journal of Molecular Sciences. 2025; 26(5):2360. https://doi.org/10.3390/ijms26052360
Chicago/Turabian StylePu, Zhuanfang, Tianyuan Qin, Yihao Wang, Xiangdong Wang, Ningfan Shi, Panfeng Yao, Yuhui Liu, Jiangping Bai, Zhenzhen Bi, and Chao Sun. 2025. "Genome-Wide Analysis of the JAZ Gene Family in Potato and Functional Verification of StJAZ23 Under Drought Stress" International Journal of Molecular Sciences 26, no. 5: 2360. https://doi.org/10.3390/ijms26052360
APA StylePu, Z., Qin, T., Wang, Y., Wang, X., Shi, N., Yao, P., Liu, Y., Bai, J., Bi, Z., & Sun, C. (2025). Genome-Wide Analysis of the JAZ Gene Family in Potato and Functional Verification of StJAZ23 Under Drought Stress. International Journal of Molecular Sciences, 26(5), 2360. https://doi.org/10.3390/ijms26052360






