Establishment of a Yeast Two-Hybrid-Based High-Throughput Screening Model for Selection of SARS-CoV-2 Spike-ACE2 Interaction Inhibitors
Abstract
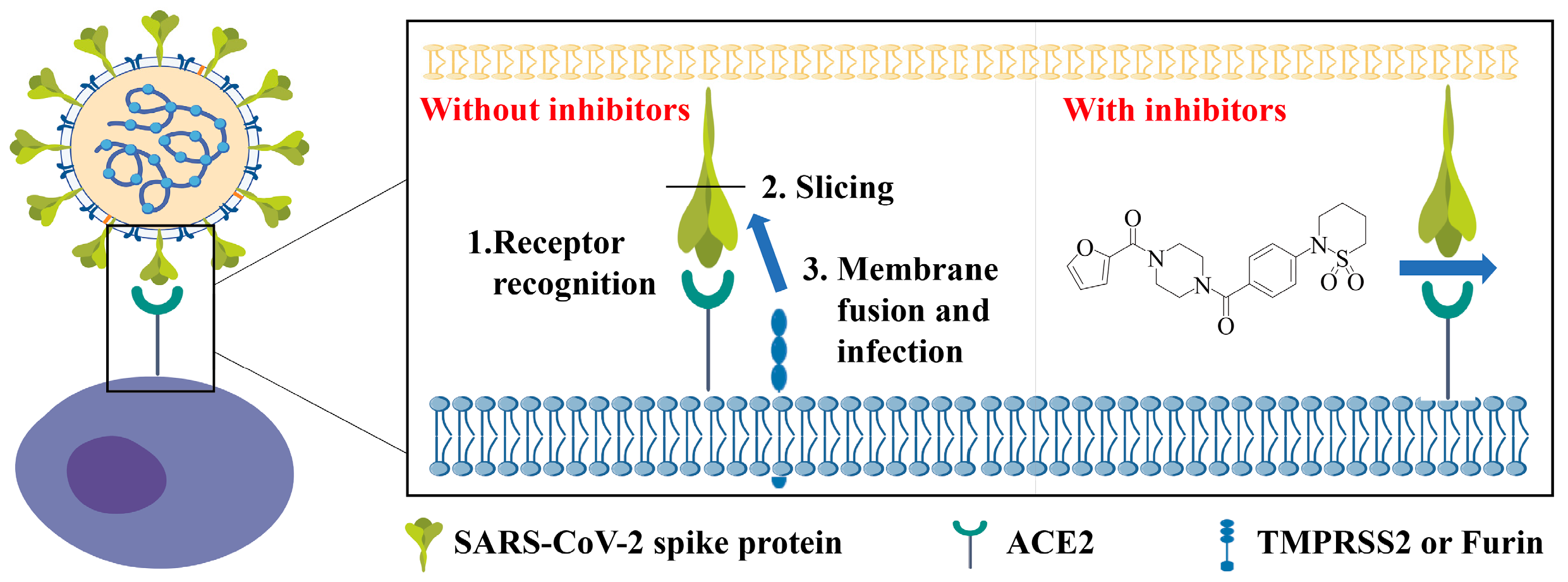
1. Introduction
2. Results
2.1. SARS-CoV-2 S-RBD Interacted with ACE2 in the Y2H Assay
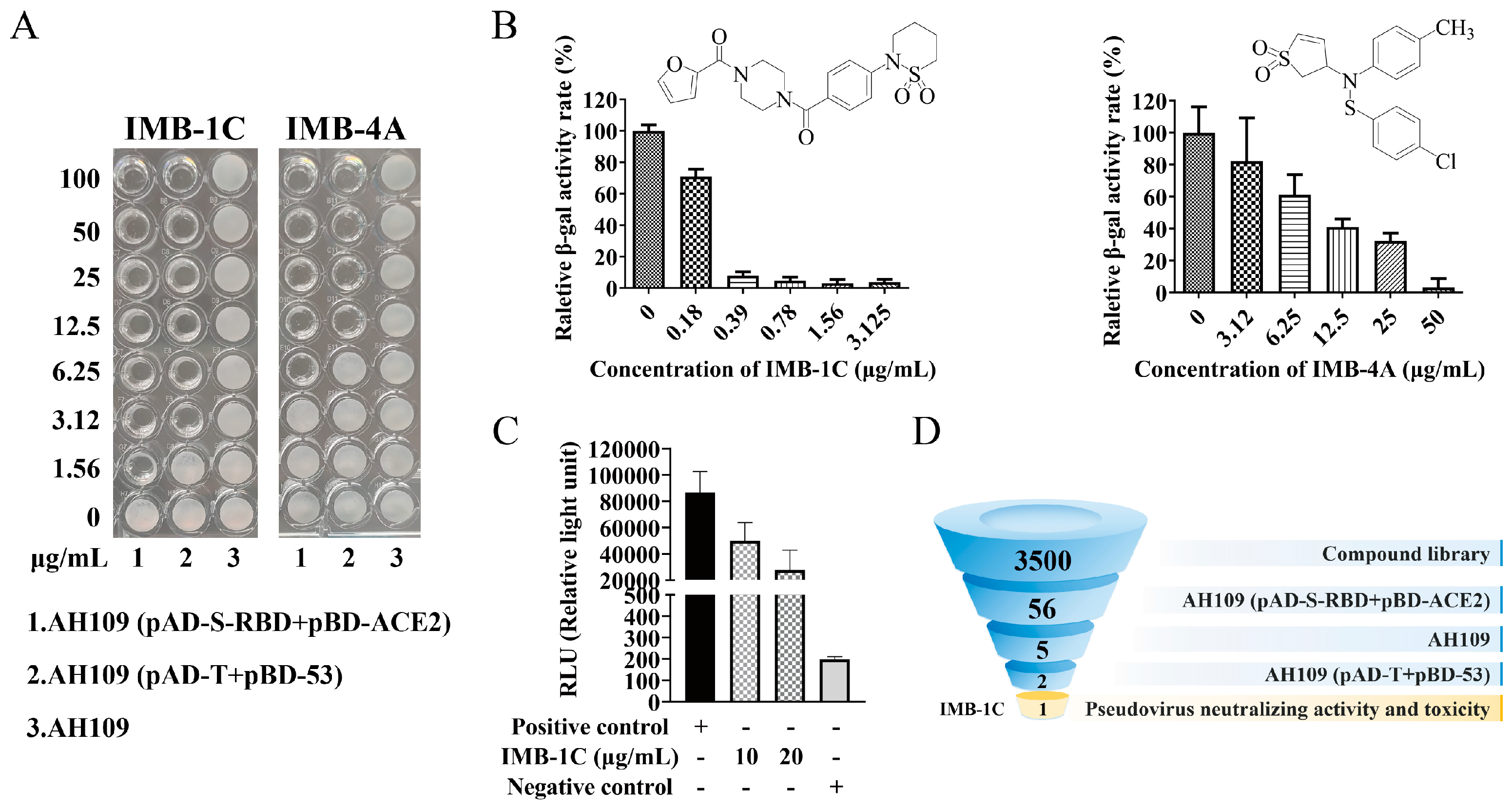
2.2. HTS for Entry Inhibitors from a Compound Library
2.3. Inhibition of SARS-CoV-2 Pseudovirus Viral Entry
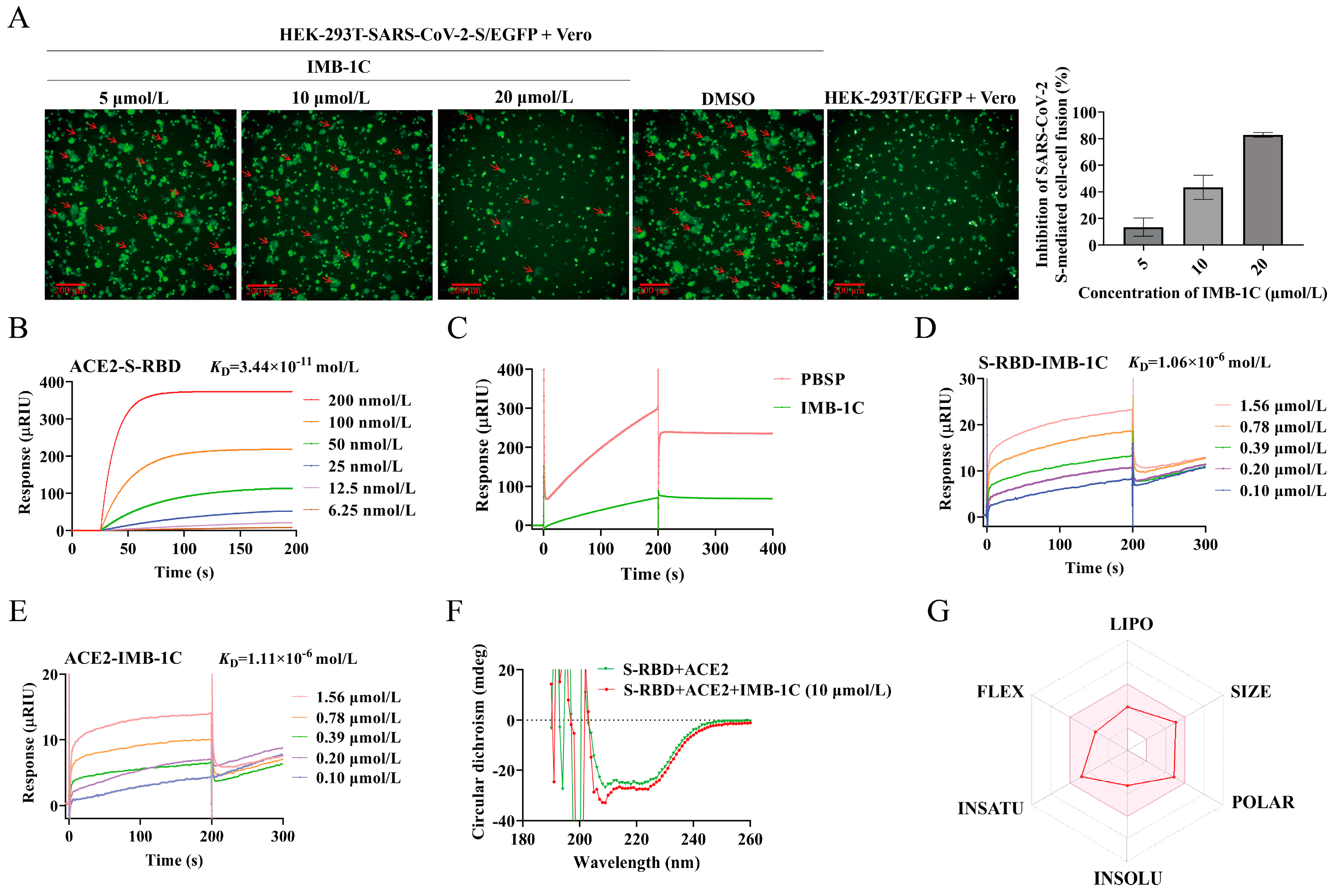
2.4. IMB-1C Effectively Inhibited SARS-CoV-2 S-Mediated Cell–Cell Fusion
2.5. IMB-1C Blocked S-RBD-ACE2 Interaction Through Binding with Both S-RBD and ACE2
2.6. Circular Dichroism (CD) Spectroscopy Assay
2.7. Druggability Evaluation of IMB-1C
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Construction of Y2H Assay
4.3. β-Gal Colony Filter Lift Assay
4.4. ONPG Assay
4.5. Western Blot Analysis
4.6. HTS Assay
4.7. Pseudovirus Entry Assay
4.8. In Vitro Cytotoxicity Test
4.9. Assessment of S Protein-Mediated Inhibition of Cell–Cell Fusion
4.10. SPR Analysis
4.10.1. Detection of ACE2-S-RBD Interaction
4.10.2. Blockage of ACE2-S-RBD Interaction by IMB-1C
4.10.3. Measurement for the Binding Affinity Between IMB-1C and ACE2 or S-RBD
4.11. CD Spectroscopy Assay
4.12. SwissADMET Prediction for IMB-1C
4.13. Statistical Analysis
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Drosten, C.; Gunther, S.; Preiser, W.; van der Werf, S.; Brodt, H.R.; Becker, S.; Rabenau, H.; Panning, M.; Kolesnikova, L.; Fouchier, R.A.; et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348, 1967–1976. [Google Scholar] [CrossRef]
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Lin, D.Y.; Gu, Y.; Wheeler, B.; Young, H.; Holloway, S.; Sunny, S.K.; Moore, Z.; Zeng, D. Effectiveness of Covid-19 Vaccines over a 9-Month Period in North Carolina. N. Engl. J. Med. 2022, 386, 933–941. [Google Scholar] [CrossRef]
- Acharya, K.P.; Ghimire, T.R.; Subramanya, S.H. Access to and equitable distribution of COVID-19 vaccine in low-income countries. npj Vaccines 2021, 6, 54. [Google Scholar] [CrossRef]
- Ekwebelem, O.C.; Yunusa, I.; Onyeaka, H.; Ekwebelem, N.C.; Nnorom-Dike, O. COVID-19 vaccine rollout: Will it affect the rates of vaccine hesitancy in Africa? Public Health 2021, 197, e18–e19. [Google Scholar] [CrossRef]
- Tortorici, M.A.; Veesler, D. Structural insights into coronavirus entry. Adv. Virus Res. 2019, 105, 93–116. [Google Scholar] [PubMed]
- Sun, H.; Xiao, Y.; Liu, J.; Wang, D.; Li, F.; Wang, C.; Li, C.; Zhu, J.; Song, J.; Sun, H.; et al. Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection. Proc. Natl. Acad. Sci. USA 2020, 117, 17204–17210. [Google Scholar] [CrossRef] [PubMed]
- Xiu, S.; Dick, A.; Ju, H.; Mirzaie, S.; Abdi, F.; Cocklin, S.; Zhan, P.; Liu, X. Inhibitors of SARS-CoV-2 Entry: Current and Future Opportunities. J. Med. Chem. 2020, 63, 12256–12274. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.Y.; Wu, S.L.; Chen, J.C.; Wei, Y.C.; Cheng, S.E.; Chang, Y.H.; Liu, H.J.; Hsiang, C.Y. Design and biological activities of novel inhibitory peptides for SARS-CoV spike protein and angiotensin-converting enzyme 2 interaction. Antivir. Res. 2006, 69, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Arabi, Y.; Balkhy, H.; Hajeer, A.H.; Bouchama, A.; Hayden, F.G.; Al-Omari, A.; Al-Hameed, F.M.; Taha, Y.; Shindo, N.; Whitehead, J.; et al. Feasibility, safety, clinical, and laboratory effects of convalescent plasma therapy for patients with Middle East respiratory syndrome coronavirus infection: A study protocol. Springerplus 2015, 4, 709. [Google Scholar] [CrossRef]
- Wu, A.; Shi, K.; Wang, J.; Zhang, R.; Wang, Y. Targeting SARS-CoV-2 entry processes: The promising potential and future of host-targeted small-molecule inhibitors. Eur. J. Med. Chem. 2024, 263, 115923. [Google Scholar] [CrossRef] [PubMed]
- Alexpandi, R.; De Mesquita, J.F.; Pandian, S.K.; Ravi, A.V. Quinolines-Based SARS-CoV-2 3CLpro and RdRp Inhibitors and Spike-RBD-ACE2 Inhibitor for Drug-Repurposing Against COVID-19: An in silico Analysis. Front. Microbiol. 2020, 11, 1796. [Google Scholar] [CrossRef] [PubMed]
- Ngoepe, M.P.; Tapala, K.C.; Clayton, H.S. Prophylactic and Therapeutic Potential Zinc Metallodrugs Drug Discovery: Identification of SARS-CoV-2 Replication and Spike/ACE2 Inhibitors. Curr. Comput. Aided Drug Des. 2022, 18, 519–534. [Google Scholar] [CrossRef]
- Hakmi, M.; Bouricha, E.L.M.; Akachar, J.; Lmimouni, B.; El Harti, J.; Belyamani, L.; Ibrahimi, A. In silico exploration of small-molecule alpha-helix mimetics as inhibitors of SARS-COV-2 attachment to ACE2. J. Biomol. Struct. Dyn. 2022, 40, 1546–1557. [Google Scholar] [CrossRef]
- Khattab, A.R.; Teleb, M. In silico discovery of non-psychoactive scaffolds in Cannabis halting SARS-CoV-2 host entry and replication machinery. Future Virol. 2022, 17, 367–386. [Google Scholar] [CrossRef] [PubMed]
- Gollapalli, P.; S, S.B.; Rimac, H.; Patil, P.; Nalilu, S.K.; Kandagalla, S.; Shetty, P. Pathway enrichment analysis of virus-host interactome and prioritization of novel compounds targeting the spike glycoprotein receptor binding domain-human angiotensin-converting enzyme 2 interface to combat SARS-CoV-2. J. Biomol. Struct. Dyn. 2022, 40, 2701–2714. [Google Scholar] [CrossRef] [PubMed]
- Bojadzic, D.; Alcazar, O.; Chen, J.; Chuang, S.T.; Condor Capcha, J.M.; Shehadeh, L.A.; Buchwald, P. Small-Molecule Inhibitors of the Coronavirus Spike: ACE2 Protein-Protein Interaction as Blockers of Viral Attachment and Entry for SARS-CoV-2. ACS Infect. Dis. 2021, 7, 1519–1534. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Gao, C.; Das, T.; Luo, S.; Tang, H.; Yao, X.; Cho, C.Y.; Lv, J.; Maravillas, K.; Jones, V.; et al. The spike-ACE2 binding assay: An in vitro platform for evaluating vaccination efficacy and for screening SARS-CoV-2 inhibitors and neutralizing antibodies. J. Immunol. Methods 2022, 503, 113244. [Google Scholar] [CrossRef] [PubMed]
- Emam, M.H.; Mahmoud, M.I.; El-Guendy, N.; Loutfy, S.A. Establishment of in-house assay for screening of anti-SARS-CoV-2 protein inhibitors. AMB Express 2024, 14, 104. [Google Scholar] [CrossRef]
- Alves, J.; Engel, L.; de Vasconcelos Cabral, R.; Rodrigues, E.L.; de Jesus Ribeiro, L.; Higa, L.M.; da Costa Ferreira Junior, O.; Castineiras, T.; de Carvalho Leitao, I.; Tanuri, A.; et al. A bioluminescent and homogeneous SARS-CoV-2 spike RBD and hACE2 interaction assay for antiviral screening and monitoring patient neutralizing antibody levels. Sci. Rep. 2021, 11, 18428. [Google Scholar] [CrossRef] [PubMed]
- Trischitta, P.; Tamburello, M.P.; Venuti, A.; Pennisi, R. Pseudovirus-Based Systems for Screening Natural Antiviral Agents: A Comprehensive Review. Int. J. Mol. Sci. 2024, 25, 5188. [Google Scholar] [CrossRef] [PubMed]
- MacFarlane, S.A.; Uhrig, J.F. Yeast two-hybrid assay to identify host-virus interactions. Methods Mol. Biol. 2008, 451, 649–672. [Google Scholar]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Serebriiskii, I.G.; Golemis, E.A. Uses of lacZ to study gene function: Evaluation of beta-galactosidase assays employed in the yeast two-hybrid system. Anal. Biochem. 2000, 285, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Guimond, S.E.; Mycroft-West, C.J.; Gandhi, N.S.; Tree, J.A.; Le, T.T.; Spalluto, C.M.; Humbert, M.V.; Buttigieg, K.R.; Coombes, N.; Elmore, M.J.; et al. Synthetic Heparan Sulfate Mimetic Pixatimod (PG545) Potently Inhibits SARS-CoV-2 by Disrupting the Spike-ACE2 Interaction. ACS Cent. Sci. 2022, 8, 527–545. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, H.; Tian, L.; Pang, Z.; Yang, Q.; Huang, T.; Fan, J.; Song, L.; Tong, Y.; Fan, H. COVID-19 vaccine development: Milestones, lessons and prospects. Signal Transduct. Target. Ther. 2022, 7, 146. [Google Scholar] [CrossRef]
- Yang, H.; Rao, Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol. 2021, 19, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Guy, R.K.; DiPaola, R.S.; Romanelli, F.; Dutch, R.E. Rapid repurposing of drugs for COVID-19. Science 2020, 368, 829–830. [Google Scholar] [CrossRef]
- Maldonado, L.L.; Bertelli, A.M.; Kamenetzky, L. Molecular features similarities between SARS-CoV-2, SARS, MERS and key human genes could favour the viral infections and trigger collateral effects. Sci. Rep. 2021, 11, 4108. [Google Scholar] [CrossRef]
- Campos, M.F.; Mendonca, S.C.; Penaloza, E.M.C.; de Oliveira, B.A.C.; Rosa, A.S.; Leitao, G.G.; Tucci, A.R.; Ferreira, V.N.S.; Oliveira, T.K.F.; Miranda, M.D.; et al. Anti-SARS-CoV-2 Activity of Ampelozizyphus amazonicus (Saracura-Mira): Focus on the Modulation of the Spike-ACE2 Interaction by Chemically Characterized Bark Extracts by LC-DAD-APCI-MS/MS. Molecules 2023, 28, 3159. [Google Scholar] [CrossRef] [PubMed]
- Bojadzic, D.; Alcazar, O.; Buchwald, P. Methylene Blue Inhibits the SARS-CoV-2 Spike-ACE2 Protein-Protein Interaction-a Mechanism that can Contribute to its Antiviral Activity Against COVID-19. Front. Pharmacol. 2020, 11, 600372. [Google Scholar] [CrossRef] [PubMed]
- Leader, B.; Baca, Q.J.; Golan, D.E. Protein therapeutics: A summary and pharmacological classification. Nat. Rev. Drug Discov. 2008, 7, 21–39. [Google Scholar] [CrossRef]
- Suntharalingam, G.; Perry, M.R.; Ward, S.; Brett, S.J.; Castello-Cortes, A.; Brunner, M.D.; Panoskaltsis, N. Cytokine storm in a phase 1 trial of the anti-CD28 monoclonal antibody TGN1412. N. Engl. J. Med. 2006, 355, 1018–1028. [Google Scholar] [CrossRef] [PubMed]
- Wadman, M. London’s disastrous drug trial has serious side effects for research. Nature 2006, 440, 388–389. [Google Scholar] [CrossRef] [PubMed]
- Huck, B.R.; Kotzner, L.; Urbahns, K. Small Molecules Drive Big Improvements in Immuno-Oncology Therapies. Angew. Chem. Int. Ed. Engl. 2018, 57, 4412–4428. [Google Scholar] [CrossRef] [PubMed]
- Downing, N.S.; Shah, N.D.; Aminawung, J.A.; Pease, A.M.; Zeitoun, J.D.; Krumholz, H.M.; Ross, J.S. Postmarket Safety Events Among Novel Therapeutics Approved by the US Food and Drug Administration Between 2001 and 2010. JAMA 2017, 317, 1854–1863. [Google Scholar] [CrossRef] [PubMed]
- Makurvet, F.D. Biologics vs. small molecules: Drug costs and patient access. Med. Drug Discov. 2020, 9, 100075. [Google Scholar] [CrossRef]
- Bekono, B.D.; Onguene, P.A.; Simoben, C.V.; Owono, L.C.O.; Ntie-Kang, F. Computational discovery of dual potential inhibitors of SARS-CoV-2 spike/ACE2 and M(pro): 3D-pharmacophore, docking-based virtual screening, quantum mechanics and molecular dynamics. Eur. Biophys. J. 2024, 53, 277–298. [Google Scholar] [CrossRef]
- Perrella, F.; Coppola, F.; Petrone, A.; Platella, C.; Montesarchio, D.; Stringaro, A.; Ravagnan, G.; Fuggetta, M.P.; Rega, N.; Musumeci, D. Interference of Polydatin/Resveratrol in the ACE2: Spike Recognition during COVID-19 Infection. A Focus on Their Potential Mechanism of Action through Computational and Biochemical Assays. Biomolecules 2021, 11, 1048. [Google Scholar] [CrossRef] [PubMed]
- da Costa, K.S.; Galucio, J.M.; da Costa, C.H.S.; Santana, A.R.; Dos Santos Carvalho, V.; do Nascimento, L.D.; Lima, E.L.A.H.; Neves Cruz, J.; Alves, C.N.; Lameira, J. Exploring the Potentiality of Natural Products from Essential Oils as Inhibitors of Odorant-Binding Proteins: A Structure- and Ligand-Based Virtual Screening Approach to Find Novel Mosquito Repellents. ACS Omega 2019, 4, 22475–22486. [Google Scholar] [CrossRef]
- Pinto, V.S.; Araujo, J.S.C.; Silva, R.C.; da Costa, G.V.; Cruz, J.N.; De, A.N.M.F.; Campos, J.M.; Santos, C.B.R.; Leite, F.H.A.; Junior, M.C.S. In Silico Study to Identify New Antituberculosis Molecules from Natural Sources by Hierarchical Virtual Screening and Molecular Dynamics Simulations. Pharmaceuticals 2019, 12, 36. [Google Scholar] [CrossRef]
- Ramos, R.S.; Macedo, W.J.C.; Costa, J.S.; da Silva, C.; Rosa, J.M.C.; da Cruz, J.N.; de Oliveira, M.S.; de Aguiar Andrade, E.H.; e Silva, R.B.L.; Souto, R.N.P.; et al. Potential inhibitors of the enzyme acetylcholinesterase and juvenile hormone with insecticidal activity: Study of the binding mode via docking and molecular dynamics simulations. J. Biomol. Struct. Dyn. 2020, 38, 4687–4709. [Google Scholar] [CrossRef]
- Araujo, P.H.F.; Ramos, R.S.; da Cruz, J.N.; Silva, S.G.; Ferreira, E.F.B.; de Lima, L.R.; Macedo, W.J.C.; Espejo-Roman, J.M.; Campos, J.M.; Santos, C.B.R. Identification of Potential COX-2 Inhibitors for the Treatment of Inflammatory Diseases Using Molecular Modeling Approaches. Molecules 2020, 25, 4183. [Google Scholar] [CrossRef] [PubMed]
- Rego, C.M.A.; Francisco, A.F.; Boeno, C.N.; Paloschi, M.V.; Lopes, J.A.; Silva, M.D.S.; Santana, H.M.; Serrath, S.N.; Rodrigues, J.E.; Lemos, C.T.L.; et al. Inflammasome NLRP3 activation induced by Convulxin, a C-type lectin-like isolated from Crotalus durissus terrificus snake venom. Sci. Rep. 2022, 12, 4706. [Google Scholar] [CrossRef] [PubMed]
- Neves Cruz, J.; da Costa, K.S.; de Carvalho, T.A.A.; de Alencar, N.A.N. Measuring the structural impact of mutations on cytochrome P450 21A2, the major steroid 21-hydroxylase related to congenital adrenal hyperplasia. J. Biomol. Struct. Dyn. 2020, 38, 1425–1434. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Yang, Y.; Wu, Y.; Huang, C.; Ding, Y.; Wang, X.; Wang, S. An optimized and robust SARS-CoV-2 pseudovirus system for viral entry research. J. Virol. Methods 2021, 295, 114221. [Google Scholar] [CrossRef]
- Lentze, N.; Auerbach, D. The yeast two-hybrid system and its role in drug discovery. Expert. Opin. Ther. Targets 2008, 12, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, C.; Zhu, N.; Lin, Y.; Jiang, J.; Wang, Y.; Li, Y.; Si, S. Identification of anti-Gram-negative bacteria agents targeting the interaction between ribosomal proteins L12 and L10. Acta Pharm. Sin. B 2018, 8, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yang, C.; Xu, X.F.; Xu, W.; Liu, S.W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, X.; Zhang, J.; Lin, Y.; You, X.; Chen, M.; Wang, Y.; Zhu, N.; Si, S. Identification of a Compound That Inhibits the Growth of Gram-Negative Bacteria by Blocking BamA-BamD Interaction. Front. Microbiol. 2020, 11, 1252. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; You, B.; Zhao, X.; Si, S.; Li, Y.; Zhang, J. Establishment, optimization and validation of a fluorescence polarization-based high-throughput screening assay targeting cathepsin L inhibitors. SLAS Discov. 2024, 29, 100153. [Google Scholar] [CrossRef] [PubMed]

| MIC (μg/mL) | IMB-1C | IMB-4A |
|---|---|---|
| AH109 (pAD-S-RBD+pBD-ACE2) | 1.56 | 6.25 |
| AH109 (pAD-T+pBD-53) | 3.125 | 12.5 |
| Survival Rate (%) | Vero | HEK-293T | Huh-7 |
|---|---|---|---|
| IMB-1C | 105 | 88 | 122 |
| IMB-4A | 71 | 18 | 25 |
| Sample | Secondary Structure Content (%) | |||
|---|---|---|---|---|
| α-Helix | β-Fold | β-Turn | Random | |
| S-RBD+ACE2 | 6.27 | 51.53 | 16.80 | 29.53 |
| S-RBD+ACE2+IMB-1C | 13.12 | 42.07 | 15.97 | 32.93 |
| Lipinski’s Rules | GI Absorption | Bioavailability | BBB Permeant | P-gb Substrate | CYP Enzymes’ Inhibitors |
|---|---|---|---|---|---|
| Yes | High | 0.55 | No | Yes | CYP2D6 (−), CYP3A4 (−), CYP2C19 (+), CYP2C9 (+), CYP3A4 (+) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, D.; You, B.; Guo, K.; Zhou, W.; Li, Y.; Wang, C.; Chen, X.; Wang, Z.; Zhang, J.; Si, S. Establishment of a Yeast Two-Hybrid-Based High-Throughput Screening Model for Selection of SARS-CoV-2 Spike-ACE2 Interaction Inhibitors. Int. J. Mol. Sci. 2025, 26, 678. https://doi.org/10.3390/ijms26020678
Li D, You B, Guo K, Zhou W, Li Y, Wang C, Chen X, Wang Z, Zhang J, Si S. Establishment of a Yeast Two-Hybrid-Based High-Throughput Screening Model for Selection of SARS-CoV-2 Spike-ACE2 Interaction Inhibitors. International Journal of Molecular Sciences. 2025; 26(2):678. https://doi.org/10.3390/ijms26020678
Chicago/Turabian StyleLi, Dongsheng, Baoqing You, Keyu Guo, Wenwen Zhou, Yan Li, Chenyin Wang, Xiaofang Chen, Zhen Wang, Jing Zhang, and Shuyi Si. 2025. "Establishment of a Yeast Two-Hybrid-Based High-Throughput Screening Model for Selection of SARS-CoV-2 Spike-ACE2 Interaction Inhibitors" International Journal of Molecular Sciences 26, no. 2: 678. https://doi.org/10.3390/ijms26020678
APA StyleLi, D., You, B., Guo, K., Zhou, W., Li, Y., Wang, C., Chen, X., Wang, Z., Zhang, J., & Si, S. (2025). Establishment of a Yeast Two-Hybrid-Based High-Throughput Screening Model for Selection of SARS-CoV-2 Spike-ACE2 Interaction Inhibitors. International Journal of Molecular Sciences, 26(2), 678. https://doi.org/10.3390/ijms26020678






