Identification of Small RNAs in Streptomyces clavuligerus Using High-Resolution Transcriptomics and Expression Profiling During Clavulanic Acid Production
Abstract
:1. Introduction
2. Results
2.1. In Silico Prediction of Small RNAs
2.1.1. General Features of Intergenic Regions in Streptomyces clavuligerus
2.1.2. Conserved IGRs in S. clavuligerus Are Enriched for sRNAs
2.2. High-Resolution Transcriptome Assembly
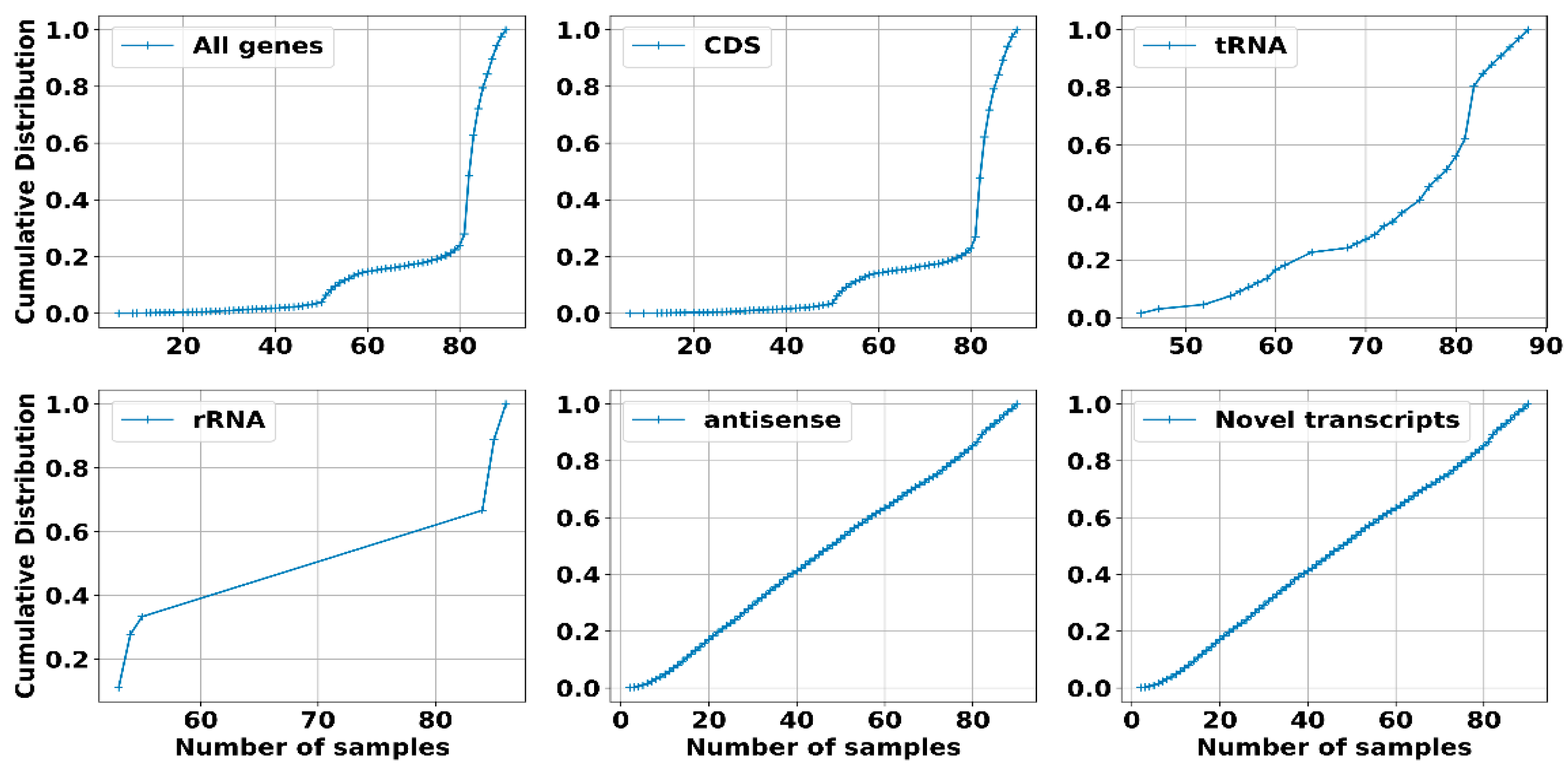
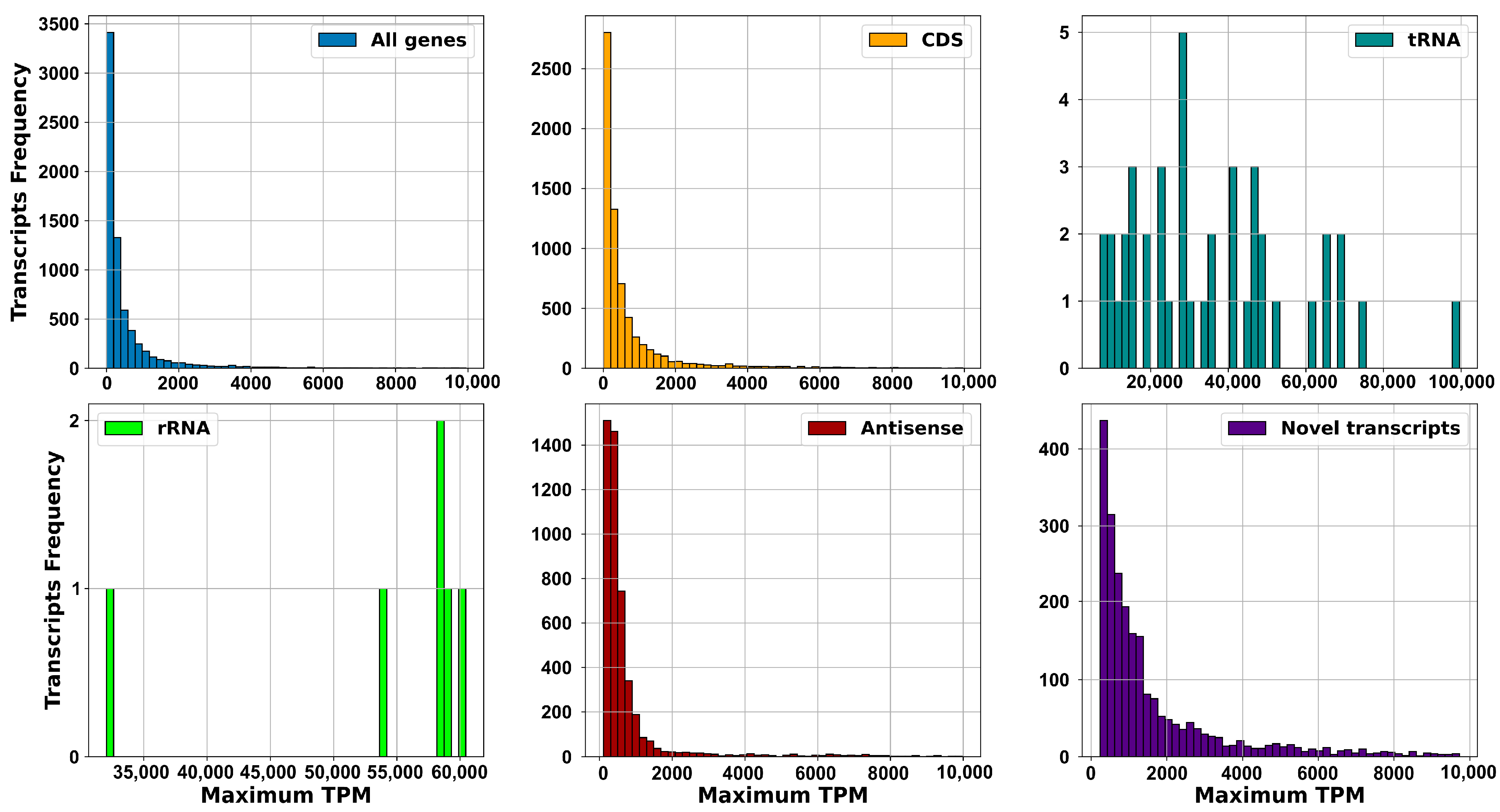
2.2.1. Dataset, Transcriptome Assembly and Expression: sRNAs Have Expression Levels That Are Strongly Dependent on the Specific Environmental Conditions in Which the RNA Was Extracted
2.2.2. Operons Prediction: The Multiple RNA-Seq Data Sets Used in This Study, Capturing the Complete Transcriptional Dynamics of S. clavuligerus
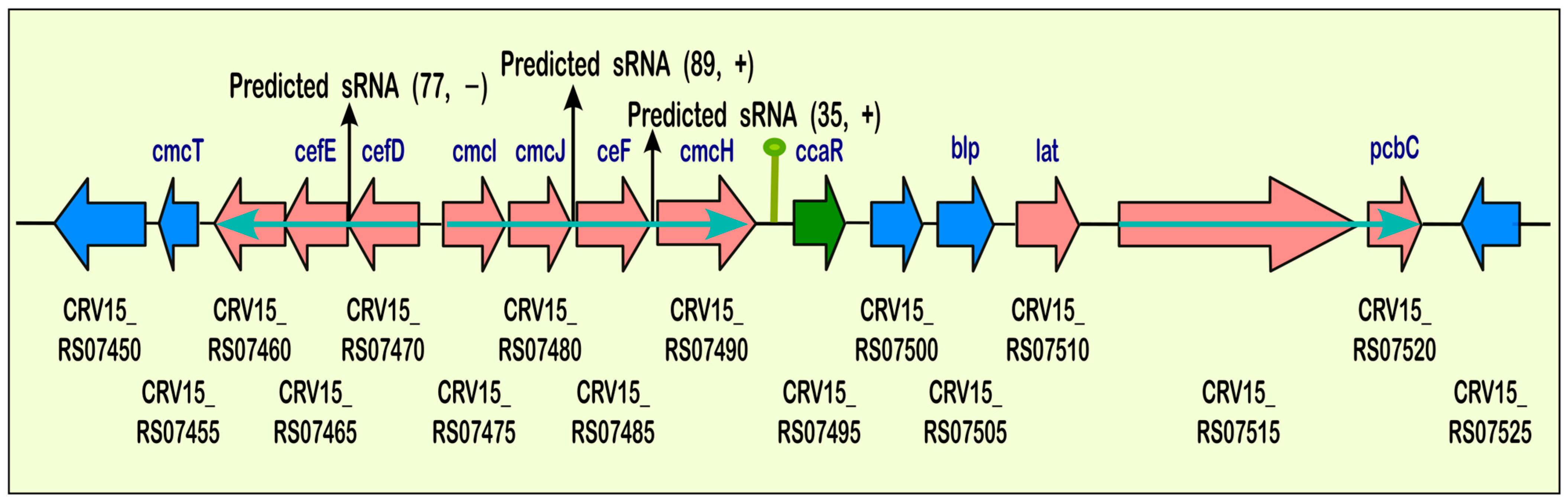
2.2.3. Small RNAs Identification: Multiple sRNAs Are Expressed in the Cephamycin C Cluster
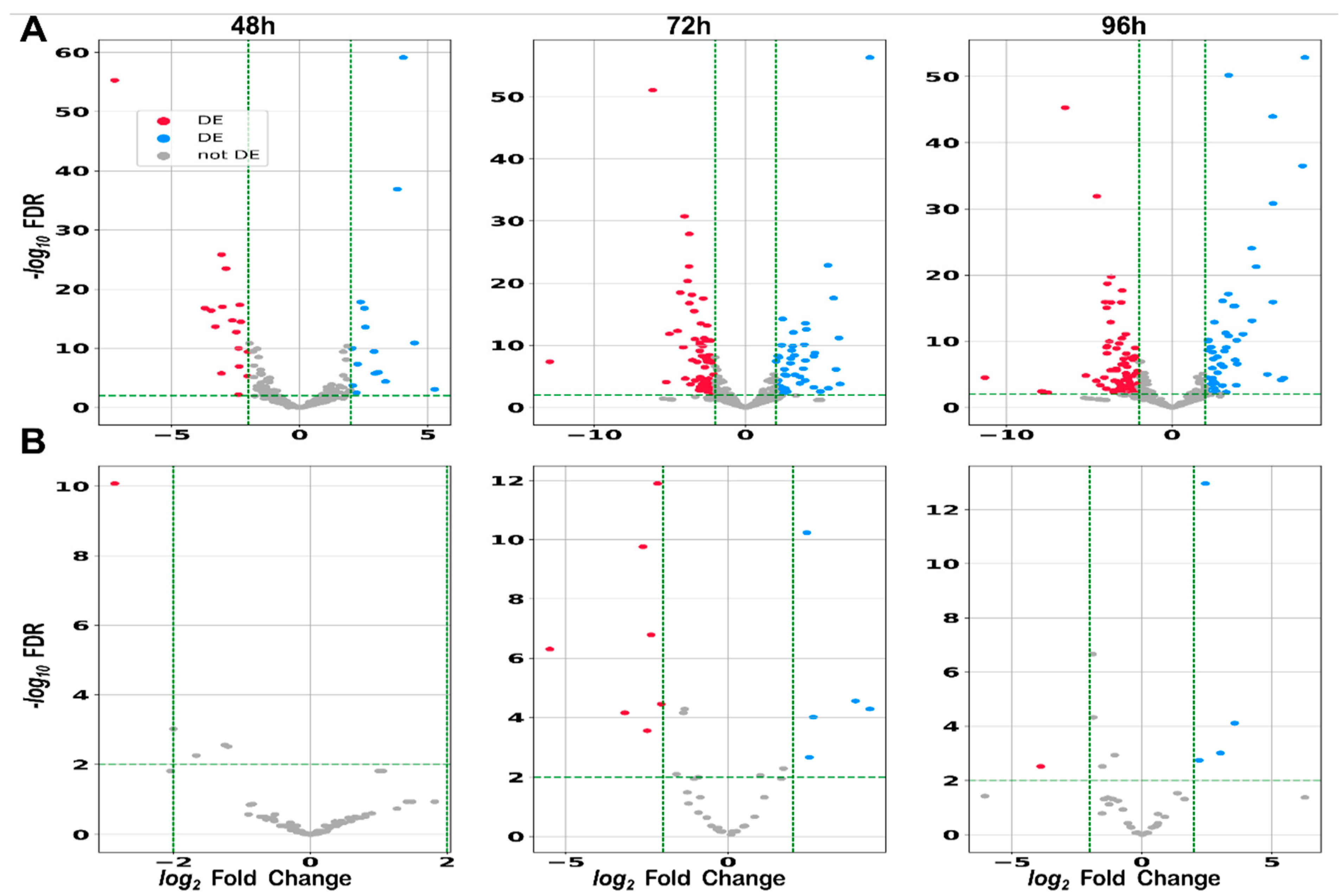
2.2.4. Differential Expression: sRNAs Can Act as Global Regulators in S. clavuligerus
3. Discussion
4. Materials and Methods
4.1. In Silico Prediction of Small RNAs
4.1.1. Intergenic Regions Extraction
4.1.2. Conservation Analysis of IGRs
4.1.3. Detection of Structured Non-Coding RNAs and Transcriptional Signals
4.1.4. Search for Conserved IGRs in the RFAM Database
4.1.5. Statistical Analysis
4.2. High-Resolution Transcriptome Assembly
4.2.1. Bacterial Strains and Culture Conditions
4.2.2. CA Determination
4.2.3. RNA Extraction and Sequencing
4.2.4. Data Collection
4.2.5. Transcriptome Characterization
4.3. Expression Profile of sRNAs During CA Production
4.3.1. Small RNA Detection by APERO
4.3.2. Differential Expression
4.3.3. Small RNAs Interactome
4.3.4. Total RNA Analysis
4.3.5. Code Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wade, J.T.; Grainger, D.C. Pervasive Transcription: Illuminating the Dark Matter of Bacterial Transcriptomes. Nat. Rev. Microbiol. 2014, 12, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Findeiß, S.; Washietl, S.; Hofacker, I.L.; Stadler, P.F. RNAz 2.0: Improved Noncoding Rna Detection. In Biocomputing 2010; World Scientific: Singapore, 2010; pp. 69–79. ISBN 978-981-4295-29-1. [Google Scholar]
- Wadler, C.S.; Vanderpool, C.K. A Dual Function for a Bacterial Small RNA: SgrS Performs Base Pairing-Dependent Regulation and Encodes a Functional Polypeptide. Proc. Natl. Acad. Sci. USA 2007, 104, 20454–20459. [Google Scholar] [CrossRef] [PubMed]
- Schnoor, S.B.; Neubauer, P.; Gimpel, M. Recent Insights into the World of Dual-function Bacterial. WIREs RNA 2024, 15, e1824. [Google Scholar] [CrossRef]
- Uguru, G.C.; Mondhe, M.; Goh, S.; Hesketh, A.; Bibb, M.J.; Good, L.; Stach, J.E.M. Synthetic RNA Silencing of Actinorhodin Biosynthesis in Streptomyces Coelicolor A3(2). PLoS ONE 2013, 8, e67509. [Google Scholar] [CrossRef] [PubMed]
- Moody, M.J.; Young, R.A.; Jones, S.E.; Elliot, M.A. Comparative Analysis of Non-Coding RNAs in the Antibiotic-Producing Streptomyces Bacteria. BMC Genom. 2013, 14, 558. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.G.H.; Romby, P. Small RNAs in Bacteria and Archaea. In Advances in Genetics; Elsevier: Amsterdam, The Netherlands, 2015; Volume 90, pp. 133–208. ISBN 978-0-12-803694-5. [Google Scholar]
- Dutta, T.; Srivastava, S. Small RNA-Mediated Regulation in Bacteria: A Growing Palette of Diverse Mechanisms. Gene 2018, 656, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Hindra; Moody, M.J.; Jones, S.E.; Elliot, M.A. Complex Intra-Operonic Dynamics Mediated by a Small RNA in Streptomyces Coelicolor. PLoS ONE 2014, 9, e85856. [Google Scholar] [CrossRef] [PubMed]
- Levine, E.; Hwa, T. Small RNAs Establish Gene Expression Thresholds. Curr. Opin. Microbiol. 2008, 11, 574–579. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Melamed, S. Small RNAs, Large Networks: Posttranscriptional Regulons in Gram-Negative Bacteria. Annu. Rev. Microbiol. 2023, 77, 23–43. [Google Scholar] [CrossRef] [PubMed]
- Richards, G.R.; Vanderpool, C.K. Molecular Call and Response: The Physiology of Bacterial Small RNAs. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2011, 1809, 525–531. [Google Scholar] [CrossRef]
- Watkins, D.; Arya, D.P. Regulatory Roles of Small RNAs in Prokaryotes: Parallels and Contrast with Eukaryotic miRNA. Non-Coding RNA Investig. 2019, 3, 28. [Google Scholar] [CrossRef]
- Boisset, S.; Geissmann, T.; Huntzinger, E.; Fechter, P.; Bendridi, N.; Possedko, M.; Chevalier, C.; Helfer, A.C.; Benito, Y.; Jacquier, A.; et al. Staphylococcus Aureus RNAIII Coordinately Represses the Synthesis of Virulence Factors and the Transcription Regulator Rot by an Antisense Mechanism. Genes Dev. 2007, 21, 1353–1366. [Google Scholar] [CrossRef] [PubMed]
- Ponath, F.; Hör, J.; Vogel, J. An Overview of Gene Regulation in Bacteria by Small RNAs Derived from mRNA 3′ Ends. FEMS Microbiol. Rev. 2022, 46, fuac017. [Google Scholar] [CrossRef] [PubMed]
- Groher, F.; Suess, B. Synthetic Riboswitches—A Tool Comes of Age. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2014, 1839, 964–973. [Google Scholar] [CrossRef] [PubMed]
- Waters, L.S.; Storz, G. Regulatory RNAs in Bacteria. Cell 2009, 136, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Nussenzweig, P.M.; Marraffini, L.A. Molecular Mechanisms of CRISPR-Cas Immunity in Bacteria. Annu. Rev. Genet. 2020, 54, 93–120. [Google Scholar] [CrossRef]
- Desnoyers, G.; Bouchard, M.-P.; Massé, E. New Insights into Small RNA-Dependent Translational Regulation in Prokaryotes. Trends Genet. 2013, 29, 92–98. [Google Scholar] [CrossRef]
- Menard, G.; Silard, C.; Suriray, M.; Rouillon, A.; Augagneur, Y. Thirty Years of sRNA-Mediated Regulation in Staphylococcus Aureus: From Initial Discoveries to In Vivo Biological Implications. Int. J. Mol. Sci. 2022, 23, 7346. [Google Scholar] [CrossRef] [PubMed]
- Pánek, J.; Bobek, J.; Mikulík, K.; Basler, M.; Vohradský, J. Biocomputational Prediction of Small Non-Coding RNAs in Streptomyces. BMC Genom. 2008, 9, 217. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.-Q.; Lv, Z.-X.; Song, X.; Liu, X.-X.; Xia, Y.-J.; Ai, L.-Z. Recent Research Advances in Small Regulatory RNAs in Streptococcus. Curr. Microbiol. 2021, 78, 2231–2241. [Google Scholar] [CrossRef]
- Tsai, C.-H.; Liao, R.; Chou, B.; Palumbo, M.; Contreras, L.M. Genome-Wide Analyses in Bacteria Show Small-RNA Enrichment for Long and Conserved Intergenic Regions. J. Bacteriol. 2015, 197, 40–50. [Google Scholar] [CrossRef]
- Wang, M.; Fleming, J.; Li, Z.; Li, C.; Zhang, H.; Xue, Y.; Chen, M.; Zhang, Z.; Zhang, X.-E.; Bi, L. An Automated Approach for Global Identification of sRNA-Encoding Regions in RNA-Seq Data from Mycobacterium Tuberculosis. Acta Biochim. Biophys. Sin. 2016, 48, 544–553. [Google Scholar] [CrossRef]
- Salwan, R.; Sharma, V. Molecular and Biotechnological Aspects of Secondary Metabolites in Actinobacteria. Microbiol. Res. 2020, 231, 126374. [Google Scholar] [CrossRef] [PubMed]
- Swiercz, J.P.; Hindra; Bobek, J.; Haiser, H.J.; Di Berardo, C.; Tjaden, B.; Elliot, M.A. Small Non-Coding RNAs in Streptomyces Coelicolor. Nucleic Acids Res. 2008, 36, 7240–7251. [Google Scholar] [CrossRef]
- Vockenhuber, M.-P.; Sharma, C.M.; Statt, M.G.; Schmidt, D.; Xu, Z.; Dietrich, S.; Liesegang, H.; Mathews, D.H.; Suess, B. Deep Sequencing-Based Identification of Small Non-Coding RNAs in Streptomyces Coelicolor. RNA Biol. 2011, 8, 468–477. [Google Scholar] [CrossRef] [PubMed]
- Tezuka, T.; Hara, H.; Ohnishi, Y.; Horinouchi, S. Identification and Gene Disruption of Small Noncoding RNAs in Streptomyces griseus. J. Bacteriol. 2009, 191, 4896–4904. [Google Scholar] [CrossRef] [PubMed]
- Heueis, N.; Vockenhuber, M.-P.; Suess, B. Small Non-Coding RNAs in Streptomycetes. RNA Biol. 2014, 11, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Lee, N.; Choe, D.; Lee, Y.; Kim, W.; Jeong, Y.; Cho, S.; Palsson, B.O.; Cho, B.-K. Elucidating the Regulatory Elements for Transcription Termination and Posttranscriptional Processing in the Streptomyces Clavuligerus Genome. mSystems 2021, 6, e01013-20. [Google Scholar] [CrossRef] [PubMed]
- Melamed, S.; Peer, A.; Faigenbaum-Romm, R.; Gatt, Y.E.; Reiss, N.; Bar, A.; Altuvia, Y.; Argaman, L.; Margalit, H. Global Mapping of Small RNA-Target Interactions in Bacteria. Mol. Cell 2016, 63, 884–897. [Google Scholar] [CrossRef]
- Hwang, S.; Lee, N.; Jeong, Y.; Lee, Y.; Kim, W.; Cho, S.; Palsson, B.O.; Cho, B.-K. Primary Transcriptome and Translatome Analysis Determines Transcriptional and Translational Regulatory Elements Encoded in the Streptomyces Clavuligerus Genome. Nucleic Acids Res. 2019, 47, 6114–6129. [Google Scholar] [CrossRef]
- Thorpe, H.A.; Bayliss, S.C.; Sheppard, S.K.; Feil, E.J. Piggy: A Rapid, Large-Scale Pan-Genome Analysis Tool for Intergenic Regions in Bacteria. GigaScience 2018, 7, giy015. [Google Scholar] [CrossRef] [PubMed]
- Caicedo-Montoya, C.; Manzo-Ruiz, M.; Ríos-Estepa, R. Pan-Genome of the Genus Streptomyces and Prioritization of Biosynthetic Gene Clusters With Potential to Produce Antibiotic Compounds. Front. Microbiol. 2021, 12, 677558. [Google Scholar] [CrossRef]
- Weinberg, Z.; Lünse, C.E.; Corbino, K.A.; Ames, T.D.; Nelson, J.W.; Roth, A.; Perkins, K.R.; Sherlock, M.E.; Breaker, R.R. Detection of 224 Candidate Structured RNAs by Comparative Analysis of Specific Subsets of Intergenic Regions. Nucleic Acids Res. 2017, 45, 10811–10823. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, Z.; Wang, J.X.; Bogue, J.; Yang, J.; Corbino, K.; Moy, R.H.; Breaker, R.R. Comparative Genomics Reveals 104 Candidate Structured RNAs from Bacteria, Archaea, and Their Metagenomes. Genome Biol. 2010, 11, R31. [Google Scholar] [CrossRef]
- Kingsford, C.L.; Ayanbule, K.; Salzberg, S.L. Rapid, Accurate, Computational Discovery of Rho-Independent Transcription Terminators Illuminates Their Relationship to DNA Uptake. Genome Biol. 2007, 8, R22. [Google Scholar] [CrossRef] [PubMed]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-Fold Faster RNA Homology Searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [PubMed]
- Katz, K.; Shutov, O.; Lapoint, R.; Kimelman, M.; Brister, J.R.; O’Sullivan, C. The Sequence Read Archive: A Decade More of Explosive Growth. Nucleic Acids Res. 2022, 50, D387–D390. [Google Scholar] [CrossRef] [PubMed]
- Patiño, L.F.; Aguirre-Hoyos, V.; Pinilla, L.I.; Toro, L.F.; Ríos-Estepa, R. Environmental Factors Modulate the Role of Orf21 Sigma Factor in Clavulanic Acid Production in Streptomyces Clavuligerus ATCC27064. Bioengineering 2022, 9, 78. [Google Scholar] [CrossRef]
- Tjaden, B. A Computational System for Identifying Operons Based on RNA-Seq Data. Methods 2020, 176, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Liras, P.; Martín, J.F. Streptomyces Clavuligerus: The Omics Era. J. Ind. Microbiol. Biotechnol. 2021, 48, kuab072. [Google Scholar] [CrossRef]
- Leonard, S.; Meyer, S.; Lacour, S.; Nasser, W.; Hommais, F.; Reverchon, S. APERO: A Genome-Wide Approach for Identifying Bacterial Small RNAs from RNA-Seq Data. Nucleic Acids Res. 2019, 47, e88. [Google Scholar] [CrossRef]
- Helmann, J.D. Where to Begin? Sigma Factors and the Selectivity of Transcription Initiation in Bacteria. Mol. Microbiol. 2019, 112, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Vasilyev, N.; Gao, A.; Serganov, A. Noncanonical Features and Modifications on the 5′-end of Bacterial sRNAs and mRNAs. WIREs RNA 2019, 10, e1509. [Google Scholar] [CrossRef] [PubMed]
- Kurt, A.; Álvarez-Álvarez, R.; Liras, P.; Özcengiz, G. Role of the cmcH–ccaR Intergenic Region and ccaR Overexpression in Cephamycin C Biosynthesis in Streptomyces Clavuligerus. Appl. Microbiol. Biotechnol. 2013, 97, 5869–5880. [Google Scholar] [CrossRef] [PubMed]
- Yepes-García, J.; Caicedo-Montoya, C.; Pinilla, L.; Toro, L.F.; Ríos-Estepa, R. Morphological Differentiation of Streptomyces Clavuligerus Exposed to Diverse Environmental Conditions and Its Relationship with Clavulanic Acid Biosynthesis. Processes 2020, 8, 1038. [Google Scholar] [CrossRef]
- Mann, M.; Wright, P.R.; Backofen, R. IntaRNA 2.0: Enhanced and Customizable Prediction of RNA–RNA Interactions. Nucleic Acids Res. 2017, 45, W435–W439. [Google Scholar] [CrossRef]
- Tjaden, B. TargetRNA3: Predicting Prokaryotic RNA Regulatory Targets with Machine Learning. Genome Biol. 2023, 24, 276. [Google Scholar] [CrossRef] [PubMed]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG Ortholog Assignment Based on Profile HMM and Adaptive Score Threshold. Bioinformatics 2020, 36, 2251–2252. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, D.; Bhasuran, B.; Natarajan, J. Genomic Analysis of RNA-Seq and sRNA-Seq Data Identifies Potential Regulatory sRNAs and Their Functional Roles in Staphylococcus Aureus. Genomics 2019, 111, 1431–1446. [Google Scholar] [CrossRef]
- Lee, N.; Hwang, S.; Lee, Y.; Cho, S.; Palsson, B.; Cho, B.-K. Synthetic Biology Tools for Novel Secondary Metabolite Discovery in Streptomyces. J. Microbiol. Biotechnol. 2019, 29, 667–686. [Google Scholar] [CrossRef] [PubMed]
- Alam, K.; Islam, M.M.; Islam, S.; Hao, J.; Abbasi, M.N.; Hayat, M.; Shoaib, M.; Zhang, Y.; Li, A. Comparative Genomics with Evolutionary Lineage in Streptomyces Bacteria Reveals High Biosynthetic Potentials. World J. Microbiol. Biotechnol. 2023, 39, 64. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, M.C.C.; Costa, C.L.L.; Esperança, M.N.; Mazziero, V.T.; Cerri, M.O.; Badino, A.C. Effect of Impeller Type on Cellular Morphology and Production of Clavulanic Acid by Streptomyces Clavuligerus. Braz. J. Microbiol. 2024, 55, 1167–1177. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, R.M.M.G.P.; Esperança, M.N.; Sousa, A.P.A.; Neto, Á.B.; Cerri, M.O. Individual Effect of Shear Rate and Oxygen Transfer on Clavulanic Acid Production by Streptomyces Clavuligerus. Bioprocess Biosyst. Eng. 2021, 44, 1721–1732. [Google Scholar] [CrossRef]
- Bilyk, O.; Luzhetskyy, A. Metabolic Engineering of Natural Product Biosynthesis in Actinobacteria. Curr. Opin. Biotechnol. 2016, 42, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Palazzotto, E.; Tong, Y.; Lee, S.Y.; Weber, T. Synthetic Biology and Metabolic Engineering of Actinomycetes for Natural Product Discovery. Biotechnol. Adv. 2019, 37, 107366. [Google Scholar] [CrossRef]
- Mingyar, E.; Mühling, L.; Kulik, A.; Winkler, A.; Wibberg, D.; Kalinowski, J.; Blin, K.; Weber, T.; Wohlleben, W.; Stegmann, E. A Regulator Based “Semi-Targeted” Approach to Activate Silent Biosynthetic Gene Clusters. Int. J. Mol. Sci. 2021, 22, 7567. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Townsend, C.A. Rational Strain Improvement for Enhanced Clavulanic Acid Production by Genetic Engineering of the Glycolytic Pathway in Streptomyces Clavuligerus. Metab. Eng. 2006, 8, 240–252. [Google Scholar] [CrossRef]
- Gottesman, S.; Storz, G. Bacterial Small RNA Regulators: Versatile Roles and Rapidly Evolving Variations. Cold Spring Harb. Perspect. Biol. 2011, 3, a003798. [Google Scholar] [CrossRef]
- Liu, W.-B.; Shi, Y.; Yao, L.-L.; Zhou, Y.; Ye, B.-C. Prediction and Characterization of Small Non-Coding RNAs Related to Secondary Metabolites in Saccharopolyspora Erythraea. PLoS ONE 2013, 8, e80676. [Google Scholar] [CrossRef]
- Sridhar, J.; Gunasekaran, P. Computational Small RNA Prediction in Bacteria. Bioinforma. Biol. Insights 2013, 7, BBI-S11213. [Google Scholar] [CrossRef]
- Martín-Sánchez, L.; Singh, K.S.; Avalos, M.; Van Wezel, G.P.; Dickschat, J.S.; Garbeva, P. Phylogenomic Analyses and Distribution of Terpene Synthases among Streptomyces. Beilstein J. Org. Chem. 2019, 15, 1181–1193. [Google Scholar] [CrossRef]
- Zhu, D.-Q.; Liu, F.; Sun, Y.; Yang, L.-M.; Xin, L.; Meng, X.-C. Genome-Wide Identification of Small RNAs in Bifidobacterium Animalis Subsp. Lactis KLDS 2.0603 and Their Regulation Role in the Adaption to Gastrointestinal Environment. PLoS ONE 2015, 10, e0117373. [Google Scholar] [CrossRef] [PubMed]
- Jackson, L.A.; Day, M.; Allen, J.; Scott, E.; Dyer, D.W. Iron-Regulated Small RNA Expression as Neisseria Gonorrhoeae FA 1090 Transitions into Stationary Phase Growth. BMC Genom. 2017, 18, 317. [Google Scholar] [CrossRef] [PubMed]
- Georg, J.; Hess, W.R. Widespread Antisense Transcription in Prokaryotes. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Tjaden, B. Escherichia Coli Transcriptome Assembly from a Compendium of RNA-Seq Data Sets. RNA Biol. 2023, 20, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Álvarez, R.; Rodríguez-García, A.; Santamarta, I.; Pérez-Redondo, R.; Prieto-Domínguez, A.; Martínez-Burgo, Y.; Liras, P. Transcriptomic Analysis of Streptomyces clavuligerus ΔccaR::tsr: Effects of the Cephamycin C-clavulanic Acid Cluster Regulator CcaR on Global Regulation. Microb. Biotechnol. 2014, 7, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Bushell, M.E.; Kirk, S.; Zhao, H.-J.; Avignone-Rossa, C.A. Manipulation of the Physiology of Clavulanic Acid Biosynthesis with the Aid of Metabolic Flux Analysis. Enzyme Microb. Technol. 2006, 39, 149–157. [Google Scholar] [CrossRef]
- Saudagar, P.S.; Singhal, R.S. Optimization of Nutritional Requirements and Feeding Strategies for Clavulanic Acid Production by Streptomyces Clavuligerus. Bioresour. Technol. 2007, 98, 2010–2017. [Google Scholar] [CrossRef] [PubMed]
- Brantl, S.; Müller, P. Cis- and Trans-Encoded Small Regulatory RNAs in Bacillus Subtilis. Microorganisms 2021, 9, 1865. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Chevez-Guardado, R.; Peña-Castillo, L. Promotech: A General Tool for Bacterial Promoter Recognition. Genome Biol. 2021, 22, 318. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Kalvari, I.; Nawrocki, E.P.; Ontiveros-Palacios, N.; Argasinska, J.; Lamkiewicz, K.; Marz, M.; Griffiths-Jones, S.; Toffano-Nioche, C.; Gautheret, D.; Weinberg, Z.; et al. Rfam 14: Expanded Coverage of Metagenomic, Viral and microRNA Families. Nucleic Acids Res. 2021, 49, D192–D200. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-J.; Yang, D.-C.; Kong, L.; Hou, M.; Meng, Y.-Q.; Wei, L.; Gao, G. CPC2: A Fast and Accurate Coding Potential Calculator Based on Sequence Intrinsic Features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Malule, H.; Junne, S.; López, C.; Zapata, J.; Sáez, A.; Neubauer, P.; Rios-Estepa, R. An Improved HPLC-DAD Method for Clavulanic Acid Quantification in Fermentation Broths of Streptomyces Clavuligerus. J. Pharm. Biomed. Anal. 2016, 120, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Lloréns-Rico, V.; Cano, J.; Kamminga, T.; Gil, R.; Latorre, A.; Chen, W.-H.; Bork, P.; Glass, J.I.; Serrano, L.; Lluch-Senar, M. Bacterial Antisense RNAs Are Mainly the Product of Transcriptional Noise. Sci. Adv. 2016, 2, e1501363. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Muzellec, B.; Teleńczuk, M.; Cabeli, V.; Andreux, M. PyDESeq2: A Python Package for Bulk RNA-Seq Differential Expression Analysis. Bioinformatics 2023, 39, btad547. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y.; Wolf, Y.I.; Makarova, K.S.; Vera Alvarez, R.; Landsman, D.; Koonin, E.V. COG Database Update: Focus on Microbial Diversity, Model Organisms, and Widespread Pathogens. Nucleic Acids Res. 2021, 49, D274–D281. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Höner Zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and Accurate Filtering of Ribosomal RNAs in Metatranscriptomic Data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef] [PubMed]




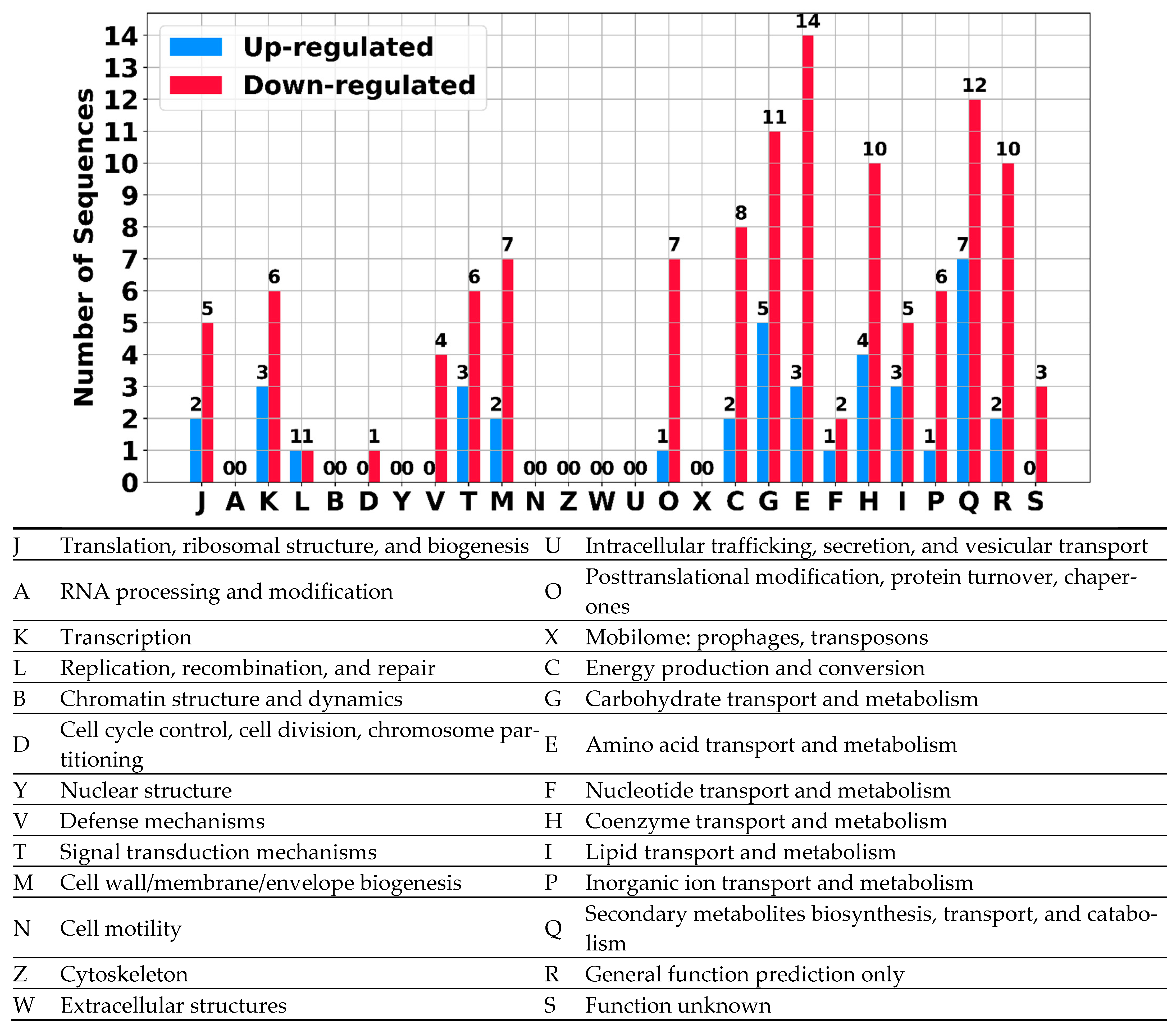
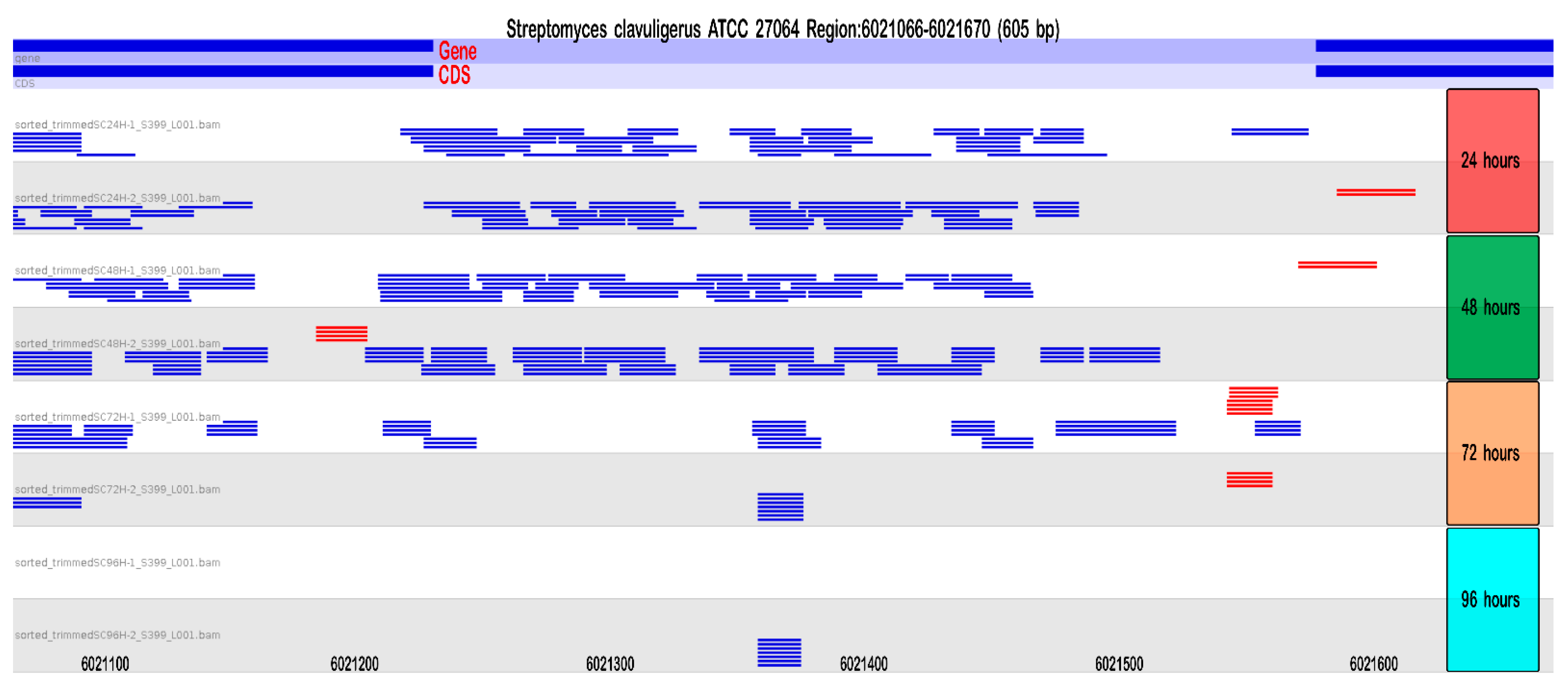
| Chromosome | Plasmid | Total | |
|---|---|---|---|
| transcripts | 11,456 | 2904 | 14,360 |
| annotated | 5546 | 1451 | 6997 |
| predicted RNA | 5910 | 1453 | 7363 |
| antisense | 4005 | 908 | 4913 |
| sRNA | 1905 | 545 | 2450 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caicedo-Montoya, C.; Patiño, L.F.; Ríos-Estepa, R. Identification of Small RNAs in Streptomyces clavuligerus Using High-Resolution Transcriptomics and Expression Profiling During Clavulanic Acid Production. Int. J. Mol. Sci. 2024, 25, 13472. https://doi.org/10.3390/ijms252413472
Caicedo-Montoya C, Patiño LF, Ríos-Estepa R. Identification of Small RNAs in Streptomyces clavuligerus Using High-Resolution Transcriptomics and Expression Profiling During Clavulanic Acid Production. International Journal of Molecular Sciences. 2024; 25(24):13472. https://doi.org/10.3390/ijms252413472
Chicago/Turabian StyleCaicedo-Montoya, Carlos, Luisa F. Patiño, and Rigoberto Ríos-Estepa. 2024. "Identification of Small RNAs in Streptomyces clavuligerus Using High-Resolution Transcriptomics and Expression Profiling During Clavulanic Acid Production" International Journal of Molecular Sciences 25, no. 24: 13472. https://doi.org/10.3390/ijms252413472
APA StyleCaicedo-Montoya, C., Patiño, L. F., & Ríos-Estepa, R. (2024). Identification of Small RNAs in Streptomyces clavuligerus Using High-Resolution Transcriptomics and Expression Profiling During Clavulanic Acid Production. International Journal of Molecular Sciences, 25(24), 13472. https://doi.org/10.3390/ijms252413472






