Advancements in Reference Gene Selection for Fruit Trees: A Comprehensive Review
Abstract
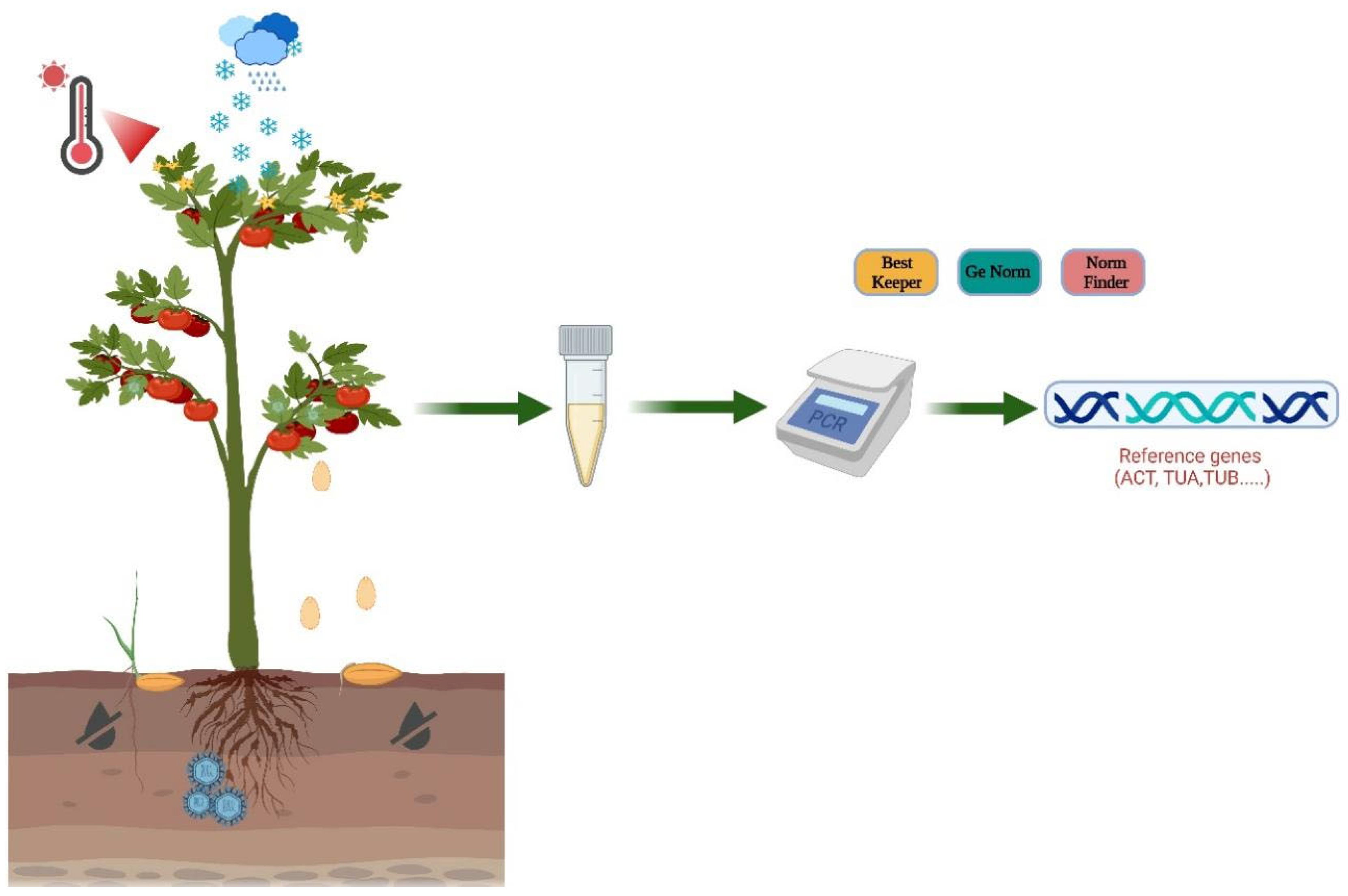
1. Introduction
2. Selection of Endogenous Reference Genes
2.1. Ideal Internal Reference Gene
2.2. Common Internal Genes
3. Methods for Analyses of Internal Reference Gene Stability
3.1. BestKeeper
3.2. geNorm
3.3. NormFinder
4. Research Progress on Internal Reference Genes in Fruit Trees
4.1. Selection of Internal Reference Genes in Vegetative Organs
| Species | Genus | Vegetative Organs | Reference Genes | References |
|---|---|---|---|---|
| Apple | Malus | Leaves, callus | UBQ | [59] |
| Leaf development process | MDH, SAND, THFS, TMp1, WD40 | [60] | ||
| Pomelo | Citrus | Leaves, Stems, root | β-Tub | [42] |
| Citrus | Leaves | ACTB, 18S rRNA, RPII | [40] | |
| Leaves, stems | IF3, Rpl35, IF5A | [41] | ||
| Leaves | FBOX, GAPC2, SAND, UPL7 | [56] | ||
| Pitaya | Hylocereus | Root, stems | ACT(1) | [44] |
| loquat | Eriobotrya | fruit setting | GAPDH, UBCE, ACT | [62] |
| floral development | GAPDH, EF1α, ACT | |||
| Durian honey | Artocarpus | Leaves, stems | β-Tub2, α-Tub1 | [49] |
| Fig | Ficus | Leaves, stems, root | 18s rRNA | [47] |
| Grape | Vitis | Branch and leaf development processes | GAPDH, UBQ-1, EF-1α1 | [51] |
| Leaves | EF-1α, RRM1 | |||
| Tendril | EF-1α and Actin combination | [52] | ||
| Leaves | RRM1 and EF-1α combination | |||
| Jackfruit | Artocarpus | Leaves | UBQ, GAPDH, β-Tub | [55] |
| Jujube | Ziziphus | Bud, fruiting branches, leaves, stem tips, root | ZjH3 | [48] |
| Kiwifruit | Actinidia | Leaves, stems, root | TUB and ACTB combination | [50] |
| Leaves | 18s rRNA, ACT 2 | [45] | ||
| Leaves, stems, root | GAPDH and UBQ combination | [46] | ||
| Longan | Dimocarpus | Somatic embryo development process | UBQ, Fe-SOD | [61] |
| Lychee | Litchi | Leaves | UBQ, RPII | [58] |
| Peach | Prunus | Leaves, stems, root | TEF2, UBQ10, RPII | [43] |
| Pear | Pyrus | Leaf blade | WDP | [57] |
| Starfruit | Averrhoa | Leaf bud, leaves, stems | α-Tub, β-Tub | [53] |
4.2. Selection of Internal Reference Genes in Reproductive Organs
| Species | Genus | Reproductive Organs | Reference Genes | References |
|---|---|---|---|---|
| Apple | Malus | Flowers, pericarp and pulp development process | MDH, SAND, THFS, TMp1, WD40 | [60] |
| Flowers, fruit development process, seeds | UBQ | [59] | ||
| Pericarp | ACT, GAPD, WD40H | [62] | ||
| Pericarp development process | EF-1α, 18s rRNA | [59] | ||
| Flesh | EF-1α, CKL, WD40 | [64] | ||
| Apricot | Prunus | Fruit postharvest | CAC and UNK or CAC, ACT and CLATH | [85] |
| Seeds | UBC | [84] | ||
| Cherry | Flower bud development process | EF-1α2, RSP3 | [81] | |
| Flower bud dormancy removal process | ACTB, UBCE | [82] | ||
| Peach | Fruit development process | ACT | [72] | |
| Flowers, fruit development process | TEF2, UBQ10, RPII | [79] | ||
| Fruit postharvest | PpeIF-1A | [88] | ||
| Plum | Fruit development process | IPGD, HAM1, SNX1 | [73] | |
| Soursop | Annona | Fruit postharvest | EF-1α | [86] |
| Banana | Musa | Fruit | CAC, SAMDC1 | [69] |
| Pulp development process | 18s rRNA, RPS2 | |||
| Pomelo | Fruit development process | β-Tub | [42] | |
| Citrus | Citrus | Flowers, flesh development process | TUA3, GAPDH | [70] |
| Flower organs (petals), pericarp | 18s rRNA, RPII | [40] | ||
| Flowers, fruit | FBOX, GAPC2, SAND, UPL7 | [56] | ||
| Durian honey | Artocarpus | Inflorescence | α-Tub1, β-Tub2 | [53] |
| Fig | Ficus | Fruit | 18s rRNA | [47] |
| Grape | Vitis | Flowers, fruit | PP2A, SAND, Sutra | [75] |
| Fruit | EF1-γ and PPR2 combination | [65] | ||
| Late development of the pericarp | β-ACT, SAND | [66] | ||
| Pericarp | EF1-α and EF1-γ combination | [65] | ||
| Jackfruit | Artocarpus | Fruit development process | UBQ, GAPDH, 18S rRNA | [35] |
| Inflorescence | UBQ, GAPDH, α-Tub | |||
| Jujube | Ziziphus | Flowers, fruit development process | ZjH3 | [46] |
| Kiwifruit | Actinidia | Flowers | Tub and ACTB combination | [45] |
| Flowers, fruit | GAPDH and UBQ combination | [46] | ||
| Fruit | ACTB | [74] | ||
| Fruit (young fruit) | ACT | [50] | ||
| Fruit postharvest | UBQ-CONJ-E2, TUB-FCB | [87] | ||
| Longan | Dimocarpus | Pericarp | GAPDH, Fe-SOD, Cu/Zn-SOD | [63] |
| Pericarp development process | EF-1α, Mn-SOD | |||
| Pulp | GAPDH, Mn-SOD | |||
| Fruit postharvest | 18S rRNA + EF-1a or 18S rRNA + ACT | |||
| Lychee | Litchi | Fruit development process | β-ACT | [58] |
| Fruit postharvest | HDAC9 | [90] | ||
| Loquat | Eriobotrya | Fruit development process | EF1α, GAPDH, eIF2B | [67] |
| Fruit setting | GAPDH, UBCE, ACT | |||
| Flowers development | GAPDH, EF1α, ACT | |||
| Pear | Pyrus | Floral organs (pollen, style) | WDP | [57] |
| Flower organs (receptacle) | ACT | [71] | ||
| Pericarp development process | Tub2 | |||
| Fruit development process | SOX2, PP2A | [78] | ||
| Pulp development process | BPS1 and ICDH1 | [76] | ||
| Pineapple | Ananas | Ovule development | RPS4 and RPL23 combination | [83] |
| Stamen development | CCR, RPS4 | |||
| Starfruit | Averrhoa | Flower buds, fruit | α-Tub, β-Tub | [53] |
| Inflorescence development process | β-Tub, UBC4 |
4.3. Selection of Internal Reference Genes under Stresses
| Species | Genus | Stress Treatments | Reference Genes | References |
|---|---|---|---|---|
| Banana | Musa | Heat and cold stresses, infection with germs (banana anthracnose) | ACT1, EIF5A-2 | [69] |
| Hormone treatment (SA/MeJA) | UBQ2, RAN | |||
| Blueberry | Vaccinium | Salt treatment, alkaline treatment, saline–alkaline treatment, drought treatment and AlCl3 treatment | EF1α, EIF, TBP | [95] |
| Citrus | Citrus | Infection with germs (citrus bacterial canker) | ATCB, 18s rRNA, RPII | [40] |
| Infection with pathogens (Alternaria alternata, Phytophthora parasitica, Xylella fastidiosa and Candidatus Liberibacter asiaticus) | FBOX, GAPC2, SAND, UPL7 | [56] | ||
| Grape | Vitis | Shoot pinching | SAND, VAG | [103] |
| Kiwifruit | Actinidia | Hormone treatment (MT/PP1/PP2/HBR1/HBR2/MT + PP/MT + HBR) | ACT1, UBQ | [46] |
| Longan | Dimocarpus | Cold stress | 18s rRNA, EF-1α, Fe-SOD | [63] |
| Hormone treatment (NAA/ETH) | GAPDH, EF-1α | |||
| Lychee | Litchi | Hormone treatment (NAA) | GAPDH | [58] |
| Hormone treatment (ABA/CPPU) | β-ACT | [90] | ||
| Shading treatment | EF-1α | [58] | ||
| Loquat | Eriobotrya | Heat stress | ACT, EF1α and UBCE | [67] |
| Freezing stress | eIF2B, UBCE and EF1α | |||
| Salt stress | EF1α, TUA and UBCE | |||
| Mango | Mangifera | Infection with germs (keratosis) | GAPDH, gyrβ | [101] |
| Mulberry | Morus | Infection with a virus (sclerotinia) | GST1, Tub | [96] |
| Cherry | Prunus | Cold and salt stresses | GAPDH | [81] |
| Hormone treatment (ABA) | ACTB, UBCE | |||
| Peach | Chilling stress | ACT and UBQ10 | [92] | |
| Infectious bacteria (tobacco crackling virus) | CYP2, Tua5 | [102] | ||
| Pear | Pyrus | Cold stress | Tub, WDP | [57] |
| Heat and salt stresses | UBQ | |||
| Salt stress | GAPDH, β-Tub | [93] | ||
| Shading treatment | EF-1α, His | [104] | ||
| hormone treatments (ABA, 6-BA and NAA) | TIP41 | [94] | ||
| Persimmon | Diospyros | Cold, heat and salt stresses | UBC, RPII, Tua | [91] |
| Hormone treatment (GA/ABA/SA) | α-Tub, PP2A | |||
| Pitaya | Hylocereus | Cold stress | ACT(1) | [44] |
| Strawberry | Fragaria | Heat stress | HISTH4 | [99] |
| Cold stress | ACTIN2 | |||
| Drought stress | DBP | |||
| Salt stress | GAPDH |
4.4. Application of Internal Reference Genes in Fruit Trees
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Savadi, S.; Mangalassery, S.; Sandesh, M.S. Advances in genomics and genome editing for breeding next generation of fruit and nut crops. Genomics 2021, 113, 3718–3734. [Google Scholar] [CrossRef]
- Wang, R.; Li, X.; Sun, M.; Xue, C.; Korban, S.S.; Wu, J. Genomic insights into domestication and genetic improvement of fruit crops. Plant Physiol. 2023, 192, 2604–2627. [Google Scholar] [CrossRef]
- Bustin, S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef]
- Chen, Y.; Tan, Z.; Hu, B.; Yang, Z.; Xu, B.; Zhuang, L.; Huang, B. Selection and validation of reference genes for target gene analysis with quantitative RT-PCR in leaves and roots of bermudagrass under four different abiotic stresses. Physiol. Plant. 2015, 155, 138–148. [Google Scholar] [CrossRef]
- Wang, X.; Fu, Y.; Ban, L.; Wang, Z.; Feng, G.; Li, J.; Gao, H. Selection of reliable reference genes for quantitative real-time RT-PCR in alfalfa. Genes Genet. Syst. 2015, 90, 175–180. [Google Scholar] [CrossRef]
- Ma, H.; Bell, K.N.; Loker, R.N. qPCR and qRT-PCR analysis: Regulatory points to consider when conducting biodistribution and vector shedding studies. Mol. Ther. Methods Clin. Dev. 2021, 20, 152–168. [Google Scholar] [CrossRef]
- Dahn, M.L.; Dean, C.A.; Jo, D.B.; Coyle, K.M.; Marcato, P. Human-specific GAPDH qRT-PCR is an accurate and sensitive method of xenograft metastasis quantification. Mol. Ther. Methods Clin. Dev. 2021, 20, 398–408. [Google Scholar] [CrossRef]
- Ban, E.; Song, E.J. Considerations and suggestions for the reliable analysis of miRNA in plasma using q RT-PCR. Genes 2022, 13, 328. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef]
- Mahoney, D.J.; Carey, K.; Fu, M.H.; Snow, R.; Cameron-Smith, D.; Parise, G.; Tarnopolsky, M.A. Real-time RT-PCR analysis of housekeeping genes in human skeletal muscle following acute exercise. Physiol. Genom. 2004, 18, 226–231. [Google Scholar] [CrossRef]
- Hasanpur, K.; Hosseinzadeh, S.; Mirzaaghayi, A.; Alijani, S. Investigation of chicken housekeeping genes using next-generation sequencing data. Front. Genet. 2022, 13, 827538. [Google Scholar] [CrossRef]
- Wei, K.; Ma, L. Concept development of housekeeping genes in the high-throughput sequencing era. Hereditas 2017, 39, 127–134. [Google Scholar] [CrossRef]
- Lin, J.; Redies, C. Histological evidence: Housekeeping genes beta-actin and GAPDH are of limited value for normalization of gene expression. Dev. Genes Evol. 2012, 222, 369–376. [Google Scholar] [CrossRef]
- Yulia, P.; Arno, G.; Shinji, M.; Watanabe, T.M. Validation of common housekeeping genes as reference for qPCR gene expression analysis during iPS reprogramming process. Sci. Rep. 2018, 8, 8716. [Google Scholar] [CrossRef]
- Dejosez, M.; Dall’Agnese, A.; Ramamoorthy, M.; Platt, J.; Yin, X.; Hogan, M.; Brosh, R.; Weintraub, A.S.; Hnisz, D.; Abraham, B.J.; et al. Regulatory architecture of housekeeping genes is driven by promoter assemblies. Cell Rep. 2023, 42, 112505. [Google Scholar] [CrossRef]
- Cao, A.; Shao, D.; Cui, B.; Tong, X.; Zheng, Y.; Sun, J.; Li, H. Screening the reference genes for quantitative gene expression by RT-qPCR during SE initial dedifferentiation in four Gossypium hirsutum cultivars that have different SE capability. Genes 2019, 10, 497. [Google Scholar] [CrossRef]
- Kumar, G.; Singh, A.K. Reference gene validation for qRT-PCR based gene expression studies in different developmental stages and under biotic stress in apple. Sci. Hortic. 2015, 197, 597–606. [Google Scholar] [CrossRef]
- Orellana, E.A.; Siegal, E.; Gregory, R. tRNA dysregulation and disease. Nat. Rev. Genet. 2022, 23, 651–664. [Google Scholar] [CrossRef]
- Kozera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef]
- Deng, L.T.; Wu, Y.L.; Li, J.C.; OuYang, K.X.; Ding, M.M.; Zhang, J.J.; Li, S.Q.; Lin, M.F.; Chen, H.B.; Hu, X.S.; et al. Screening reliable reference genes for RT-qPCR analysis of gene expression in Moringa oleifera. PLoS ONE 2016, 11, e0159458. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Higgins, P.J.; Crawford, D.R. Control selection for RNA quantitation. Biotechniques 2000, 29, 332–337. [Google Scholar] [CrossRef]
- Remans, T.; Keunen, E.; Bex, G.J.; Smeets, K.; Vangronsveld, J.; Cuypers, A. Reliable gene expression analysis by reverse transcription-quantitative PCR: Reporting and minimizing the uncertainty in data accuracy. Plant Cell 2014, 26, 3829–3837. [Google Scholar] [CrossRef] [PubMed]
- Quackenbush, J. Microarray data normalization and transformation. Nat. Genet. 2002, 32, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Thellin, O.; Zorzi, W.; Lakaye, B.; De Borman, B.; Coumans, B.; Hennen, G.; Grisar, T.; Igout, A.; Heinen, E. Housekeeping genes as internal standards: Use and limits. J. Biotechnol. 1999, 75, 291–295. [Google Scholar] [CrossRef]
- Dheda, K.; Huggett, J.F.; Bustin, S.A.; Johnson, M.A.; Rook, G.; Zumla, A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 2004, 37, 112–114. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Miao, Y.; Wang, Y.; Shan, Q. Hprt serves as an ideal reference gene for qRT-PCR normalization in rat DRG neurons. J. Integr. Neurosci. 2023, 22, 125. [Google Scholar] [CrossRef]
- Ferreira, M.J.; Silva, J.; Pinto, S.C.; Coimbra, S. I choose you: Selecting accurate reference genes for qPCR expression analysis in reproductive tissues in Arabidopsis thaliana. Biomolecules 2023, 13, 463. [Google Scholar] [CrossRef]
- Skiljaica, A.; Jagic, M.; Vuk, T.; Leljak, L.D.; Bauer, N.; Markulin, L. Evaluation of reference genes for RT-qPCR gene expression analysis in Arabidopsis thaliana exposed to elevated temperatures. Plant Biol. 2022, 24, 367–379. [Google Scholar] [CrossRef]
- Chang, M.M.; Li, A.; Feissner, R.; Ahmad, T. RT-qPCR demonstrates light-dependent AtRBCS1A and AtRBCS3B mRNA expressions in Arabidopsis thaliana leaves. Biochem. Mol. Biol. Educ. 2016, 44, 405–411. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Wang, Q.; Ishikawa, T.; Michiue, T.; Zhu, B.L.; Guan, D.W.; Maeda, H. Stability of endogenous reference genes in postmortem human brains for normalization of quantitative real-time PCR data: Comprehensive evaluation using geNorm, NormFinder, and BestKeeper. Int. J. Leg. Med. 2012, 126, 943–952. [Google Scholar] [CrossRef]
- Wan, Q.; Chen, S.; Shan, Z.; Yang, Z.; Chen, L.; Zhang, C.; Yuan, S.; Hao, Q.; Zhang, X.; Qiu, D.; et al. Stability evaluation of reference genes for gene expression analysis by RT-qPCR in soybean under different conditions. PLoS ONE 2017, 12, e0189405. [Google Scholar] [CrossRef] [PubMed]
- Auler, P.A.; Benitez, L.C.; Do, A.M.; Vighi, I.L.; Dos, S.R.G.; Da, M.L.; Braga, E.J. Evaluation of stability and validation of reference genes for RT-qPCR expression studies in rice plants under water deficit. J. Appl. Genet. 2017, 58, 163–177. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: Amodel-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, Y.; Fang, H.; Shi, H.; Chen, K.; Zhang, Z.; Tan, X. Selection of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in Brassica napus under various stress conditions. Mol. Genet. Genom. 2014, 289, 1023–1035. [Google Scholar] [CrossRef]
- Perez, S.; Royo, L.J.; Astudillo, A.; Escudero, D.; Alvarez, F.; Rodriguez, A.; Gomez, E.; Otero, J. Identifying the most suitable endogenous control for determining gene expression in hearts from organ donors. BMC Mol. Biol. 2007, 8, 114. [Google Scholar] [CrossRef]
- De Spiegelaere, W.; Dern-Wieloch, J.; Weigel, R.; Schumacher, V.; Schorle, H.; Nettersheim, D.; Bergmann, M.; Brehm, R.; Kliesch, S.; Vandekerckhove, L.; et al. Reference gene validation for RT-qPCR, a note on different available software packages. PLoS ONE 2015, 10, e0122515. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Lu, L.; Li, Y.; Zhen, C.; Hu, G.; Jiang, K.; Yan, Y.; Xu, Y.; Wang, G.; Shi, M.; et al. Reference gene selection for quantitative real-time PCR (qRT-PCR) expression analysis in Galium aparine L. PLoS ONE 2020, 15, e0226668. [Google Scholar] [CrossRef]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef]
- Yan, J.; Yuan, F.; Long, G.; Qin, L.; Deng, Z. Selection of reference genes for quantitative real-time RT-PCR analysis in citrus. Mol. Biol. Rep. 2012, 39, 1831–1838. [Google Scholar] [CrossRef]
- Pinheiro, T.T.; Nishimura, D.S.; De Nadai, F.B.; Figueira, A.; Latado, R.R. Selection of reference genes for expression analyses of red-fleshed sweet orange (Citrus sinensis). Genet. Mol. Res. 2015, 14, 18440–18451. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.H.; Pan, Y.G.; Yang, L.; Cai, S.H.; Huang, X.Z. Validation of internal reference genes for qRT-PCR normalization in ‘Guanxi Sweet Pummelo’ (Citrus grandis). J. Fruit Sci. 2013, 30, 48–54. [Google Scholar] [CrossRef]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10, 71. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, J.; Hua, Q.; Tel-Zur, N.; Xie, F.; Zhang, Z.; Chen, J.; Zhang, R.; Hu, G.; Zhao, J.; et al. Identification of reliable reference genes for quantitative real-time PCR normalization in pitaya. Plant Methods 2019, 15, 70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Q.; Xie, J.; Qiao, J.P.; Zhou, L.Q.; Song, G.H. Screening of reference genes for real-time quantitative PCR in kiwifruit. Acta Agric. Zhejiangensis 2015, 27, 567–573. [Google Scholar]
- Zhou, Y.; Xia, H.; Liu, X.; Lin, Z.; Guo, Y.; Deng, H.; Wang, J.; Lin, L.; Deng, Q.; Lv, X.; et al. Identification of suitable reference genes for qRT-PCR normalization in kiwifruit. Horticulturae 2022, 8, 170. [Google Scholar] [CrossRef]
- Liu, J.Q.; Chen, L.Y.; Chen, S.W.; Zhang, W.; Ma, H.Q. RNA isolation and internal reference gene selection for semi-quantitative RT-PCR of fig (Ficus carica). J. China Agric. Univ. 2012, 17, 54–58. [Google Scholar]
- Meng, Y.P.; Cao, Q.F.; Sun, H.F. Cloning and selection of housekeeping gene ZjH3 for Ziziphus jujuba. Biotechnol. Bull. 2010, 11, 101–107. [Google Scholar]
- Guo, Q.Y.; Hu, F.C.; Wu, F.Z.; Wang, X.H.; Fan, H.Y.; Feng, X.J.; Chen, Z. Cloning and selection of reference genes for real-time PCR in Artocarpus integer (Thunb.) Merr. South China Fruits 2022, 51, 42–49+54. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Huang, S.N.; Wang, T.; Pan, D.L.; Zhai, M.; Guo, Z.R. Screening of RT-qPCR reference genes in Jinkui kiwifruit. J. Shanghai Jiaotong Univ. Agric. Sci. 2018, 34, 84–88. [Google Scholar] [CrossRef]
- Wei, T.L.; Wang, H.; Pei, M.S.; Liu, H.N.; Yu, Y.H.; Jiang, J.F.; Guo, D.L. Identification of optimal and novel reference genes for quantitative real-time polymerase chain reaction analysis in grapevine. Aust. J. Grape Wine Res. 2021, 27, 325–333. [Google Scholar] [CrossRef]
- Ren, F.; Zhang, Z.P.; Fan, X.D.; Hu, G.J.; Li, C.; Dong, Y.F. Screening and validation of reference genes for real-time fluorescence quantitative PCR in Grapevine. Mol. Plant Breed. 2019, 17, 7801–7810. [Google Scholar] [CrossRef]
- Li, X.P.; Zhu, Y.T.; Zhao, Y.M.; Chen, L.; Ren, H.; Wu, S.S.; Zhai, J.W. Selection and validation of reference genes of Averrhoa carambola by quantitative real-time PCR. Mol. Plant Breed. 2023, 3, 1–17. [Google Scholar]
- Ferradas, Y.; Rey, L.; Martinez, O.; Rey, M.; Gonzalez, M.V. Identification and validation of reference genes for accurate normalization of real-time quantitative PCR data in kiwifruit. Plant Physiol. Biochem. 2016, 102, 27–36. [Google Scholar] [CrossRef]
- Wang, Y.B.; Yu, Q.; Li, Y.Z.; Li, H.B.; Ye, C.H. Screening of reference genes for real-time fluorescence quantitative PCR in jackfruit. Chin. J. Trop. Crops 2014, 35, 1374–1381. [Google Scholar]
- Mafra, V.; Kubo, K.S.; Alves-Ferreira, M.; Ribeiro-Alves, M.; Stuart, R.M.; Boava, L.P.; Rodrigues, C.M.; Machado, M.A. Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. PLoS ONE 2012, 7, e31263. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Wu, X.; Gu, C.; Yin, H.; Zhang, S.L. Selection of reference genes in qRT-PCR of pear ‘Dangshansuli’. China Fruit 2018, 1, 16–22+35. [Google Scholar] [CrossRef]
- Zhong, H.Y.; Chen, J.W.; Li, C.Q.; Chen, L.; Wu, J.Y.; Chen, J.Y.; Lu, W.J.; Li, J.G. Selection of reliable reference genes for expression studies by reverse transcription quantitative real-time PCR in litchi under different experimental conditions. Plant Cell Rep. 2011, 30, 641–653. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, L.Y.; Zhang, C.X.; Kang, G.D.; Tian, Y.; Cong, P.H. Screening of reference genes for real-time fluorescence quantitative PCR in apple (Malus × domestica). J. Fruit Sci. 2012, 29, 965–970. [Google Scholar] [CrossRef]
- Perini, P.; Pasquali, G.; Margis-Pinheiro, M.; Oliviera, P.R.D.D.; Rever, L.F. Reference genes for transcriptional analysis of flowering and fruit ripening stages in apple (Malus × domestica Borkh.). Mol. Breed. 2014, 34, 829–842. [Google Scholar] [CrossRef]
- Lin, Y.L.; Lai, Z.X. Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Sci. 2010, 178, 359–365. [Google Scholar] [CrossRef]
- Zhu, L.; Yang, C.; You, Y.; Liang, W.; Wang, N.; Ma, F.; Li, C. Validation of reference genes for qRT-PCR analysis in peel and flesh of six apple cultivars (Malus domestica) at diverse stages of fruit development. Sci. Hortic. 2019, 244, 165–171. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, H.; Liu, L.; Li, W.; Wei, Y.; Shi, S. Validation of reference genes for RT-qPCR studies of gene expression in preharvest and postharvest longan fruits under different experimental conditions. Front. Plant Sci. 2016, 7, 780. [Google Scholar] [CrossRef]
- Fan, L.M.; Wang, C.; Liu, G.S.; Yuan, Y.B. Screening and validation of reference genes for real-time fluorescence quantitative PCR during coloring period in apple (Malus domestica). Plant Physiol. J. 2014, 50, 1903–1911. [Google Scholar] [CrossRef]
- Cha, Q.; Xi, X.J.; Jiang, A.L.; Tian, Y.H.; Wang, S.P. Screening of stable internal reference genes in real-time quantitative PCR of grapes. J. Fruit Trees 2016, 33, 268–274. [Google Scholar] [CrossRef]
- Dai, H.J.; Qin, C.L.; Xu, W.R. Screening and validation of reference genes for real-time fluorescence quantitative PCR during grape berry development of Cabernet Sauvianon. Jiangsu J. Agric. Sci. 2016, 32, 668–673. [Google Scholar]
- Lin, S.; Xu, S.; Huang, L.; Qiu, F.; Zheng, Y.; Liu, Q.; Ma, S.; Wu, B.; Wu, J. Selection and validation of reference genes for normalization of RT-qPCR analysis in developing or abiotic-stressed tissues of loquat (Eriobotrya japonica). Phyton Int. J. Exp. Bot. 2023, 92, 1185–1201. [Google Scholar] [CrossRef]
- Su, W.; Yuan, Y.; Zhang, L.; Jiang, Y.; Gan, X.; Bai, Y.; Peng, J.; Wu, J.; Liu, Y.; Lin, S. Selection of the optimal reference genes for expression analyses in different materials of Eriobotrya japonica. Plant Methods 2019, 15, 7. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhong, H.; Kuang, J.; Li, J.; Lu, W.; Chen, J. Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 2011, 234, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Su, S.; Fu, L.; Zhang, Y.; Chai, L.; Yi, H. Selection of reliable reference genes for gene expression studies using quantitative real-time PCR in navel orange fruit development and pummelo floral organs. Sci. Hortic. 2014, 176, 180–188. [Google Scholar] [CrossRef]
- Pu, X.Q.; Tian, J.; Li, J.; Zhang, Y.; Li, P.; Qin, W.M.; Jing, C.Z. Analysis on expression stability of internal reference genes at cell division stage of pear fruits. Nonwood For. Res. 2020, 38, 66–74. [Google Scholar] [CrossRef]
- You, S.; Cao, K.; Chen, C.; Li, Y.; Wu, J.; Zhu, G.; Fang, W.; Wang, X.; Wang, L. Selection and validation reference genes for qRT-PCR normalization in different cultivars during fruit ripening and softening of peach (Prunus persica). Sci. Rep. 2021, 11, 7302. [Google Scholar] [CrossRef]
- Kim, H.; Saha, P.; Farcuh, M.; Li, B.; Sadka, A.; Blumwald, E. RNA-Seq analysis of spatiotemporal gene expression patterns during fruit development revealed reference genes for transcript normalization in Plums. Plant Mol. Biol. Rep. 2015, 33, 1634–1649. [Google Scholar] [CrossRef]
- Zhao, S.P.; Hu, B.L.; Hu, H.L.; Zhou, H.S.; Liu, H.Y.; Li, P.X. Optimization of RNA extraction conditions and reference genes of ‘Xuxiang’ kiwifruit. Jiangsu Agric. Sci. 2017, 45, 44–48. [Google Scholar] [CrossRef]
- Upadhyay, A.; Jogaiah, S.; Maske, S.R.; Kadoo, N.Y.; Gupta, V.S. Expression of stable reference genes and SPINDLY gene in response to gibberellic acid application at different stages of grapevine development. Biol. Plant. 2015, 59, 436–444. [Google Scholar] [CrossRef]
- Chen, C.; Yuan, M.; Song, J.; Liu, Y.; Xia, Z.; Yuan, Y.; Wang, W.; Xie, Q.; Guan, X.; Chen, Q.; et al. Genome-wide identification and testing of superior reference genes for transcript normalization during analyses of flesh development in Asian pear cultivars. Sci. Hortic. 2020, 271, 109459. [Google Scholar] [CrossRef]
- Ye, X.; Zhang, F.; Tao, Y.; Song, S.; Fang, J. Reference gene selection for quantitative real-time PCR normalization in different cherry genotypes, developmental stages and organs. Sci. Hortic. 2015, 181, 182–188. [Google Scholar] [CrossRef]
- Wang, Y.; Dai, M.; Cai, D.; Shi, Z. Screening for quantitative real-time PCR reference genes with high stable expression using the mRNA-sequencing data for pear. Tree Genet. Genomes 2019, 15, 54. [Google Scholar] [CrossRef]
- Tong, J.; Hu, M.; Han, B.; Ji, Y.; Wang, B.; Liang, H.; Liu, M.; Wu, Z.; Liu, N. Determination of reliable reference genes for gene expression studies in Chinese chive (Allium tuberosum) based on the transcriptome profiling. Sci. Rep. 2021, 11, 16558. [Google Scholar] [CrossRef]
- Liu, Z.; Ge, X.; Wu, X.; Kou, S.; Chai, L.; Guo, W. Selection and validation of suitable reference genes for mRNA qRT-PCR analysis using somatic embryogenic cultures, floral and vegetative tissues in citrus. Plant Cell Tissue Organ Cult. 2013, 113, 469–481. [Google Scholar] [CrossRef]
- Zhu, Y.Y.; Wang, Y.; Zhang, H.; Shao, X.; Li, Y.Q.; Guo, W.D. Selection and characterization of reliable reference genes in Chinese cherry (Prunus pseudocerasus) using quantitative real-time PCR (qRT-PCR). J. Agric. Biotechnol. 2015, 23, 690–700. [Google Scholar]
- Qiu, Z.L.; He, M.Q.; Wen, Z.; Yang, K.; Hong, Y.; Wen, X.P. Selection and validation of reference genes in sweet cherry flower bud at different development stages. Seed 2020, 39, 37–43. [Google Scholar]
- Jin, X.; Hou, Z.; Zhao, L.; Liu, L.; Priyadarshani, S.V.G.N.; Wang, L.; Huang, Y.; Chen, F.; Qin, Y. Genome-wide identification and evaluation of new reference genes in pineapple (Ananas comosus L.) during stamen and ovule development. Trop. Plant Biol. 2020, 13, 371–381. [Google Scholar] [CrossRef]
- Niu, J.; Zhu, B.; Cai, J.; Li, P.; Wang, L.; Dai, H.; Qiu, L.; Yu, H.; Ha, D.; Zhao, H.; et al. Selection of reference genes for gene expression studies in Siberian Apricot (Prunus sibirica L.) germplasm using quantitative real-time PCR. PLoS ONE 2014, 9, e103900. [Google Scholar] [CrossRef]
- You, Y.; Zhang, L.; Li, P.; Yang, C.; Ma, F. Selection of reliable reference genes for quantitative real-time PCR analysis in plum (Prunus salicina Lindl.) under different postharvest treatments. Sci. Hortic. 2016, 210, 285–293. [Google Scholar] [CrossRef]
- Berumen-Varela, G.; Palomino-Hermosillo, Y.A.; Bautista-Rosales, P.U.; Pena-Sandoval, G.R.; Lopez-Guzman, G.G.; Balois-Morales, R. Identification of reference genes for quantitative real-time PCR in different developmental stages and under refrigeration conditions in soursop fruits (Annona muricata L.). Sci. Hortic. 2020, 260, 108893. [Google Scholar] [CrossRef]
- Liu, J.; Huang, S.; Niu, X.; Chen, D.; Chen, Q.; Tian, L.; Xiao, F.; Liu, Y. Genome-wide identification and validation of new reference genes for transcript normalization in developmental and post-harvested fruits of Actinidia chinensis. Gene 2018, 645, 12. [Google Scholar] [CrossRef]
- Kou, X.; Zhang, L.; Yang, S.; Li, G.; Ye, J. Selection and validation of reference genes for quantitative RT-PCR analysis in peach fruit under different experimental conditions. Sci. Hortic. 2017, 225, 195–203. [Google Scholar] [CrossRef]
- Zhu, X.; Li, X.; Chen, W.; Chen, J.; Lu, W.; Chen, L.; Fu, D. Evaluation of new reference genes in papaya for accurate transcript normalization under different experimental conditions. PLoS ONE 2012, 7, e44405. [Google Scholar] [CrossRef]
- Wei, Y.Z.; Lai, B.; Hu, F.C.; Li, X.J.; Hu, G.B.; Wang, H.C. Cloning and stability analysis of reference genes for expression studies by quantitative real-time PCR in litchi. J. South China Agric. Univ. 2012, 33, 301–306. [Google Scholar]
- Wang, P.; Xiong, A.; Gao, Z.; Yu, X.; Li, M.; Hou, Y.; Sun, C.; Qu, S. Selection of suitable reference genes for RT-qPCR normalization under abiotic stresses and hormone stimulation in persimmon (Diospyros kaki Thunb). PLoS ONE 2017, 11, e0160885. [Google Scholar] [CrossRef] [PubMed]
- Marini, N.; Bevilacqua, C.B.; Büttow, M.V.; Raseira, M.C.B.; Bonow, S. Identification of reference genes for RT-qPCR analysis in peach genotypes with contrasting chilling requirements. Genet. Mol. Res. 2017, 16, gmr16029666. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.Y.; Liu, C.L.; Yu, X.J.; Yang, J.D.; Feng, C.N. Screening of reference genes for differentially expressed genes in Pyrus betulaefolia plant under salt stress by qRT-PCR. Acta Hortic. Sin. 2022, 49, 1557–1570. [Google Scholar] [CrossRef]
- Xu, Y.; Li, H.; Li, X.; Lin, J.; Wang, Z.; Yang, Q.; Chang, Y. Systematic selection and validation of appropriate reference genes for gene expression studies by quantitative real-time PCR in pear. Acta Physiol. Plant. 2015, 37, 40. [Google Scholar] [CrossRef]
- Deng, Y.; Li, Y.; Sun, H. Selection of reference genes for RT-qPCR normalization in blueberry (Vaccinium corymbosum × angustifolium) under various abiotic stresses. FEBS Open Bio 2020, 10, 1418–1435. [Google Scholar] [CrossRef] [PubMed]
- Kormula, Y.L.; Chao, N.; Liu, N.Y.; Wang, Y.Q.; Gao, X.C.; Zhang, H.; Liu, L. Evaluation of the expression stability of qRT-PCR candidate internal reference genes in mulberry fruit. Acta Sericologica Sin. 2021, 47, 387–392. [Google Scholar] [CrossRef]
- Borges, A.F.; Fonseca, C.; Ferreira, R.B.; Lourenco, A.M.; Monteiro, S. Reference gene validation for quantitative RT-PCR during biotic and abiotic stresses in Vitis vinifera. PLoS ONE 2014, 9, e111399. [Google Scholar] [CrossRef]
- Monteiro, F.; Sebastiana, M.; Pais, M.S.; Figueiredo, A. Reference gene selection and validation for the early responses to downy mildew infection in susceptible and resistant Vitis vinifera cultivars. PLoS ONE 2017, 8, e72998. [Google Scholar] [CrossRef]
- Ye, Y.; Lu, Y.; Wang, G.; Liu, Y.; Zhang, Y.; Tang, H. Stable reference gene selection for qRT-PCR normalization in strawberry (Fragaria × ananassa) leaves under different stress and light-quality conditions. Horticulturae 2021, 7, 452. [Google Scholar] [CrossRef]
- Galimba, K.; Tosetti, R.; Loerich, K.; Michael, L.; Pabhakar, S.; Dove, C.; Dardick, C.; Callahan, A. Identification of early fruit development reference genes in plum. PLoS ONE 2020, 15, e0230920. [Google Scholar] [CrossRef]
- Yao, Q.S.; Yang, Q.; Liu, F.; Zhan, R.L. Screening of reference genes in Xanthomonas citri pv. mangiferaeindicae during the infection of mango leaf. Mol. Plant Breed. 2021, 19, 6088–6095. [Google Scholar] [CrossRef]
- Xu, Z.; Dai, J.; Su, W.; Wu, H.; Shah, K.; Xing, L.; Ma, J.; Zhang, D.; Zhao, C. Selection and validation of reliable reference genes for gene expression studies in different genotypes and TRV-infected fruits of peach (Prunus persica L. Batsch) during ripening. Genes 2022, 13, 160. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.C.; Pan, H.; Zhang, J.; Wang, Q.; Gao, H.Y.; Chen, Y.; Huang, X.G. Selection and validation of reference genes for quantitative real-time polymerase chain reaction (qRT-PCR) after different shoot pinching treatments on grape (Vitis vinifera L.). J. Jiangxi Agric. Univ. 2019, 41, 890–900. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, L.; Qu, F.; Yang, S.J. Reference gene screening for real-time quantitative PCR in red pear (Pyrus pyrifolia). J. Agric. Biotechnol. 2019, 27, 361–370. [Google Scholar]
- Zdarska, I.; Cmejla, R. Effect of long-term storage on the change in the expression of selected Mald1 gene isoforms in the apple cultivar Opal. Czech J. Genet. Plant Breed. 2023, 59, 141–147. [Google Scholar] [CrossRef]
- Feng, S.; Yan, C.; Zhang, T.; Ji, M.; Tao, R.; Gao, H. Comparative Study of volatile compounds and expression of related genes in fruit from two apple cultivars during different developmental stages. Molecules 2021, 26, 1553. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, J.; Li, H.; Luo, D.; Xie, J.; Li, H.; Liu, S.; Zhang, Y.; Chen, L.; Xie, X.; et al. A preliminary study on the root-knot nematode resistance of the cherry plum cultivar Mirabolano 29C. Czech J. Genet. Plant Breed. 2023, 59, 133–140. [Google Scholar] [CrossRef]
- Li, D.; Li, X.; Cheng, Y.; Guan, J. Effect of 1-methylcyclopropene on peel greasiness, yellowing, and related gene expression in postharvest ‘Yuluxiang’ pear. Front. Plant Sci. 2023, 13, 1082041. [Google Scholar] [CrossRef]
- Liu, D.; Ma, Q.; Yang, L.; Hu, W.; Guo, W.; Wang, M.; Zhou, R.; Liu, Y. Comparative analysis of the cuticular waxes and related gene expression between ‘Newhall’ and ‘Ganqi 3′ navel orange during long-term cold storage. Plant Physiol. Biochem. 2021, 167, 1049–1060. [Google Scholar] [CrossRef]
- Jiang, Z.; Huang, Q.; Jia, D.; Zhong, M.; Tao, J.; Liao, G.; Huang, C.; Xu, X. Characterization of organic acid metabolism and expression of related genes during fruit development of Actinidia eriantha ‘Ganmi 6’. Plants 2020, 9, 332. [Google Scholar] [CrossRef]
- Lee, K.; Lee, J.G.; Min, K.; Choi, J.H.; Lim, S.; Lee, E.J. Transcriptome analysis of the fruit of two strawberry cultivars “Sunnyberry” and “Kingsberry” that show different susceptibility to Botrytis cinerea after harvest. Int. J. Mol. Sci. 2021, 22, 1518. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Deng, H.; Xiong, B.; Li, S.; Yang, L.; Yang, Y.; Huang, S.; Tan, L.; Sun, G.; Wang, Z. Rootstock effects on anthocyanin accumulation and associated biosynthetic gene expression and enzyme activity during fruit development and ripening of blood oranges. Agriculture 2022, 12, 342. [Google Scholar] [CrossRef]
- Sachin, A.J.; Rao, D.V.S.; Ravishankar, K.; Ranjitha, K.; Vasugi, C.; Narayana, C.K.; Reddy, S.V.R. 1-MCP treatment modulated physiological, biochemical and gene expression activities of guava during low-temperature storage. Acta Physiol. Plant. 2022, 44, 125. [Google Scholar] [CrossRef]
- Tatmala, N.; Ma, G.; Zhang, L.; Kato, M.; Kaewsuksaen, S. Characterization of carotenoid accumulation and carotenogenic gene expression during fruit ripening in red colored pulp of ‘Siam Red Ruby’ pumelo (Citrus grandis) cultivated in Thailand. Hortic. J. 2020, 89, 237–243. [Google Scholar] [CrossRef]
- Gonzalez-Aguilera, K.L.; Saad, C.F.; Chavez, M.R.; Alves-Ferreira, M.; de Folter, S. Selection of reference genes for quantitative real-time RT-PCR studies in tomato fruit of the genotype MT-Rg1. Front. Plant Sci. 2016, 7, 1386. [Google Scholar] [CrossRef]
| Gene Symbols | Full Names | Functions |
|---|---|---|
| ACT | Actin | An important skeleton protein of the cell |
| EF-1α | Elongation factor-1α | Elongation of transcription |
| GAPDH | Glyceraldehyde-3-phosphate-dehydrogenase | Key enzymes in the carbon fixation cycle of glycolysis, gluconeogenesis, and photosynthesis |
| His | Histone | Formation of higher chromosome structures |
| β-ACT | Beta actin | Maintenance of cellular structure, intracellular movement, and cell division |
| 18s rRNA | 18s ribosomal RNA | Cytoplasmic ribosome small subunit, translation |
| α-Tub | Alpha tubulin | Cytoskeletal structural proteins |
| β-Tub | Beta tubulin | Cells grow and participate in light-stimulating responses |
| UBC | Ubiquitin conjugating enzyme | Label proteins that need to be broken down, causing them to hydrolyze |
| UBQ | Ubiquitin | Protein modification, binding, and degradation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, S.; Ali Sabir, I.; Hu, X.; Chen, J.; Qin, Y. Advancements in Reference Gene Selection for Fruit Trees: A Comprehensive Review. Int. J. Mol. Sci. 2024, 25, 1142. https://doi.org/10.3390/ijms25021142
Peng S, Ali Sabir I, Hu X, Chen J, Qin Y. Advancements in Reference Gene Selection for Fruit Trees: A Comprehensive Review. International Journal of Molecular Sciences. 2024; 25(2):1142. https://doi.org/10.3390/ijms25021142
Chicago/Turabian StylePeng, Shujun, Irfan Ali Sabir, Xinglong Hu, Jiayi Chen, and Yonghua Qin. 2024. "Advancements in Reference Gene Selection for Fruit Trees: A Comprehensive Review" International Journal of Molecular Sciences 25, no. 2: 1142. https://doi.org/10.3390/ijms25021142
APA StylePeng, S., Ali Sabir, I., Hu, X., Chen, J., & Qin, Y. (2024). Advancements in Reference Gene Selection for Fruit Trees: A Comprehensive Review. International Journal of Molecular Sciences, 25(2), 1142. https://doi.org/10.3390/ijms25021142





