A Narrative Review on Oral and Periodontal Bacteria Microbiota Photobiomodulation, through Visible and Near-Infrared Light: From the Origins to Modern Therapies
Abstract
:1. Light Is Life: The Origin
2. Light Is Life: The Evolution
3. Light Is Life: The Therapy
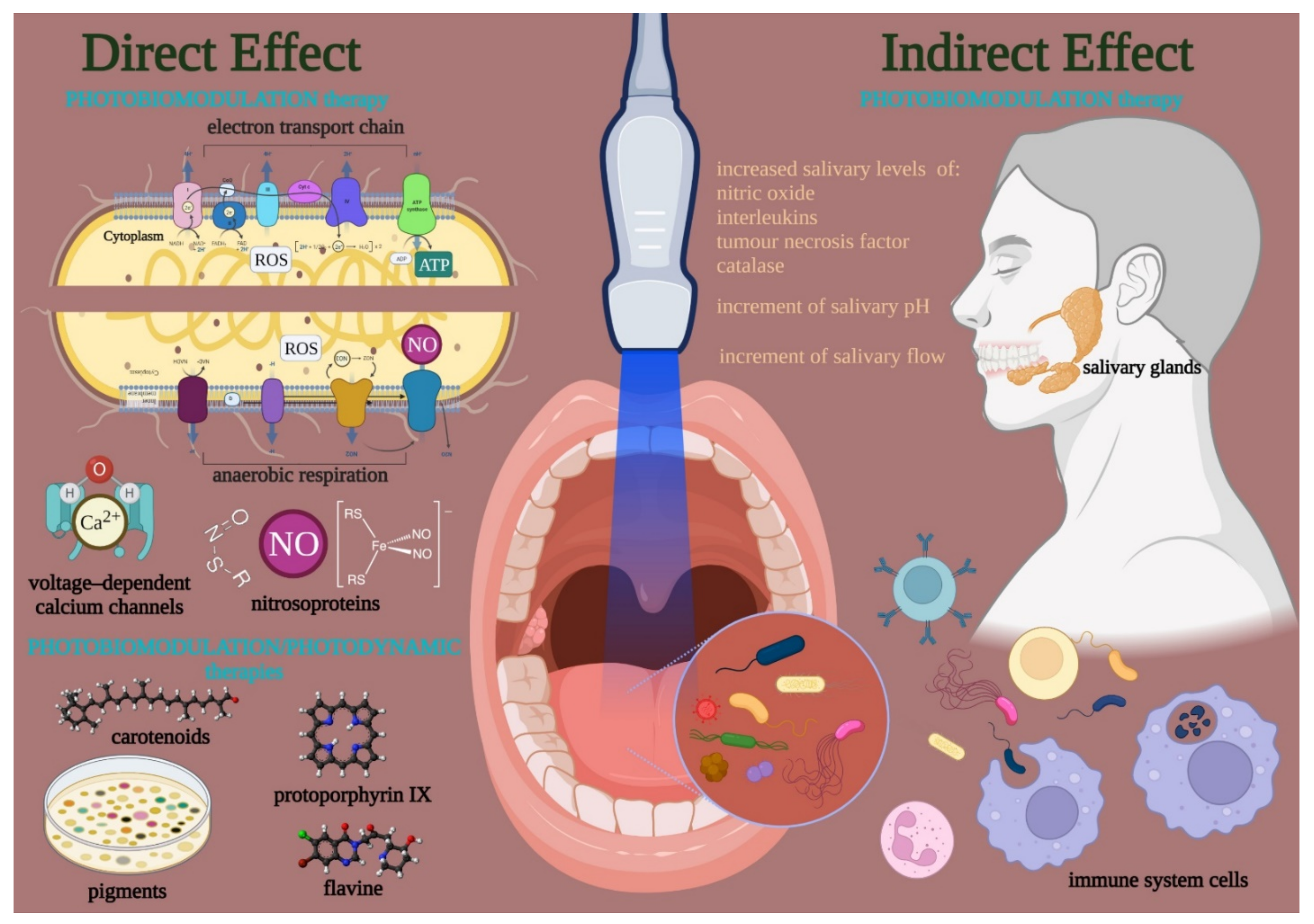
3.1. Photobiomodulation
3.2. The Oral Microbiota in Health and Disease
- gram-positive: cocci—Abiotrophia, Peptostreptococcus, Streptococcus, and Stomatococcus; rods—Actinomyces, Bifidobacterium, Corynebacterium, Eubacterium, Lactobacillus, Propionibacterium, Pseudoramibacter, and Rothia;
- gram-negative: cocci—Moraxella, Neisseria, and Veillonella; rods—Campylobacter, Capnocytophaga, Desulfobacter, Desulfovibrio, Eikenella, Fusobacterium, Hemophilus, Leptotrichia, Prevotella, Selemonas, Simonsiella, Treponema, and Wolinella;
- the uncultured divisions GN02, SR1, and TM7 [89].
3.3. Photobiomodulation on Bacterial Microbiota
3.3.1. Evidence-Based Literature
3.3.2. Possible Mechanism of Action
4. Light Is Life: Outlook for the Future
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Witzany, G. Crucial steps to life: From chemical reactions to code using agents. Biosystems 2016, 140, 49–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marijuán, P.C.; Navarro, J. From Molecular Recognition to the “Vehicles” of Evolutionary Complexity: An Informational Approach. Int. J. Mol. Sci. 2021, 22, 11965. [Google Scholar] [CrossRef] [PubMed]
- Trevors, J.T. Origin of life: Hypothesized roles of high-energy electrical discharges, infrared radiation, thermosynthesis and pre-photosynthesis. Theory Biosci. 2012, 131, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Barge, L.M. Considering planetary environments in origin of life studies. Nat. Commun. 2018, 9, 5170. [Google Scholar] [CrossRef] [PubMed]
- Oparin, A.I. Genesis and Evolutionary Development of Life; Academic Press: New York, NY, USA, 1968. [Google Scholar]
- Urey, H.C. The Planets: Their Origin and Development; Yale University Press: New Haven, CT, USA, 1952. [Google Scholar]
- Rubey, W.W. Geological history of sea water: An attempt to state the problem. Bull. Geol. Soc. Am. 1951, 62, 1111–1148. [Google Scholar] [CrossRef]
- Holland, H.D. Model for the evolution of the Earth’s atmosphere. In Petrologic Studies: A Volume to Honour A.F. Buddington; Engel, A.E., James, H.L., Leonard, B.F., Eds.; Geological Society of America: New York, NY, USA, 1962; pp. 447–477. [Google Scholar]
- Luisi, P.L. The Emergence of Life; Cambridge University Press: Cambridge, UK, 2006. [Google Scholar]
- Dalai, P.; Sahai, N. Mineral-Lipid Interactions in the Origins of Life. Trends Biochem. Sci. 2019, 44, 331–341. [Google Scholar] [CrossRef]
- Baur, M.E. Thermodynamics of heterogeneous iron·carbon systems. Implications for the terrestrial primitive reducing atmosphere. Chem. Ceol. 1978, 22, 189–206. [Google Scholar]
- Lu, A.; Li, Y.; Ding, H. Photoelectric conversion on Earth’s surface via widespread Fe- and Mn-mineral coatings. Proc. Natl. Acad. Sci. USA 2019, 116, 9741–9746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, S.W.; Dose, K. Molecular Evolution and the Origins of Life; W.H. Freeman: San Francisco, CA, USA, 1972. [Google Scholar]
- Kritsky, M.S.; Telegina, T.A.; Buglak, A.A. Modeling of abiotic ATP synthesis in the context of problems of early biosphere evolution. Geochem. Int. 2014, 52, 1227–1238. [Google Scholar] [CrossRef]
- Kolesnikov, M.P.; Kritsky, M.S. Study of chemical structure and of photochemical activity of biogenic flavin pigment. J. Evol. Biochem. Physiol. 2001, 37, 507–514. [Google Scholar] [CrossRef]
- Losi, A. Flavinbased bluelight photosensors: A photobiophysics update. Photochem. Photobiol. 2007, 83, 1283–1300. [Google Scholar] [CrossRef] [PubMed]
- Bahn, P.R.; Fox, S.W. Models for protocellular photophosphorylation. BioSystems 1981, 14, 3–14. [Google Scholar] [CrossRef]
- Kritsky, M.S.; Kolesnikov, M.P.; Telegina, T.A. Modeling of abiogenic synthesis of ATP. Dokl. Biochem. Biophys. 2007, 417, 313–315. [Google Scholar] [CrossRef]
- Krasnovsky, A.A.; Umrikhina, A.V. On abiogenic formation of porphin and its role in the process of photochemical electron transport. Dokl. ANSSSR 1964, 155, 691. [Google Scholar]
- Szutka, A. porphine-like substances: Probable synthesis during chemical evolution. Nature 1964, 202, 1231–1232. [Google Scholar] [CrossRef]
- Olson, J.M.; Pierson, B.K. Origin and evolution of photosynthetic reaction centers. Orig. Life Evol. Biosph. 1987, 17, 419–430. [Google Scholar] [CrossRef]
- Lozovaya, G.I.; Masinovsky, Z.; Sivash, A.A. Protoporphyrin ix as a possible ancient photosensitizer: Spectral and photochemical studies. Orig. Life Evol. Biosph. 1990, 20, 321–330. [Google Scholar] [CrossRef]
- Kolesnikov, M.P.; Telegina, T.A.; Lyudnikova, T.A.; Kritsky, M.S. Abiogenic photophosphorylation of ADP to ATP sensitized by flavoproteinoid microspheres. Orig. Life Evol. Biosph. 2008, 38, 243–255. [Google Scholar] [CrossRef]
- Nelson, K.E.; Levy, M.; Miller, S.L. Peptide nucleic acids rather than RNA may have been the first genetic molecule. Proc. Natl. Acad. Sci. USA 2000, 97, 3868–3871. [Google Scholar] [CrossRef] [Green Version]
- Tamulis, A. Quantum mechanical interpretation of the origin of life. In Science in the Faculty of Natural Sciences of Vilnius University; Ruksenas, O., Ed.; Publishing house of Vilnius University: Vilnius, Lithuania, 2008; pp. 7–19. [Google Scholar]
- Dzieciol, A.J.; Mann, S. Designs for life: Protocell models in the laboratory. Chem. Soc. Rev. 2012, 41, 79–85. [Google Scholar] [CrossRef]
- Soo, R.M.; Hemp, J.; Parks, D.H.; Fischer, W.W.; Hugenholtz, P. On the origins of oxygenic photosynthesis and aerobic respiration in Cyanobacteria. Science 2017, 355, 1436–1440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sessions, A.L.; Doughty, D.M.; Welander, P.V.; Summons, R.E.; Newman, D.K. The continuing puzzle of the great oxidation event. Curr. Biol. 2009, 19, 567–574. [Google Scholar] [CrossRef] [Green Version]
- Schäfer, G.; Engelhard, M.; Müller, V. Bioenergetics of the Archaea. Microbiol. Mol. Biol. Rev. 1999, 63, 570–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esposti, M.D. On the evolution of cytochrome oxidases consuming oxygen. Biochim. Biophys. Acta Bioenerg. 2020, 1861, 148304. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, G.; Purschke, W.; Schmidt, C.L. On the origin of respiration: Electron transport proteins from archaea to man. FEMS Microbiol. Rev. 1996, 18, 173–188. [Google Scholar] [CrossRef]
- Castresana, J.; Saraste, M. Evolution of energetic metabolism: The respiration-early hypothesis. Trends Biochem. Sci. 1995, 20, 443–448. [Google Scholar] [CrossRef]
- Woese, C.R.; Kandler, O.; Wheelis, M.L. Towards a natural system of organisms, proposal for the domains archaea, bacteria and eucarya. Proc. Natl. Acad. Sci. USA 1990, 87, 4576–4579. [Google Scholar] [CrossRef] [Green Version]
- Gogarten, J.P.; Iaiz, L. Evolution of proton pumping ATPases: Rooting the tree of life. Photosynth. Res. 1992, 33, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Kracke, F.; Vassilev, I.; Krömer, J.O. Microbial electron transport and energy conservation—The foundation for optimizing bioelectrochemical systems. Front. Microbiol. 2015, 6, 575. [Google Scholar] [CrossRef] [Green Version]
- Sousa, F.L.; Alves, R.J.; Ribeiro, M.A.; Pereira-Leal, J.B.; Teixeira, M.; Pereira, M.M. The superfamily of heme-copper oxygen reductases: Types and evolutionary considerations. Biochim. Biophys. Acta 2012, 1817, 629–637. [Google Scholar] [CrossRef] [Green Version]
- Hernandez, M.; Newman, D. Extracellular electron transfer. Cell. Mol. Life Sci. 2001, 58, 1562–1571. [Google Scholar] [CrossRef]
- John, P.; Whatley, F. Paracoccus denitrificans and the evolutionary origin of the mitochondrion. Nature 1975, 254, 495–498. [Google Scholar] [CrossRef] [PubMed]
- Van Spanning, R.J.; de Boer, A.P.; Reijnders, W.N.; De Gier, J.W.; Delorme, C.O.; Stouthamer, A.H.; Westerhoff, H.V.; Harms, N.; van der Oost, J. Regulation of oxidative phosphorylation: The flexible respiratory network of Paracoccus denitrificans. J. Bioenerg. Biomembr. 1995, 27, 499–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hino, T.; Matsumoto, Y.; Nagano, S.; Sugimoto, H.; Fukumori, Y.; Murata, T.; Iwata, S.; Shiro, Y. Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science 2010, 330, 1666–1670. [Google Scholar] [CrossRef] [PubMed]
- Moodie, A.D.; Ingledew, W.J. Microbial anaerobic respiration. Adv. Microb. Physiol. 1990, 31, 225–269. [Google Scholar] [PubMed]
- O’Malley, M.A. Endosymbiosis and its implications for evolutionary theory. Proc. Natl. Acad. Sci. USA 2015, 112, 10270–10277. [Google Scholar] [CrossRef] [Green Version]
- Martin, W.F.; Garg, S.; Zimorski, V. Endosymbiotic theories for eukaryote origin. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140330. [Google Scholar] [CrossRef] [Green Version]
- Zimorski, V.; Ku, C.; Martin, W.F.; Gould, S.B. Endosymbiotic theory for organelle origins. Curr. Opin. Microbiol. 2014, 22, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 2010, 11, 209. [Google Scholar] [CrossRef]
- Berry, S. Endosymbiosis and the design of eukaryotic electron transport. Biochim. Biophys. Acta 2003, 1606, 57–72. [Google Scholar] [CrossRef] [Green Version]
- Archibald, J.M. Endosymbiosis and Eukaryotic Cell Evolution. Curr. Biol. 2015, 25, 911–921. [Google Scholar] [CrossRef] [Green Version]
- Bewley, M.C.; Marohnic, C.C.; Barber, M.J. The structure and biochemistry of NADH-dependent cytochrome b5 reductase are now consistent. Biochemistry 2001, 40, 13574–13582. [Google Scholar] [CrossRef] [PubMed]
- Niklas, K.J. The Evolutionary Biology of Plant; University of Chicago: Chicago, IL, USA, 1997; p. 449. [Google Scholar]
- Anders, J.J.; Lanzafame, R.J.; Arany, P.R. Low-level light/laser therapy versus photobiomodulation therapy. Photomed. Laser Surg. 2015, 33, 183–184. [Google Scholar] [CrossRef] [Green Version]
- Albini, A. Some remarks on the first law of photochemistry. Photochem. Photobiol. Sci. 2016, 15, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Pastore, D.; Greco, M.; Passarella, S. Specific helium-neon laser sensitivity of the purified cytochrome c oxidase. Int. J. Radiat. Biol. 2000, 76, 863–870. [Google Scholar] [CrossRef]
- Karu, T.I. Multiple roles of cytochrome c oxidase in mammalian cells under action of red and IR-A radiation. IUBMB Life 2010, 62, 607–610. [Google Scholar] [CrossRef] [PubMed]
- Amaroli, A.; Ravera, S.; Parker, S.; Panfoli, I.; Benedicenti, A.; Benedicenti, S. An 808-nm Diode Laser with a Flat-Top Handpiece Positively Photobiomodulates Mitochondria Activities. Photomed. Laser Surg. 2016, 34, 564–571. [Google Scholar] [CrossRef]
- Amaroli, A.; Pasquale, C.; Zekiy, A.; Utyuzh, A.; Benedicenti, S.; Signore, A.; Ravera, S. Photobiomodulation and oxidative stress: 980 nm diode-laser light regulates mitochondria activity and reactive oxygen species production. Oxid. Med. Cell. Longev. 2021, 3, 6626286. [Google Scholar] [CrossRef]
- Ravera, S.; Ferrando, S.; Agas, D.; De Angelis, N.; Raffetto, M.; Sabbieti, M.G.; Signore, A.; Benedicenti, S.; Amaroli, A. 1064 nm Nd:YAG laser light affects transmembrane mitochondria respiratory chain complexes. J. Biophotonics 2019, 12, 201900101. [Google Scholar] [CrossRef]
- Swartz, T.E.; Corchnoy, S.B.; Christie, J.M.; Lewis, J.W.; Szundi, I.; Briggs, W.R.; Bogomolni, R.A. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J. Biol. Chem. 2001, 276, 36493–36500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buravlev, E.A.; Zhidkova, T.V.; Vladimirov, Y.A.; Osipov, A.N. Effects of low-level laser therapy on mitochondrial respiration and nitrosyl complex content. Lasers Med. Sci. 2014, 29, 861–1866. [Google Scholar] [CrossRef] [PubMed]
- Koren, K.; Borisov, S.M.; Saf, R.; Klimant, I. Strongly Phosphorescent Iridium(III) Porphyrins—New Oxygen Indicators with Tuneable Photophysical Properties and Functionalities. Eur. J. Inorg. Chem. 2011, 10, 1531–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werck-Reichhart, D.; Feyereisen, R. Cytochromes P450: A success story. Genome Biol. 2000, 1, 9. [Google Scholar] [CrossRef] [Green Version]
- Colombo, E.; Signore, A.; Aicardi, S.; Zekiy, A.; Utyuzh, A.; Benedicenti, S.; Amaroli, A. Experimental and Clinical Applications of Red and Near-Infrared Photobiomodulation on Endothelial Dysfunction: A Review. Biomedicines 2021, 9, 274. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, Y.-Y.; Wang, Y.; Lyu, P.; Hamblin, M.R. Photobiomodulation of human adipose-derived stem cells using 810 nm and 980 nm lasers operates via different mechanisms of action. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 441–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jansen, K.; Wu, M.; van der Steen, A.F. Photoacoustic imaging of human coronary atherosclerosis in two spectral bands. Photoacoustics 2013, 5, 12–20. [Google Scholar]
- Castellano-Pellicena, I.; Uzunbajakava, N.E.; Mignon, C.; Raafs, B.; Botchkarev, V.A.; Thornton, M.J. Does blue light restore human epidermal barrier function via activation of Opsin during cutaneous wound healing? Lasers Surg. Med. 2019, 51, 370–382. [Google Scholar] [CrossRef] [Green Version]
- Verbon, E.H.; Post, J.A.; Boonstra, J. The influence of reactive oxygen species on cell cycle progression in mammalian cells. Gene 2012, 511, 1–6. [Google Scholar] [CrossRef]
- Whitaker, M.; Patel, R. Calcium and cell cycle control. Development 1990, 108, 525–542. [Google Scholar] [CrossRef]
- Villalobo, A. Nitric oxide and cell proliferation. FEBS J. 2006, 273, 2329–2344. [Google Scholar] [CrossRef]
- Ravera, S.; Colombo, E.; Pasquale, C.; Benedicenti, S.; Solimei, L.; Signore, A.; Amaroli, A. Mitochondrial Bioenergetic, Photobiomodulation and Trigeminal Branches Nerve Damage, What’s the Connection? A Review. Int. J. Mol. Sci. 2021, 22, 4347. [Google Scholar] [CrossRef] [PubMed]
- Amaroli, A.; Ferrando, S.; Benedicenti, S. Photobiomodulation Affects Key Cellular Pathways of all Life-Forms: Considerations on Old and New Laser Light Targets and the Calcium Issue. Photochem. Photobiol. 2019, 95, 455–459. [Google Scholar] [CrossRef] [Green Version]
- Amaroli, A.; Colombo, E.; Zekiy, A.; Aicardi, S.; Benedicenti, S.; De Angelis, N. Interaction between Laser Light and Osteoblasts: Photobiomodulation as a Trend in the Management of Socket Bone Preservation—A Review. Biology 2020, 9, 409. [Google Scholar] [CrossRef] [PubMed]
- Agas, D.; Hanna, R.; Benedicenti, S.; De Angelis, N.; Sabbieti, M.G.; Amaroli, A. Photobiomodulation by Near-Infrared 980-nm Wavelengths Regulates Pre-Osteoblast Proliferation and Viability through the PI3K/Akt/Bcl-2 Pathway. Int. J. Mol. Sci. 2021, 22, 7586. [Google Scholar] [CrossRef] [PubMed]
- Pasquale, C.; Utyuzh, A.; Mikhailova, M.V.; Colombo, E.; Amaroli, A. Recovery from Idiopathic Facial Paralysis (Bell’s Palsy) Using Photobiomodulation in Patients Non-Responsive to Standard Treatment: A Case Series Study. Photonics 2021, 8, 341. [Google Scholar] [CrossRef]
- Cassano, P.; Petrie, S.R.; Hamblin, M.R.; Henderson, T.A.; Iosifescu, D.V. Review of transcranial photobiomodulation for major depressive disorder: Targeting brain metabolism, inflammation, oxidative stress, and neurogenesis. Neurophotonics 2016, 3, 031404. [Google Scholar] [CrossRef] [Green Version]
- Hanna, R.; Dalvi, S.; Benedicenti, S.; Amaroli, A.; Sălăgean, T.; Pop, I.D.; Todea, D.; Bordea, I.R. Photobiomodulation Therapy in Oral Mucositis and Potentially Malignant Oral Lesions: A Therapy Towards the Future. Cancers 2020, 12, 1949. [Google Scholar] [CrossRef]
- Zadik, Y.; Arany, P.R.; Fregnani, E.R.; Bossi, P.; Antunes, H.S.; Bensadoun, R.J.; Gueiros, L.A.; Majorana, A.; Nair, R.G.; Ranna, V. Mucositis Study Group of the Multinational Association of Supportive Care in Cancer/International Society of Oral Oncology (MASCC/ISOO). Systematic review of photobiomodulation for the management of oral mucositis in cancer patients and clinical practice guidelines. Support. Care Cancer 2019, 27, 3969–3983. [Google Scholar]
- Ravera, S.; Bertola, N.; Pasquale, C.; Bruno, S.; Benedicenti, S.; Ferrando, S.; Zekiy, A.; Arany, P.; Amaroli, A. 808-nm Photobiomodulation Affects the Viability of a Head and Neck Squamous Carcinoma Cellular Model, Acting on Energy Metabolism and Oxidative Stress Production. Biomedicines 2021, 9, 1717. [Google Scholar] [CrossRef]
- De Pauli Paglioni, M.; Araújo, A.L.D.; Arboleda, L.P.A.; Palmier, N.R.; Fonsêca, J.M.; Gomes-Silva, W.; Madrid-Troconis, C.C.; Silveira, F.M.; Martins, M.D.; Faria, K.M. Tumor safety and side effects of photobiomodulation therapy used for prevention and management of cancer treatment toxicities. A systematic review. Oral Oncol. 2019, 93, 21–28. [Google Scholar] [CrossRef]
- Bensadoun, R.J.; Epstein, J.B.; Nair, R.G.; Barasch, A.; Raber-Durlacher, J.E.; Migliorati, C.; Genot-Klastersky, M.T.; Treister, N.; Arany, P.; Lodewijckx, J. Safety and efficacy of photobiomodulation therapy in oncology: A systematic review. Cancer Med. 2020, 9, 8279–8300. [Google Scholar] [CrossRef] [PubMed]
- Jensen, S.B.; Mouridsen, H.T.; Bergmann, O.J.; Reibel, J.; Breunner, N.; Nauntofte, B. Oral mucosal lesions, microbial changes, and taste disturbances induced by adjuvant chemotherapy in breast cancer patients. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2008, 106, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Liebert, A.; Bicknell, B.; Johnstone, D.M.; Gordon, L.C.; Kiat, H.; Hamblin, M.R. “Photobiomics”: Can Light, Including Photobiomodulation, Alter the Microbiome? Photobiomodul. Photomed. Laser Surg. 2019, 37, 681–693. [Google Scholar] [CrossRef] [Green Version]
- Pflughoeft, K.J.; Versalovic, J. Human microbiome in health and disease. Annu. Rev. Pathol. 2012, 7, 99–122. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Hord, N.G. Eukaryotic-microbiota cross talk: Potential mechanisms for health benefits of prebiotics and probiotics. Annu. Rev. Nutr. 2008, 28, 215–231. [Google Scholar] [CrossRef]
- Cho, I.; Blaser, M. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef] [Green Version]
- Di Spirito, F.; La Rocca, M.; De Bernardo, M.; Rosa, N.; Sbordone, C.; Sbordone, L. Possible Association of Periodontal Disease and Macular Degeneration: A Case-Control Study. Dent. J. 2021, 9, 1. [Google Scholar] [CrossRef]
- Di Spirito, F.; Toti, P.; Pilone, V.; Carinci, F.; Lauritano, D.; Sbordone, L. The Association between Periodontitis and Human Colorectal Cancer: Genetic and Pathogenic Linkage. Life 2020, 10, 211. [Google Scholar] [CrossRef]
- Deo, P.N.; Deshmukh, R. Oral microbiome: Unveiling the fundamentals. J. Oral Maxillofac. Pathol. 2019, 23, 122–128. [Google Scholar]
- Wade, W.G. The oral microbiome in health and disease. Pharmacol. Res. 2013, 69, 137–143. [Google Scholar] [CrossRef]
- Sharma, N.; Bhatia, S.; Sodhi, A.S.; Batra, N. Oral microbiome and health. AIMS Microbiol. 2018, 4, 42–66. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, Y.; Takeshita, T. The oral microbiome and human health. J. Oral Sci. 2017, 59, 201–206. [Google Scholar] [CrossRef] [Green Version]
- Irfan, M.; Delgado, R.Z.R.; Frias-Lopez, J. The Oral Microbiome and Cancer. Front. Immunol. 2020, 11, 591088. [Google Scholar] [CrossRef] [PubMed]
- Barone, A.; Chatelain, S.; Derchi, G.; Di Spirito, F.; Martuscelli, R.; Porzio, M.; Sbordone, L. Antibiotic’s effectiveness after erupted tooth extractions: A retrospective study. Oral Dis. 2020, 26, 967–973. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Costerton, J.W. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
- Bunce, J.; Hellyer, P. Antibiotic resistance and antibiotic prescribing by dentists in England 2007–2016. Br. Dent. J. 2018, 225, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Lighthart, B. Solar radiation is shown to select for pigmented bacteria in the ambient outdoor atmosphere. Photochem. Photobiol. 1997, 65, 103–106. [Google Scholar] [CrossRef]
- Trushin, M.V. Studies on distant regulation of bacterial growth and light emission. Microbiology 2003, 149, 363–368. [Google Scholar] [CrossRef] [Green Version]
- Lubart, R.; Lipovski, A.; Nitzan, Y.; Friedmann, H. A possible mechanism for the bactericidal effect of visible light. Laser Ther. 2011, 20, 17–22. [Google Scholar] [CrossRef] [Green Version]
- Bordea, I.R.; Hanna, R.; Chiniforush, N.; Grădinaru, E.; Câmpian, R.S.; Sîrbu, A.; Amaroli, A.; Benedicenti, S. Evaluation of the outcome of various laser therapy applications in root canal disinfection: A systematic review. Photodiagn. Photodyn. Ther. 2020, 29, 101611. [Google Scholar] [CrossRef]
- Bicknell, B.; Liebert, A.; Johnstone, D.; Kiat, H. Photobiomodulation of the microbiome: Implications for metabolic and inflammatory diseases. Lasers Med. Sci. 2019, 34, 317–327. [Google Scholar] [CrossRef]
- Thomé Lima, A.M.C.; da Silva Sergio, L.P.; da Silva Neto Trajano, L.A.; de Souza, B.P.; da Motta Mendes, J.P.; Cardoso, A.F.R.; Figueira, C.P.; Dos Anjos Tavares, B.; Figueira, D.S.; Mencalha, A.L.; et al. Photobiomodulation by dualwavelength low-power laser effects on infected pressure ulcers. Lasers Med. Sci. 2020, 35, 651–660. [Google Scholar] [CrossRef]
- Amaroli, A.; Ferrando, S.; Pozzolini, M.; Gallus, L.; Parker, S.; Benedicenti, S. The earthworm Dendrobaena veneta (Annelida): A new experimental-organism for photobiomodulation and wound healing. Eur. J. Histochem. 2018, 62, 2867. [Google Scholar] [PubMed]
- Feuerstein, O.; Persman, N.; Weiss, E.I. Phototoxic effect of visible light on Porphyromonas gingivalis and Fusobacterium nucleatum: An in vitro study. Photochem. Photobiol. 2004, 80, 412–415. [Google Scholar] [CrossRef]
- Henry, C.A.; Judy, M.; Dyer, B.; Wagner, M.; Matthews, J.L. Sensitivity of Porphyromonas and Prevotella species in liquid media to argon laser. Photochem. Photobiol. 1995, 61, 410–413. [Google Scholar] [CrossRef]
- Nussbaum, E.L.; Lilge, L.; Mazzulli, T. Effects of 630-, 660-, 810-, and 905-nm laser irradiation delivering radiant exposure of 1–50 J/cm2 on three species of bacteria in vitro. J. Clin. Laser Med. Surg. 2002, 20, 325–333. [Google Scholar] [CrossRef]
- Nussbaum, E.L.; Lilge, L.; Mazzulli, T. Effects of 810 nm laser irradiation on in vitro growth of bacteria: Comparison of continuous wave and frequency modulated light. Lasers Surg. Med. 2002, 31, 343–351. [Google Scholar] [CrossRef]
- De Sousa, N.T.; Gomes, R.C.; Santos, M.F.; Brandino, H.E.; Martinez, R.; de Jesus Guirro, R.R. Red and infrared laser therapy inhibits in vitro growth of major bacterial species that commonly colonize skin ulcers. Lasers Med. Sci. 2016, 31, 549–556. [Google Scholar] [CrossRef]
- Dixit, S.; Ahmad, I.; Hakami, A.; Gular, K.; Tedla, J.S.; Abohashrh, M. Comparison of Anti-Microbial Effects of Low-Level Laser Irradiation and Microwave Diathermy on Gram-Positive and Gram-Negative Bacteria in an In Vitro Model. Medicina 2019, 55, 330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Sousa, N.T.; Guirro, R.R.; Santana, H.F.; Silva, C.C. In vitro analysis of bacterial morphology by atomic force microscopy of low level laser therapy 660, 830 and 904 nm. Photomed. Laser Surg. 2012, 30, 281–285. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, N.T.; Santos, M.F.; Gomes, R.C.; Brandino, H.E.; Martinez, R.; de Jesus Guirro, R.R. Blue Laser Inhibits Bacterial Growth of Staphylococcus aureus, Escherichia coli, and Pseudomonas aeruginosa. Photomed. Laser Surg. 2015, 33, 278–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinothkumar, T.S.; Apathsakayan, R.; El-Shamy, F.M.M.; Homeida, H.E.; Hommedi, A.I.M.; Safhi, M.Y.A.; Alsalhi, H.A.M. Antibacterial effect of diode laser on different cariogenic bacteria: An In-vitro study. Niger. J. Clin. Pract. 2020, 23, 1578–1582. [Google Scholar] [CrossRef] [PubMed]
- Basso, F.G.; Oliveira, C.F.; Fontana, A.; Kurachi, C.; Bagnato, V.S.; Spolidório, D.M.; Hebling, J.; de Souza Costa, C.A. In vitro effect of low-level laser therapy on typical oral microbial biofilms. Braz. Dent. J. 2011, 22, 502–510. [Google Scholar] [CrossRef]
- Plavskii, V.Y.; Mikulich, A.V.; Tretyakova, A.I.; Leusenka, I.A.; Plavskaya, L.G.; Kazyuchits, O.A.; Dobysh, I.I.; Krasnenkova, T.P. Porphyrins and flavins as endogenous acceptors of optical radiation of blue spectral region determining photoinactivation of microbial cells. J. Photochem. Photobiol. B 2018, 183, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Hope, C.K.; Strother, M.; Creber, H.K.; Higham, S.M. Lethal photosensitisation of Prevotellaceae under anaerobic conditions by their endogenous porphyrins. Photodiagn. Photodyn. Ther. 2016, 13, 344–346. [Google Scholar] [CrossRef]
- Yoshida, A.; Sasaki, H.; Toyama, T.; Araki, M.; Fujioka, J.; Tsukiyama, K.; Hamada, N.; Yoshino, F. Antimicrobial effect of blue light using Porphyromonas gingivalis pigment. Sci. Rep. 2017, 7, 5225. [Google Scholar] [CrossRef]
- Dai, T.; Gupta, A.; Huang, Y.Y.; Yin, R.; Murray, C.K.; Vrahas, M.S.; Sherwood, M.E.; Tegos, G.P.; Hamblin, M.R. Blue light rescues mice from potentially fatal Pseudomonas aeruginosa burn infection: Efficacy, safety, and mechanism of action. Antimicrob. Agents Chemother. 2013, 57, 1238–1245. [Google Scholar] [CrossRef] [Green Version]
- Fukui, M.; Yoshioka, M.; Satomura, K.; Nakanishi, H.; Nagayama, M. Specific-wavelength visible light irradiation inhibits bacterial growth of Porphyromonas gingivalis. J. Periodont. Res. 2008, 43, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Dadras, S.; Mohajerani, E.; Eftekhar, F.; Hosseini, M. Different photoresponses of Staphylococcus aureus and Pseudomonas aeruginosa to 514, 532, and 633 nm low level lasers in vitro. Curr. Microbiol. 2006, 53, 282–286. [Google Scholar] [CrossRef]
- Karu, T.I.; Tiphlova, O.A.; Letokhov, V.S.; Lobko, V.V. Stimulation of E. coli growth by laser and incoherent red light. Il Nuovo Cimento D 1983, 2, 1138–1144. [Google Scholar] [CrossRef]
- Karu, T.; Tiphlova, O.; Samokhina, M.; Diamantopoulos, C.; Sarantsev, V.P.; Shveikin, V. Effects of near-infrared laser and superluminous diode irradiation on Escherichia coli division rate. IEEE J. Quantum Eletron. 1990, 26, 2162–2165. [Google Scholar] [CrossRef]
- Karu, T.; Tiphlova, O.; Esenaliev, R.; Letokhov, V. Two different mechanisms of low-intensity laser photobiological effects on Escherichia coli. J. Photochem. Photobiol. B 1994, 24, 155–161. [Google Scholar] [CrossRef]
- Bertoloni, G.; Sacchetto, R.; Baro, E.; Ceccherelli, F.; Jori, G. Biochemical and morphological changes in Escherichia coli irradiated by coherent and non-coherent 632.8 nm light. J. Photochem. Photobiol. B 1993, 18, 191–196. [Google Scholar] [CrossRef]
- Theodoro, L.H.; Longo, M.; Ervolino, E.; Duque, C.; Ferro-Alves, M.L.; Assem, N.Z.; Louzada, L.M.; Garcia, V.G. Effect of low-level laser therapy as an adjuvant in the treatment of periodontitis induced in rats subjected to 5-fluorouracil chemotherapy. J. Periodont. Res. 2016, 51, 669–680. [Google Scholar] [CrossRef]
- Leanse, L.G.; Dos Anjos, C.; Mushtaq, S.; Dai, T. Antimicrobial blue light: A ‘Magic Bullet’ for the 21st century and beyond? Adv. Drug Deliv. Rev. 2022, 180, 114057. [Google Scholar] [CrossRef] [PubMed]
- De Souza da Fonseca, A.; da Silva Sergio, L.P.; Mencalha, A.L.; de Paoli, F. Low-power lasers on bacteria: Stimulation, inhibition, or effectless? Lasers Med. Sci. 2021, 36, 1791–1805. [Google Scholar] [CrossRef] [PubMed]
- Lushchak, V.I. Oxidative stress and mechanisms of protection against it in bacteria. Biochemistry 2001, 66, 476–489. [Google Scholar]
- Ezraty, B.; Gennaris, A.; Barras, F. Oxidative stress, protein damage and repair in bacteria. Nat. Rev. Microbiol. 2017, 15, 385–396. [Google Scholar] [CrossRef]
- Das, A.; Silaghi-Dumitrescu, R.; Ljungdahl, L.G.; Kurtz, D.M., Jr. Cytochrome bd oxidase, oxidative stress, and dioxygen tolerance of the strictly anaerobic bacterium Moorella thermoacetica. J. Bacteriol. 2005, 187, 2020–2029. [Google Scholar] [CrossRef] [Green Version]
- Verkhratsky, A.; Parpura, V. Calcium signalling and calcium channels: Evolution and general principles. Eur. J. Pharmacol. 2014, 739, 1–3. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.Z.; Chen, J.Y.; Yu, J.T.; Zhou, L.W. Effects of low power laser irradiation on intracellular calcium and histamine release in RBL-2H3 mast cells. Photochem. Photobiol. 2007, 83, 979–984. [Google Scholar] [CrossRef]
- Smalley, J.W.; Silver, J.; Birss, A.J.; Withnall, R.; Titler, P.J. The haem pigment of the oral anaerobes Prevotella nigrescens and Prevotella intermedia is composed of iron(III) protoporphyrin IX in the monomeric form. Microbiology 2003, 149, 1711–1718. [Google Scholar] [CrossRef] [Green Version]
- Soukos, N.S.; Som, S.; Abernethy, A.D.; Ruggiero, K.; Dunham, J.; Lee, C.; Doukas, A.G.; Goodson, J.M. Phototargeting oral black-pigmented bacteria. Antimicrob. Agents Chemother. 2005, 49, 1391–1396. [Google Scholar] [CrossRef] [Green Version]
- Amaroli, A.; Barbieri, R.; Signore, A.; Marchese, A.; Parker, S.; De Angelis, N.; Benedicenti, S. Simultaneous photoablative and photodynamic 810-nm diode laser therapy as an adjunct to non-surgical periodontal treatment: An in-vitro study. Minerva Stomatol. 2020, 69, 1–7. [Google Scholar] [CrossRef]
- Naziya, N.; Rehman, M.A.; Dixit, P.P. Influence of light wavelengths, light intensity, temperature, and pH on biosynthesis of extracellular and intracellular pigment and biomass of Pseudomonasaeruginosa NR1. J. King Saud Univ. Sci. 2020, 32, 745–752. [Google Scholar]
- Marsh, P.D.; Do, T.; Beighton, D.; Devine, D.A. Influence of saliva on the oral microbiota. Periodontology 2000 2016, 70, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Rusthen, S.; Kristoffersen, A.K.; Young, A. Dysbiotic salivary microbiota in dry mouth and primary Sjögren’s syndrome patients. PLoS ONE 2019, 14, 0218319. [Google Scholar] [CrossRef] [PubMed]
- Lynge Pedersen, A.M.; Belstrøm, D. The role of natural salivary defences in maintaining a healthy oral microbiota. J. Dent. 2019, 80, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Sousa, A.S.; Silva, J.F.; Pavesi, V.C.S. Photobiomodulation and salivary glands: A systematic review. Lasers Med. Sci. 2020, 35, 777–788. [Google Scholar] [CrossRef]
- Li, H.; Sun, T.; Liu, C.; Cao, Y.; Liu, X. Photobiomodulation (450 nm) alters the infection of periodontitis bacteria via the ROS/MAPK/mTOR signaling pathway. Free Radic. Biol. Med. 2020, 152, 838–853. [Google Scholar] [CrossRef] [PubMed]
- Ailioaie, L.M.; Litscher, G. Probiotics, Photobiomodulation, and Disease Management: Controversies and Challenges. Int. J. Mol. Sci. 2021, 22, 4942. [Google Scholar] [CrossRef] [PubMed]
- De Castro, M.S.; Miyazawa, M.; Nogueira, E.S.C.; Chavasco, J.K.; Brancaglion, G.A.; Cerdeira, C.D.; Nogueira, D.A.; Ionta, M.; Hanemann, J.A.C.; Brigagão, M.R.P.L.; et al. Photobiomodulation enhances the Th1 immune response of human monocytes. Lasers Med. Sci. 2020. [Google Scholar] [CrossRef] [PubMed]
- Agogué, H.; Joux, F.; Obernosterer, I.; Lebaron, P. Resistance of marine bacterioneuston to solar radiation. Appl. Environ. Microbiol. 2005, 71, 5282–5289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontes, M.; Ruiz-Vázquez, R.; Murillo, F.J. Growth phase dependence of the activation of a bacterial gene for carotenoid synthesis by blue light. EMBO J. 1993, 12, 1265–1275. [Google Scholar] [CrossRef] [PubMed]

| Reference | Bacteria | Parameters | Authors Conclusions |
|---|---|---|---|
| [103] | Porphyromonas gingivalis, Fusobacteriurn nucleatun, Streptococcus mutans, Streptococcus (Enterococcus) faecalis | Halogen lamps (400–500 nm), 0.260–0.416 W/cm2, 16–75 J/cm2, 1 cm2, 60–90–120–150–180 s. Plasma-arc (450–490 nm), 1.144 W/cm2, 69–206 J/cm2, 1 cm2, 60–90–120–150–180 s, CW. LED (450–480 nm), 0.520 W/cm2, 31–94 J/cm2, 1 cm2, 60–90–120–150–180 s, CW Diode laser (830 nm), dose described above | Visible light sources without exogenous photosensitizers have a phototoxic effect mainly on Gram-negative periodontal pathogens. 830-nm did not affect the bacteria. |
| [104] | Porphyromonas endodontalis, P. gingivalis, Prevotella intermedia, Prevotella denticola | Argon laser (488 nm and 514 nm), 0.58 W, 20–200 J/cm2, 3.5–4 cm2, 120–1380 s, CW | Protoporphyrin IX content in black-pigmented bacteria is not the principal factor determining photosensitivity. Oxygen is required during irradiation for black-pigmented bacteria species to be affected. Non-black-pigmented bacteria are much less sensitive to irradiation than black-pigmented bacteria. |
| [105] | Pseudomonas aeruginosa, Escherichia coli, Staphylococcus aureus | Argon-ion pumped tunable dye laser (630–660 nm); Diode lasers (810–905 nm), 0.015 W/cm2, 1–2–5–10–20–50 J/cm2, 66–132–330–658–1320–3300 s | Photobiomodulation applied to wounds in the range of 1–20 J/cm2 may produce changes in bacterial growth of considerable importance for wound healing. A wavelength of 630 nm is most commonly associated with bacterial inhibition. |
| [106] | S. aureus, E. coli, P. aeruginosa | Diode laser (810 nm), 0.015 W/cm2; 1–50 J/cm2; 66–3290 s, CW or 50% duty cycle; 26–5000 Hz | Modulation frequency and radiant exposure of 810 nm laser irradiation significantly influence the effect on particular bacteria. Pulsed laser, at least at a wavelength of 810 nm and high pulse frequency, seems to have the potential to induce growth effects in P. aeruginosa. |
| [107] | S. aureus, E. coli, P. aeruginosa | Diode laser (660, 830 nm), 0.03 W, 3–6–12–18–24 J/cm2, 100–200–400–600–800 s, CW Diode laser (904 nm), 0.04 W, 3–6–12–18–24 J/cm2, 75–150–300–450–600 s | Laser irradiation inhibits the growth of S. aureus at all wavelengths and fluences higher than 12 J/cm2. However, for P. aeruginosa, photobiomodulation inhibits growth at all wavelengths only at a fluence of 24 J/cm2. E. coli has similar growth inhibition at a wavelength of 830 nm at fluences of 3, 6, 12, and 24 J/cm2. At wavelengths of 660 and 904 nm, growth inhibition is only observed at fluences of 12 and 18 J/cm2, respectively. |
| [108] | P. aeruginosa, E. coli, E. faecalis, Staphylococcus epidermidis, Streptococcus pyogenes, Staphylococcus saprophyticus, S. aureus | Ga-Al-As laser (810 nm), 0.36 W/cm2, 13 J/cm2 for 36 s, 18 J/cm2 for 60 s, 30 J/cm2 for 80 s, 0.5 cm2 500 Hz, duty cycle of 50% and voltage of 240 V | Photobiomodulation appears to be an effective treatment for Gram-negative and Gram-positive bacterial strains. |
| [109] | S. aureus | Diode laser (830 nm), 0.03 W, 1, 2, 3, 4, 5, and 16 J/cm2, 4–8–12–16–20–64 s, CW Diode laser (660 nm), 0.03 W, 1, 2, 3, 4, 5, and 16 J/cm2, 2–4–6–8–10–32 s, CW Diode laser (904 nm), 0.04 W, 1, 2, 3, 4, 5, and 16 J/cm2, 3, 6, 9, 12, 15, and 48 s, 9500 Hz, duty cycle of 0.1% | Photobiomodulation at 830 and 904 nm wavelengths reduces the growth of S. aureus. Specifically, the most evident topographical change of the cell structure occurred irradiating at 904 nm at a fluence of 3 J/cm2. |
| [110] | S. aureus, P. aeruginosa, E. coli | Laser (450 nm), 0.07 W, 3–6–12–18–24 J/cm2, 43–86–172–257–343 s, 1 cm2, CW | Blue laser light is capable of inhibiting bacterial growth at low fluences over time, thus presenting no time-dependent effect. |
| [111] | S. mutans, Lactobacillus casei, Actinomyces naeslundii | Diode laser (810 nm), 1–1.5 W, 30 s, flexible the optic fiber of 200 µm diameter | Diode lasers exert an antibacterial effect of varying levels against all three cariogenic bacteria. |
| [112] | S. mutans | InGaAsP diode laser (780 nm), 0.04 W, 5–10–20 J/cm2, 250–500–1000 s, CW | Photobiomodulation has an inhibitory effect on the microorganisms, and this capacity can be altered according to the interactions between different microbial species. |
| [113] | S. aureus, E. coli | Semiconductor lasers (405 nm and 445 nm), power in the range of 0.05–0.50 W, 0.050 W/cm2, 0–10800 s, CW | S. aureus and E. coli are inhibited in growth by a dose-dependent modality. 405 nm is more effective than 445 nm. |
| [114] | P. intermedia, Prevotella nigrescens | Light-emitting diode (405 nm), 0.019 W/cm2, 0.19–0.57–1.14–5.73 J/cm2, 10–30–60–300 s, 0.79 cm2 | Lethal photosensitivity is demonstrated in two species of Prevotella spp. under anaerobic conditions. |
| [115] | P. gingivalis | LED (460 nm), 0.40 W/cm2, 1–10–100 J | Photobiomodulation has a bactericidal effect on potential multiple drug-resistant bacteria. |
| [116] | P. aeruginosa | LED (415 nm), 19.5 mW/cm2, 14–28–56.1–84.2–109.9 J/cm2, from 720 to 5760 s | Blue light therapy might offer an effective and safe alternative to conventional antimicrobial therapy for P. aeruginosa burn infections. |
| [117] | P. gingivalis | LED (from 400 to 700 nm), 0.05 W/cm2 for 300 s, 0.20 W/cm2 for 75 s, 0.40 W/cm2 for 38 s, 15 J/cm2 | P. gingivalis growth is specifically suppressed by 405 nm light irradiation, suggesting that visible blue light irradiation is a promising means for eradicating periodontal pathogenic bacteria from periodontal lesions. |
| [118] | P. aeruginosa, S. aureus | Ar ion laser (514 nm); SHG Nd:YAG (532 nm); He-Ne laser (633 nm), 0.3–19 mW; 0.015–1.130 J/cm2; 350 and 420 s; CW | The laser can affect cell growth. The three wavelengths induce a proliferative effect on P. aeruginosa and an inhibitory effect on S. aureus. |
| [119] | E. coli | He-Ne laser (632.8 nm); Filament-lamp (631 nm), 4–40 mW; 0.01–10 J/cm2; CW | The laser increases cell growth. The wavelengths increase DNA synthesis and proliferation. |
| [120] | E. coli | InGaAsP-InP laser diode (1300 nm), 0.03 W/cm2, 0.9–9 J/cm2, 30–300 s, CW GaAs semiconductor diode (950 nm), 120 W/m2, 2–26–700–1000–5000 Hz and an equivalent pulse duration of 410–31.1–1.15–0.82–0.16 ms, respectively | The laser increases cell growth. 1300 nm laser diode increases the division of E. coli with an influence range of 0.9–9 J/cm2. 950 nm pulsed irradiation increases or inhibits the division rate of bacteria depending on the pulsing frequency and/or pulse duration. |
| [121] | E.coli | He-Ne laser (632.8 nm); semiconductor lasers (1066 and 1286 nm), from 0.03 to 30 W/cm2, from 0.05 to 2000 J/cm2, from 10−2 to 900 s | The laser increase cell growth. Irradiation times produce two maxima effects in the growth stimulation. First at 0.5 J/cm2 and then at 60–3000 J/cm2. |
| [122] | E. coli (different genotypes) | He-Ne laser (632.8 nm); 15 mW; 0.4–4.0 J/cm2; CW | The laser increases cell growth. The growth and protein synthesis of E. coli are affected by laser light based on the cell cycle phase and the strain genotype. |
| [123] | Aggregatibacter actinomycetemcomitans, P. gingivalis, P. nigrescens, F. nucleatum | InGaAlP laser (660 nm), 0.035 W, 74.2 J/cm2, 2.1 J/point, 60 s | Inhibition of cell growth. |
| Bacteria | Best Therapy Effect (Inhibition of Cell Growth, Death) Reported in the Selected Literature | Reference |
|---|---|---|
| Porphyromonas gingivalis Gram-negative, anaerobic, black-pigmented | Type of cell culture: medium Plasma-arc (450–490 nm), 1.144 W/cm2, 138 J/cm2, 1 cm2, 120 s, CW LED (450–480 nm), 0.520 W/cm2, 62 J/cm2, 1 cm2, 120, CW Halogen lamps (model 1, 400–500 nm), 0.416 W/cm2, 75 J/cm2, 1 cm2, 180 s Killed bacteria: ~100% Halogen lamps (model 2, 400–500 nm), 0.260 W/cm2, 47 J/cm2, 1 cm2, 180 s Killed bacteria: ~80–90% Type of cell culture: Agar Similar effects described above | [103] |
| Type of cell culture: medium Argon laser (488 nm + 514 nm), 0.58 W, 0.15 W/cm2, 18 J/cm2, 3.5–4 cm2, 120 s, CW Killed bacteria: ~25–30% | [104] | |
| Type of cell culture: medium LED (460 nm), 0.40 W/cm2, 100 J Killed bacteria: ~50% | [115] | |
| Type of cell culture: medium LED 405 nm, 0.05 W/cm2 for 300 s, 0.20 W/cm2 for 75 s, 0.40 W/cm2 for 38 s, 15 J/cm2 Inhibition bacteria growth: ~75% | [117] | |
| Organism: rat InGaAlP laser (660 nm), 0.035 W, 74.2 J/cm2, 2.1 J/point, 60 s; 4 applications Killed bacteria: ~50% respect to baseline | [123] | |
| Prevotella intermedia Gram-negative, anaerobic, black-pigmented | Type of cell culture: medium Argon laser (488 nm + 514 nm), 0.58 W, 0.15 W/cm2, 120 J/cm2, 3.5–4 cm2, 800 s, CW Killed bacteria (strain 15033): ~100% Killed bacteria (strain 49046): ~0% | [104] |
| Type of cell culture = Agar Light-emitting diode (405 nm), 0.019 W/cm2, 1.14 J/cm2, 60 s, 0.79 cm2 Killed bacteria: ~40% | [114] | |
| Prevotella nigrescens Gram-negative, anaerobic, black-pigmented | Type of cell culture: Agar Light-emitting diode (405 nm), 0.019 W/cm2, 5.7 J/cm2, 300 s, 0.79 cm2 Killed bacteria: ~13% | [114] |
| Organism: rat InGaAlP laser (660 nm), 0.035 W, 74.2 J/cm2, 2.1 J/point, 60 s; 4 applications Surviving bacteria: ~0% respect to baseline | [123] | |
| Fusobacteriurn nucleatun Gram-negative, anaerobic | Type of cell culture: medium Halogen lamps (model 2, 400–500 nm), 0.260 W/cm2, 39 J/cm2, 1 cm2, 150 s Killed bacteria: ~90–100% LED (450–480 nm), 0.520 W/cm2, 93 J/cm2, 1 cm2, 120, CW Killed bacteria: ~50% Halogen lamps (model 2, 400–500 nm), 0.416 W/cm2, 62 J/cm2, 1 cm2, 150 s Killed bacteria: ~100% Plasma-arc (450–490 nm), 1.144 W/cm2, 207 J/cm2, 1 cm2, 180 s, CW Killed bacteria: ~40% Type of cell culture: Agar Killed bacteria: they were almost totally killed through 150 s of irradiation under all experimental conditions | [103] |
| Organism: rat InGaAlP laser (660 nm), 0.035 W, 74.2 J/cm2, 2.1 J/point, 60 s; 4 applications Killed bacteria: ~40–45% with respect to baseline | [123] | |
| Escherichia coli Gram-negative, facultative anaerobic | Type of cell culture: medium Argon-ion pumped tunable dye laser (630 nm); 0.015 W/cm2, 1 J/cm2, 66 s, CW Inhibition bacteria growth: ~25% | [105] |
| Type of cell culture: medium Diode laser (830 nm), 0.03 W, 24 J/cm2, 800 s, CW Inhibition bacteria growth: ~50% | [107] | |
| Type of cell culture: medium Ga-Al-As laser 810 nm, 0.36 W/cm2, 13 J/cm2 for 36 s, 30 J/cm2 for 80 s, 0.5 cm2 500 Hz, duty Cycle of 50% and voltage of 240 V Large effect on inhibition bacteria growth | [108] | |
| Type of cell culture: medium Laser (450 nm), 0.07 W, 6 and 12 J/cm2, 86 and 172 s, 1 cm2, CW Killed bacteria: ~40% | [110] | |
| Type of cell culture: Agar Semiconductor lasers (405 nm), power in the range of 0.05–0.50 W, 0.050 W/cm2, 180 J/cm2, 3600 s, CW Killed bacteria: ~100% | [113] | |
| Pseudomonas aeruginosa Gram-negative, aerobic | Type of cell culture: medium Argon-ion pumped tunable dye laser (630); 0.015 W/cm2, 1 J/cm2, 66 s, CW Diode lasers (810), 0.015 W/cm2, 5 J/cm2, 330 s, CW Inhibition bacteria growth: ~25% | [105] |
| Type of cell culture: medium Diode lasers (810), 0.015 W/cm2, 5 J/cm2, 330 s, CW Inhibition bacteria growth: ~25% | [106] | |
| Type of cell culture: medium Diode laser (660 or 830 nm), 0.03 W, 24 J/cm2, 800 s, CW Diode laser (904 nm), 0.04 W, 24 J/cm2, 600 s CW Inhibition bacteria growth: ~50% | [107] | |
| Type of cell culture: medium Ga-Al-As laser 810 nm, 0.36 W/cm2, 13 J/cm2 for 36 s, 0.5 cm2 500 Hz, duty cycle of 50% and voltage of 240 V Large effect on inhibition bacteria growth | [108] | |
| Type of cell culture: medium Laser (450 nm), 0.07 W, 18 and 24 J/cm2, 257 and 343 s, 1 cm2, CW Killed bacteria: ~60% | [110] | |
| Type of cell culture: medium LED (415 nm), 0.0195 W/cm2, 109.9 J/cm2, 5760 s, Killed bacteria: ~100% | [116] | |
| Staphylococcus aureus Gram-positive, facultative anaerobic | Type of cell culture: medium Argon-ion pumped tunable dye laser (630 nm); 0.015 W/cm2, 5 J/cm2, 330 s, CW Diode lasers (810 nm and 905 nm), 0.015 W/cm2, 5 J/cm2, 330 s, CW Inhibition bacteria growth: ~10 | [105] |
| Type of cell culture: medium Diode laser (810 nm), 0.015 W/cm2; 1 and 2 J/cm2; 66 ans 132 s, 50% duty cycle; 292 Hz Inhibition bacteria growth: ~10% | [106] | |
| Type of cell culture: medium Diode laser (660 nm), 0.03 W, 24 J/cm2, 800 s, CW Inhibition bacteria growth: ~78% | [107] | |
| Type of cell culture: medium Diode laser (904 nm), 0.04 W, 3 J/cm2, 9 s, 9500 Hz, duty cycle of 0.1% Killed bacteria: ~80% | [109] | |
| Type of cell culture: medium Laser (450 nm), 0.07 W, 6, 12, 18, and 24 J/cm2, 86–172–257 and 343 s, 1 cm2, CW Killed bacteria: ~60% | [110] | |
| Type of cell culture: Agar Semiconductor lasers (405 nm), power in the range of 0.05–0.50 W, 0.050 W/cm2, 180 J/cm2, 3600 s, CW Killed bacteria: ~92% | [113] | |
| Type of cell culture: medium He-Ne laser (633 nm), 0.019 W; 1.130 J/cm2; 420 s; CW Surviving bacteria: ~33% | [118] | |
| Streptococcus mutans Gram-positive, facultative anaerobic | Type of cell culture: medium Plasma-arc (450–490 nm), 1.144 W/cm2, 159 J/cm2, 1 cm2, 138 s, CW Killed bacteria: inhibition of cell growth (not specified) | [103] |
| Type of cell culture: medium Diode laser (810 nm), 1.5 W, 30 s, flexible optic fiber of 200 µm diameter Killed bacteria: ~70% | [111] | |
| Type of cell culture: medium InGaAsP diode laser (780 nm), 0.04 W, 0.02 W/cm2, 20 J/cm2, 1000 s, CW Biofilm reduction: ~90% | [112] | |
| Streptococcus pyogenes Gram-positive, facultative anaerobic | Type of cell culture: medium Ga-Al-As laser (810 nm), 0.36 W/cm2, 18 J/cm2 for 60 s, 30 J/cm2 for 80 s, 0.5 cm2, 500 Hz, duty cycle of 50% and voltage of 240 V Large effect on inhibition bacteria growth | [108] |
| Enterococcus faecalis Gram-positive, facultative anaerobic | Type of cell culture: medium Plasma-arc (450–490 nm), 1.144 W/cm2, 212 J/cm2, 1 cm2, 184 s, CW Surviving bacteria: inhibition (not specified) | [103] |
| Type of cell culture: medium Ga-Al-As laser (810 nm), 0.36 W/cm2, 13 J/cm2 for 36 s, 30 J/cm2 for 80 s, 0.5 cm2, 500 Hz, duty cycle of 50% and voltage of 240 V Large effect on inhibition bacteria growth | [108] | |
| Staphylococcus epidermidis Gram-positive, facultative anaerobic | Type of cell culture: medium Ga-Al-As laser 810 nm, 0.36 W/cm2, 18 J/cm2 for 60 s, 30 J/cm2 for 80 s, 0.5 cm2 500 Hz, duty cycle of 50% and voltage of 240 V Large effect on inhibition bacteria growth | [108] |
| Staphylococcus saprophyticus Gram-positive, facultative anaerobic | Type of cell culture: medium Ga-Al-As laser 810 nm, 0.36 W/cm2, 18 J/cm2 for 60 s, 30 J/cm2 for 80 s, 0.5 cm2 500 Hz, duty cycle of 50% and voltage of 240 V Large effect on inhibition bacteria growth | [108] |
| Lactobacillus casei Gram-positive, facultative anaerobic | Type of cell culture: medium Diode laser (810 nm), 1.5 W, 30 s, flexible optic fiber of 200 µm diameter Killed bacteria: ~50% | [111] |
| Actinomyces naeslundii Gram-positive, anaerobic | Type of cell culture: medium Diode laser (810 nm), 1.5 W, 30 s, flexible optic fiber of 200 µm diameter Killed bacteria: ~38% | [111] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amaroli, A.; Ravera, S.; Zekiy, A.; Benedicenti, S.; Pasquale, C. A Narrative Review on Oral and Periodontal Bacteria Microbiota Photobiomodulation, through Visible and Near-Infrared Light: From the Origins to Modern Therapies. Int. J. Mol. Sci. 2022, 23, 1372. https://doi.org/10.3390/ijms23031372
Amaroli A, Ravera S, Zekiy A, Benedicenti S, Pasquale C. A Narrative Review on Oral and Periodontal Bacteria Microbiota Photobiomodulation, through Visible and Near-Infrared Light: From the Origins to Modern Therapies. International Journal of Molecular Sciences. 2022; 23(3):1372. https://doi.org/10.3390/ijms23031372
Chicago/Turabian StyleAmaroli, Andrea, Silvia Ravera, Angelina Zekiy, Stefano Benedicenti, and Claudio Pasquale. 2022. "A Narrative Review on Oral and Periodontal Bacteria Microbiota Photobiomodulation, through Visible and Near-Infrared Light: From the Origins to Modern Therapies" International Journal of Molecular Sciences 23, no. 3: 1372. https://doi.org/10.3390/ijms23031372







