Abstract
Diabetic retinopathy (DR) is one of the main causes of vision loss in middle-aged economically active people. Modifiable (i.e., hyperglycaemia, hypertension, hyperlipidaemia, obesity, and cigarette smoke) and non-modifiable factors (i.e., duration of diabetes, puberty, pregnancy and genetic susceptibility) are involved in the development of DR. Epigenetic mechanisms, modulating the oxidative stress, inflammation, apoptosis, and aging, could influence the course of DR. Herein, we conducted a systematic review of observational studies investigating how epigenetics affects type 2 diabetes retinopathy (T2DR). A total of 23 epidemiological studies were included: 14 studies focused on miRNA, 4 studies on lnc-RNA, one study on both miRNA and lnc-RNA, and 4 studies on global or gene-specific DNA methylation. A direct relation between the dysregulation of miR-21, miR-93, and miR-221 and FPG, HbA1c, and HOMA-IR was identified. A panel of three miRNAs (hsa-let-7a-5p, hsa-miR-novel-chr5_15976, and hsa-miR-28-3p) demonstrated a good sensitivity and specificity for predicting T2DR. Little evidence is available regarding the possible role of the long non-coding MALAT1 dysregulation and MTHFR gene promoter hypermethylation. Despite these initial, encouraging findings potentially suggesting a role of epigenetics in T2DR, the use in clinical practice for the diagnosis and staging of this complication encounters several difficulties and further targeted investigations are still necessary.
Keywords:
retinopathy; type 2 diabetes; diabetes complications; epigenetic; miRNA; lnc-RNA; DNA methylation 1. Introduction
Diabetic retinopathy (DR) is a specific microvascular complication of diabetes mellitus (DM) which results in the damage of small blood vessels and neurons of the retina. It is one of the leading causes of vision loss in middle-aged economically active people, accounting for 4.8% of the number of cases of blindness (37 million) worldwide [1]. It is worth underlining that, with the increasing incidence of DM, the number of people DR has been estimated to rise to 191 million by 2030 [2]. A European multicentre study reported that DR prevalence among patients with type 1 diabetes (T1D) ranges from 25% to 60% [3]. The burden of DR appears to be lower in type 2 diabetes (T2D) patients, with a prevalence that ranges from 25% in United Kingdom to 40% in Italy [4,5,6]. The risk factors of DR can be broadly classified into modifiable (i.e., hyperglycaemia, hypertension, hyperlipidaemia, obesity, and cigarette smoke) and non-modifiable factors (i.e., duration of diabetes, puberty, pregnancy and genetic susceptibility). These risk factors are also involved in the development of both diabetic nephropathy, neuropathy and macrovascular complications [7]. Recent strides in comprehension and awareness of risk factors for DR significantly improved its prevention and the management of DR patients. Specifically, the increasing access to community screening programs has led to a decline in the prevalence and incidence of DR, especially in developed countries [8]. Moreover, several randomised controlled trials have shown that early treatment of DR might reduce the risk of severe visual loss by 57% [9]. However, emerging evidence also suggests that a complex gene–environment interaction is involved in the pathogenesis of diabetes-related microvascular complications [10]. Epigenetic mechanisms—including DNA methylation, histone modifications, and miRNAs and long non-coding RNA (lnc-RNA) regulation—contribute to the dysregulation of signalling pathways involved in oxidative stress, inflammation, apoptosis, and aging, and modulate the expression of several key genes in DM [11,12]. Both preclinical and clinical studies in diabetic patients provided strong evidence concerning the contribution of histone modifications, post-transcriptional RNA regulation, and DNA methylation in diabetes-related microvascular complications by regulating molecular pathways involved in the pathogenesis of these complications. Here, we conducted a systematic review to summarise current evidence from observational studies investigating the relationship between DR and epigenetic mechanisms.
2. Materials and Methods
The present systematic review was carried out in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) statements [13] and the Cochrane Handbook’s guidelines [14].
2.1. Literature Search
Epidemiological studies evaluating the association between DR and epigenetic mechanisms were systematically searched on PubMed-Medline (PubMed.gov: available online: https://pubmed.ncbi.nlm.nih.gov/, accessed on 1 June 2020), and Web of Science databases (https://www.webofscience.com/wos/woscc/basic-search, accessed on 1 June 2020), from inception to June 2020. The search strategy applied the following combination of Mesh terms: (“MicroRNAs” [Mesh] OR “DNA Methylation” [Mesh]) AND “Diabetic Retinopathy” [Mesh]. Reference lists of potentially eligible articles were also screened.
2.2. Selection Criteria
The following inclusion criteria must have been satisfied: (1) epidemiological observational studies; (2) evaluating the association of DR; (3) with epigenetic mechanisms (i.e., DNA methylation and miRNA or lnc-RNA expression); (4) in patients with T1D or T2D. Only studies published in peer-reviewed journals were included, with no limitations with regard to publication date or language. Conversely, previous systematic reviews and meta-analyses, commentary articles, and editorials were excluded.
2.3. Data Extraction and Quality Assessment
Titles and abstracts of all identified articles were independently reviewed by two authors, applying the selection criteria described above. The full texts of all eligible articles were further reviewed to assess whether selection criteria were fully met. Controversies were resolved by consultation with a third author for obtaining consensus. The following information was extracted by two investigators: first author, study year, location, study design, sample size, type of DM, level of DNA methylation and/or miRNA/lnc-RNA expression, main findings.
3. Results
3.1. Characteristics of the Included Studies
Figure 1 illustrates the literature selection process. A total of 275 articles were screened once duplicates had been removed. According to selection criteria, 202 articles were excluded after reading article title and abstract: of these, 129 were not conducted on human, 72 were literature reviews, meta-analyses or microarray/network analyses, and one study did not focus on DR. Out of the 73 articles that underwent the full-text screening, 50 were excluded for the reasons detailed in Figure 1; therefore, 23 epidemiological studies investigating the role of epigenetic in the pathogenesis of T2D retinopathy (T2DR) were included in the systematic review (Figure 1).
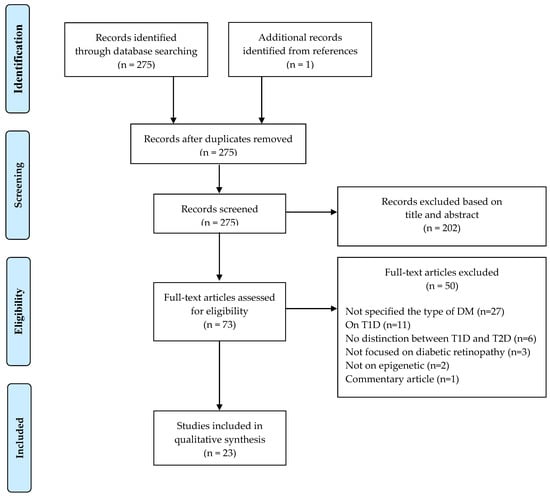
Figure 1.
PRISMA 2009 flow diagram of study selection.
Regarding the epigenetic biomarkers, 14 studies investigated the expression of miRNA in plasma (n = 6), serum (n = 6), aqueous humour (n = 1), or tears (n = 1) samples. Another four studies investigated the plasma (n = 3) or serum (n = 1) level of different lnc-RNA, while one study focused on both miRNA and lnc-RNA serum expression. The global or gene-specific DNA methylation was evaluated in four studies.
The majority of the included studies compared the level of the investigated epigenetic biomarkers in T2D patients also affected by DR with a group without DR and a healthy control group (HC) (n = 17). Among them, one study also enrolled a group of patients affected by impaired glucose tolerance (IGT). Instead, in six studies the control group was represented only by T2D patients without DR (n = 5) or healthy subjects (n = 1). No studies examined histone modifications.
3.2. miRNA Profiling
In the last decade, several investigations have been carried out to study the influence of epigenetic on T2DR. Most of these studies focused on the analysis of miRNA expression in different human samples, mostly plasma or serum (Table 1). MiR-93, miR-126, and mirR-221 have been studied in cohorts of both T2D and T1D patients [15,16,17,18,19,20].

Table 1.
Summary of the selected studies focusing on miRNA profiling in type 2 diabetes retinopathy.
Mir-93 influences the progression of DR by regulating angiogenesis, although the exact mechanism remains unclear [33,34]. In a cohort of 140 T2D patients, Zou and colleagues found a higher expression of plasma miR-93 and VEGF in the group with DR compared to the group with no ocular complications [15]. Therefore, the expression of miR-93 was directly associated with course of disease, the level of HbA1c and FPG. More recently, a study including patients with both type 1 and type 2 diabetes, confirmed an up-regulation of miR-93 in patients with DR, in particular with in those affected by severe forms and recurrent vitreous haemorrhages [18].
Confirming the role of miRNA dysregulation in the pathogenesis of DR, Liu and colleagues observed a significant overexpression of serum miR-221, VEGF, and angiotensin II (Ang-II) in T2D patients compared to healthy subjects and a progressive up-regulation in diabetic patients without DR, non-proliferative diabetic retinopathy (NPDR), proliferative diabetic retinopathy (PDR) [17]. In addition, the level of miR-221 was positively related to HbA1c, HOMA-IR, VEGF, and Ang-II. In vitro observations, performed using human umbilical vein endothelial cells (HUVECs), showed that a hyperglycaemic environment up-regulates the expression of miR-221 inducing retinal cells proliferation, migration, apoptosis, vascular endothelial hyperplasia, ischemia and neovascularisation [35]. These data suggest the potentially utility of miR-221 in predicting both the occurrence and progression of DR. Similar results were obtained by García de la Torre and colleagues in T1D patients [19]. They found an increased level of miR-221 in endothelial progenitor cells (EPCs) of T1D patients compared to HC. Moreover, T1D patients with DR had higher expression of miR-221 than those without DR.
In the study by Rezk and colleagues, the reduced expression of miR-126 was related to the onset of both macro-vascular complications and DR [16]. A similar reduction of miR-126 expression was also observed in the large cohort of the EURODIAB study among T1D patients with diabetes-related vascular complications, particularly with PDR [20]. MiR-126 is highly represented in endothelial cells playing a crucial role in endothelial homeostasis and angiogenesis influencing VEGF signalling by blocking two negative regulators of the VEGF pathway, Sprouty-related protein (SPRED1) and phosphoinositol-3 kinase regulatory subunit 2 (PIK3R2/p85-b) [16,36].
In 2017, Jiang and colleagues evaluated plasma expression of miR-21 in T2D patients with different severity of DR, showing a positive relation between miR-21 expression and the course of retinopathy [21]. In particular, they observed a significant increase of miR-21 level in the PDR group compared to both the NPDR and control groups. These data suggest the influence of miR-21 as an indicator for the severity of T2DR. Although the authors demonstrated that the role of miR-21 in the pathogenesis of DR was related to T2D course, HbA1c, fasting plasma glucose, and the homeostasis model assessment of insulin resistance (HOMA-IR), further investigations are necessary to clarify the molecular mechanisms determining the involvement of miR-21 on T2DR. In a previous study, the aberrant expression of miR-21 in aqueous humour samples of diabetic patients was associated with retinal fibrosis mediated by the transforming growth factor beta (TGF-β) [37]. Moreover, a possible angiogenetic role of miR-21 could be speculated: Liu and colleagues found that miR-21 induces vascular endothelial growth factor (VEGF)-mediated tumour angiogenesis through targeting phosphatase and tensin homolog (PTEN) and activation of AKT/ERK signalling [38,39].
A very interesting study by Kamalden and colleagues demonstrated that miR-15a was increased in the plasma of T2D patients with DR, although a correlation with DR grade was not found [22]. Moreover, the authors observed a similar expression of miR-15a in the human Müller cells (MIO-M1), human retinal endothelial cells (HRECs), human retinal pigment epithelial cells (HRPEs), and rat Müller cells (rMC-1) exposed to high glucose or advanced glycated end-product environment, thus excluding a role of hyperglycaemia. Notably, by culturing rat pancreatic β-cells (INS-1) cells in high-glucose media, they demonstrated an exosome transfer of miR-15a from pancreatic to retinal cells. These findings support the finding that miR-15a, produced in pancreatic β-cells, is transported in blood exosomes towards retinal cells, where could promote DR, increasing oxidative stress. In a previous study, Hirota and colleagues observed an increased expression of miR-15a in aqueous humour of diabetic patients with PDR [33]. It has been shown that miR-15a controls the insulin synthesis in pancreatic β-cells [40]. In T2D, the increased insulin production due to insulin-resistance, could led to an increase of miR-15 synthesis. MiR-15a is also known to regulate angiogenesis by suppressing fibroblast growth factor (FGF2) and VEGF [33].
Conflicting data are available regarding the role of miR-423, miR-29b, and miR-200b on the course of DR. Mir-423 seems to be inversely related with DR. Blum and colleagues measured the plasma level of miR-423 in T2D patient with different retinal conditions and healthy subjects, showing an overall not statistically significant negative trend in correlation to diabetic retinopathy progression [23]. In contrast, other studies, carried out in patients with different types of diabetes, reported an increased expression of miR-423 in diabetic patients affected by retinopathy [33,41]. These studies speculated on a possible cross talk between miR-423 and VEGF signalling and NOS function influencing miR-423 influencing vascular retinal proliferation [23]. However, further investigations are necessary to clarify these findings.
An inverse relation with T2DR was also observed for the plasma level of miR-29b and, most of all, miR-200b [24]. Furthermore, in another study, Zeng and colleagues observed a higher expression of Mir-29b in diabetic patients with DR. The authors identified the dysregulation of miR-29b-3p/SIRT1 as a potential mechanism of human retinal microvascular endothelial cells (HRMECs) apoptosis in DR [42].
In a recent study, three miRNAs (hsa-let-7a-5p, hsa-miR-novel-chr5_15976, and hsa-miR-28-3p) were shown to be significantly associated with DR of patients with T2D [25]. This panel presented a sensitivity and specificity of 0.92 and 0.94, respectively, for predicting DR and 0.93 sensitivity and 0.86 specificity for differentiating early stage NPDR from late-stage PDR, representing a potential diagnostic biomarker for DR. The authors, studying the in vitro proliferation rates of HRMECs with overexpression of hsa-let-7a-5p, described an increased proliferation of these cells demonstrating how miRNAs may be involved in the pathogenesis of DR.
Yang and colleagues analysed the expression of miR-155, regulatory T (Treg) cells (CD4+ CD25+ Foxp3+), and TGF-β in peripheral blood of T2D patients with different severity of DR in comparison with T2D patients not affected by DR and HC [26]. MiR-155 is a multifunctional miRNA essential to the immune response by regulating the Treg cells cytokines secretion [43]. This study observed a significantly higher expression of miR-155 and a significantly lower percentage of Treg cells in the PDR and background diabetic retinopathy (BDR) groups compared to not diabetic retinopathy (NDR) group and in the PDR group compared to the BDR group. These data suggest a possible role of miR-155 in the pathogenesis of T2DR by regulating the Treg cells with TGF-β.
Additionally, miR-122 seemed to be related with the severity of DR. A prospective study by Pastukh and colleagues showed that the serum expression of miR-122 increased from HC to NDR group and from NDR to NPDR groups [27]. Instead, the miR-122 was significantly reduced in patients with PDR. Notably, a positive trend was observed between miR-122 levels and the number of serum endothelial progenitor cells. The increase of miR-122 in patients with DR is explained by its role in preventing angiogenesis and proliferation, while the decline in patients with more severe grade of DR may represent an inhibition or exhaustion of the anti-angiogenic anti-proliferative defence system.
Another miRNA influencing the onset and progression of DR is miR-20b. Recently, Shaker and colleagues observed a significant down-regulation of miR-20b in T2D patients with progressive severity of DR [28]. The authors indicated this miRNA as promising novel biomarkers for prediction of DR severity, distinguishing PDR from NPDR. Nevertheless, these findings should be confirmed by larger studies and the pathogenesis of miRNAs involvement in DR should be clarified.
The role of miR-31, miR-3939, miR-1910-3p was investigated in different serum samples of different cohorts of T2D patients [29,30]. Nevertheless, these studies excluded their involvement in the development of T2DR.
While most of the studies analysed the expression of miRNA in plasma or serum samples of T2D patients, several studies have investigated these epigenetic biomarkers in vitreous humour or tears [31,32]. In 2019, Chen and colleagues explored the miRNA and piwi-interacting RNA (piRNA) profile in the aqueous humour of nine PDR and nine cataract control patients [31]. In addition, a mice retinopathy model supported the investigation of the relation between miRNA expression and angiogenetic processes. Among the eight miRNAs and thirty piRNAs analysed, the relative expression patterns of miR-93-5p (confirmed in the mice model), -184 and -150-5p in aqueous humour were differently expressed in patients with PDR compared to cataract controls, suggesting a role in the in the pathogenesis of PDR in T2D. This differential expression of miRNA was predicted to regulate Rho protein signal transduction, neurotransmitter uptake and histone lysine methylation.
A case–control study by Pinazo-Durán and colleagues studied the RNA concentration and the miRNAs expression in tears samples of 77 T2D patients and 55 healthy subjects. A significant difference in both total RNA and miRNAs concentration between T2D and HC groups was observed. Moreover, the authors found a direct correlation between a panel of 14 miRNAs and age, obesity, T2D duration, and a negative correlation with visual acuity. Nevertheless, other studies are necessary to better investigate the role of miRNA expression in tears as molecular biomarkers for DR.
3.3. lnc-RNA Profiling
The need to improve the knowledge about the molecular basis of DR has directed, in recent years, the efforts of researchers toward the identification of novel molecular biomarkers involved in the pathogenesis of this condition. Lnc-RNA is a functional non-protein-coding RNA of at least 200 nucleotides which modulating both transcriptional and post-transcriptional regulation plays pivotal functions in several human diseases [44]. However, very little evidence is currently available regarding the influence of lnc-RNA on DR (Table 2).

Table 2.
Summary of the selected studies focusing on long non-coding RNA profiling in type 2 diabetes retinopathy.
Recently, Shaker and colleagues evaluated the serum expression of homebox antisense intergenic RNA (HOTAIR) and metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) in T2D patients with various degree of DR in comparison to those not affected by this complication [28]. A significant increase in HOTAIR and MALAT1 was observed in both NPDR and PDR groups compared to the NDR group, indicating a possible role of HOTAIR and MALAT1 as promising novel biomarkers for prediction DR. In contrast with these results, Toraih and colleagues observed a reduced expression of circulating MALAT1—also called nuclear-enriched abundant transcript 2 (NEAT2)—and retinal non-coding RNA2 (RNCR2) in T2D patients with DR compared to those without DR [45]. Nevertheless, the expression of MALAT1 and RNCR2 did not correlate with the severity of retinopathy and with the response to aflibercept therapy [45].
Two different studies evaluated the expression of MALAT1 in aqueous humour of diabetic patients [48,49]. In both of them, the level of MALAT1 was significantly increased in patients with DR confirming the role of this lnc-RNA as a potential biomarker for DR. MALAT1 was shown to regulate retinal endothelial cell function and microvascular growth in diabetic patients. Liu and colleagues observed that the knockdown of MALAT1 improves DR in vivo and regulates the proliferation of endothelial cells in vitro through the p38-mitogen-activated protein kinase pathway [50]. Furthermore, Zhang and colleagues found an increased secretion of VEGF up-regulating MALAT1, confirming the influence of this lnc-RNA on the regulation of the angiogenetic process [51]. Further molecular studies may aid in the clarification of the exact roles of these lncRNAs in T2DR.
In 2019, Zha and colleagues published the results of their investigation regarding the role of long intergenic non-protein coding RNA p53 induced transcript (LINC-PINT), lnc-RNA known to be involved in tumour cell invasion in human cancers, in T2D retinopathy [44]. The authors analysed the expression of LINC-PINT in 244 T2D patients with different chronic diabetes-related complications (nephropathy, retinopathy, cardiomyopathy, diabetic foot) and 126 healthy subjects, and in ARPE-19 and AC16 cells. LINC-PINT was downregulated in patients with retinopathy, cardiomyopathy or both. The in vitro experiments showed that the treatment with high glucose limited the LINC-PINT expression in the ARPE-19 and AC16 cells, while the overexpression of LINC-PINT increased the viability of ARPE-19 and AC16 cells. Instead, the siRNA-mediated silencing of LINC-PINT elicited the opposite effect. Although further confirmations are necessary, these results suggest a role of LINC-PINT in inhibiting the progression of both retinopathy and cardiomyopathy in T2D patients.
No correlation with T2DR was found for circulating levels of cancer susceptibility candidate 2 (CASC2), growth arrest-specific transcript 5 (GAS5), and RNAs H19 [46,47].
3.4. DNA Methylation
Both global and gene-specific DNA methylation are involved in the epigenetic regulation of gene transcription and expression by modulating binding factor or promoting the binding of methyl binding proteins [52]. Emerging evidence suggests a relationship between a hyperglycaemic environment and changes in DNA methylation, identifying these mechanisms as a possible biomarker of diabetic complications [53]. However, the association between DNA methylation and T2DR has been poorly explored (Table 3).

Table 3.
Summary of the selected studies focusing on global and gene-specific DNA methylation in type 2 diabetes retinopathy.
To the best of our knowledge, Maghbooli and colleagues were the first suggesting that differences of the global DNA methylation profile in T2D patients with or without DR could be predictive of this complication [53]. Patients with DR compared to those not affected by DR had a significantly higher content of 5-methylcytosine, assessed to evaluate the global DNA methylation, also after correction for dyslipidaemia, hypertension, hyperglycaemia and duration of diabetes. Moreover, the authors observed a significant increasing trend of global DNA methylation in parallel with DR progression (no DR, 4.22 ± 0.13; NPDR, 4.62 ± 0.17; PDR, 5.07 ± 0.21, p = 0.006).
The methylation of the of 5,10-methylenetetrahydrofolate reductase (MTHFR) gene promoter has been studied in relation to the onset of DR [54,55]. MTHFR is involved in the methionine–homocysteine cycle. The methyl group bound to a cytosine that precedes a guanine can be methylated by MTHFR to produce 5-methyltetrahydrofolate which produces methyl donor for the conversion of homocysteine to methionine. Several studies have observed a correlation between MTHFR gene polymorphisms and the risk of developing cancer, vascular diseases, diabetes and its complications [57,58]. Dos Santos Nunes and colleagues investigated the methylation profile of MTHFR gene promoter and its relationship with biochemical (glycemia, glycated haemoglobin, and lipid level), inflammatory (C-reactive protein and alpha-1 acid glycoprotein) and oxidative stress markers (total antioxidant and malonaldehyde) in Brazilian T2D patients affected or not by DR [56]. They found a significant association between the hypermethylation of the MTHFR gene promoter, DR, higher total cholesterol and LDL levels. In 2019, the same group confirmed that the hypermethylated MTFHR gene profile, associated with the 1298AA polymorphism of this gene, was related to higher values of glycaemia, total and LDL cholesterol in T2D patients [55]. These results suggest that changes in the methylation of MTHFR gene promoter are involved in T2DR onset influencing both homocysteine and lipid metabolism. Dos Santos Nunes and colleagues also studied the methylation profiles of specific miRNA gene promoter in T2D patients [56]. They found that the methylated profile of miR-9-3 was related to an increased risk for DR, while methylated miR-137 could be protective from microvascular diabetes complications. These data showed that the methylation in miRNA promoters may differently affect the course of DR.
4. Discussion
DR is a highly specific microvascular complication of diabetes and its main risk factors are long diabetes duration and poor glucose, lipid, and blood pressure control [59,60,61]. In addition, unhealthy lifestyles—low physical activity level, unbalanced diet and tobacco consumption—could directly contribute to the development of DR and other diabetes-related vascular complications [7,62,63]. Nevertheless, it has also been observed that some diabetic patients without these traditional risk factors for DR can likewise develop this complication, suggesting the involvement of other, less well-known, pathogenetic elements [61,64,65]. Thus, over the last decade, researchers have increasingly directed their efforts towards understanding how epigenetics contributes to the initiation and progression of DR. DNA methylation, histone modifications, and miRNAs and lnc-RNA dysregulation have been studied to be proposed for predicting the course of DR in both T1D and T2D. In vitro, in vivo and clinical studies, ruled out in patients with diabetes, have been performed to explore how epigenetic dysregulation contributes, by regulating molecular pathways, to the pathogenesis of diabetes-related microvascular complications. Our study provides a systematic review of epidemiological studies investigating the predictive value of epigenetic biomarkers in T2DR. Most of this research analysed the role of several miRNAs in patients with T2D (Table 1). The expression of miRNAs in human tissues changes as a result of physio-pathological responses. Moreover, due to the properties of stability in biological samples, miRNAs are potentially useful as disease biomarkers [66]. This systematic review summarised the different possible roles of miRNAs in the development of T2DR (Table 1). While the dysregulation of some specific miRNAs (miR-15a, miR-93, miR-93-5p, miR-126, miR-150-5p, miR-184, hsa-let-7a-5p, hsa-miR-novel-chr5_15976, hsa-miR-28-3p) was only related with a higher risk to develop T2DR, for other miRNAs (miR-20b, miR-21, miR-29b, miR-122, miR-155, miR-221, miR-423) an association with the grade of DR was also identified (Table 1). Moreover, the identification of a direct relation between the dysregulation of miR-21, miR-93, and miR-221 and glycometabolic parameters, such as FPG, HbA1c, and HOMA-IR, confirms the crucial role of glucose control and insulin resistance in the development of T2DR, by providing an epigenetic explication of this mechanism [15,17,21,67]. Nevertheless, despite the broad number of miRNAs that have been investigated in relation to T2DR, their application in clinical practice is not currently feasible. Indeed, the majority of the reported results is not reproduced in multiple independent cohorts of T2D patients or, in some cases, conflicting and not univocal. Only in the case of miR-93, miR-126, and miR-221 dysregulation has the evidence obtained from T2D patients been confirmed in cohorts of T1D patients [15,16,17,18,19,20]. To overcome these critical issues, and also due to the heterogeneity of the biological samples and laboratory methodologies, a standardisation of both sample collection and analysis methodologies is mandatory [66]. Certainly, the usage of combinations of multiple biomarkers could improve their predictive value in detecting DR; recently, a panel of three miRNAs (hsa-let-7a-5p, hsa-miR-novel-chr5_15976, and hsa-miR-28-3p) presented a good level of sensitivity and specificity (about 90%) for predicting DR and differentiate early-stage NPDR from late-stage PDR [25].
5. Conclusions
Our systematic review described the main epigenetic biomarkers known to be involved in T2DR. Nevertheless, the aim of their application in clinical practice encounters several challenges because of the insufficient level of the evidence available. There is currently an unresolved need to standardise the biological samples and laboratory procedures and to confirm the obtained data in independent cohort. In clinical practice, the early detection of DR is based on instrumental retinal examinations (fundus oculi, fluorangiography, optical coherence tomography), although the timing and modality of these screening procedures are not often correctly applied on the territory. Nevertheless, the possibility of a tele-retinal evaluation, in particularly in this period of the COVID-19 pandemic, could improve patients’ adhesion to screening programs [68,69].
Of course, an integrated approach including different epigenetic biomarkers, possibly matched with patients’ clinical, instrumental and biochemical features, will be useful to identify accurate panels for the prediction of T2DR. Further prospective investigations are necessary to achieve this aim.
Author Contributions
Conceptualisation, A.M. (Agostino Milluzzo), A.M. (Andrea Maugeri), M.B., L.S. and A.A.; methodology, A.M. (Agostino Milluzzo) and A.M. (Andrea Maugeri); investigation, A.M. (Agostino Milluzzo) and A.M. (Andrea Maugeri); writing—original draft preparation, A.M. (Agostino Milluzzo) and A.M. (Andrea Maugeri); writing—review and editing, all authors; supervision, L.S. and A.A. All authors have read and agreed to the published version of the manuscript.
Funding
This work is part of the PhD research program (Clinical and Translational Biomedicine, University of Catania) of Agostino Milluzzo, funded by MIUR—Research and Innovation Plan 2014–2020. This research was partially funded by “fondi di ateneo 2020–2022, Università di Catania, linea Open Access”.
Data Availability Statement
Data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Resnikoff, S.; Pascolini, D.; Etya’ale, D.; Kocur, I.; Pararajasegaram, R.; Pokharel, G.P.; Mariotti, S.P. Global data on visual impairment in the year 2002. Bull. World Health Organ. 2004, 82, 844–851. [Google Scholar] [PubMed]
- Resnikoff, S.; Keys, T.U. Future trends in global blindness. Indian J. Ophthalmol. 2012, 60, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, J.; Fuller, J.H. Microvascular and acute complications in IDDM patients: The EURODIAB IDDM Complications Study. Diabetologia 1994, 37, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Stratton, I.M.; Kohner, E.M.; Aldington, S.J.; Turner, R.C.; Holman, R.R.; Manley, S.E.; Matthews, D.R. UKPDS 50: Risk factors for incidence and progression of retinopathy in Type II diabetes over 6 years from diagnosis. Diabetologia 2001, 44, 156–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giuffrè, G.; Lodato, G.; Dardanoni, G. Prevalence and risk factors of diabetic retinopathy in adult and elderly subjects: The Casteldaccia Eye Study. Graefe’s Arch. Clin. Exp. Ophthalmol. 2004, 242, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, L.L.; Lervang, H.H.; Lundbye-Christensen, S.; Gorst-Rasmussen, A. The North Jutland County Diabetic Retinopathy Study: Population characteristics. Br. J. Ophthalmol. 2006, 90, 1404–1409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasso, F.C.; Pafundi, P.C.; Simeon, V.; De Nicola, L.; Chiodini, P.; Galiero, R.; Rinaldi, L.; Nevola, R.; Salvatore, T.; Sardu, C.; et al. Efficacy and durability of multifactorial intervention on mortality and MACEs: A randomized clinical trial in type-2 diabetic kidney disease. Cardiovasc. Diabetol. 2021, 20, 145. [Google Scholar] [CrossRef]
- Wong, T.Y.; Mwamburi, M.; Klein, R.; Larsen, M.; Flynn, H.; Hernandez-Medina, M.; Ranganathan, G.; Wirostko, B.; Pleil, A.; Mitchell, P. Rates of progression in diabetic retinopathy during different time periods: A systematic review and meta-analysis. Diabetes Care 2009, 32, 2307–2313. [Google Scholar] [CrossRef] [Green Version]
- Grading diabetic retinopathy from stereoscopic color fundus photographs—An extension of the modified Airlie House classification. ETDRS report number 10. Early Treatment Diabetic Retinopathy Study Research Group. Ophthalmology 1991, 98, 786–806. [Google Scholar]
- Khullar, M.; Cheema, B.S.; Raut, S.K. Emerging Evidence of Epigenetic Modifications in Vascular Complication of Diabetes. Front. Endocrinol. 2017, 8, 237. [Google Scholar] [CrossRef] [Green Version]
- Ling, C.; Groop, L. Epigenetics: A molecular link between environmental factors and type 2 diabetes. Diabetes 2009, 58, 2718–2725. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, E.R.; Liu, D. Epigenetics: The missing link to understanding β-cell dysfunction in the pathogenesis of type 2 diabetes. Epigenetics 2012, 7, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Group, P. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higgins, T.J.; Chandler, J.; Cumpston, M.; Li, T.; Page, M.J.; Welch, V.A. Cochrane Handbook for Systematic Reviews Interventions, 2nd ed.; John Wiley & Sons: Chichester, UK, 2019. [Google Scholar]
- Zou, H.L.; Wang, Y.; Gang, Q.; Zhang, Y.; Sun, Y. Plasma level of miR-93 is associated with higher risk to develop type 2 diabetic retinopathy. Graefe’s Arch. Clin. Exp. Ophthalmol. 2017, 255, 1159–1166. [Google Scholar] [CrossRef] [PubMed]
- Rezk, N.A.; Sabbah, N.A.; Saad, M.S. Role of MicroRNA 126 in screening, diagnosis, and prognosis of diabetic patients in Egypt. IUBMB Life 2016, 68, 452–458. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.N.; Li, X.; Wu, N.; Tong, M.M.; Chen, S.; Zhu, S.S.; Qian, W.; Chen, X.L. Serum microRNA-221 as a biomarker for diabetic retinopathy in patients associated with type 2 diabetes. Int. J. Ophthalmol. 2018, 11, 1889–1894. [Google Scholar] [CrossRef]
- Mammadzada, P.; Bayle, J.; Gudmundsson, J.; Kvanta, A.; André, H. Identification of Diagnostic and Prognostic microRNAs for Recurrent Vitreous Hemorrhage in Patients with Proliferative Diabetic Retinopathy. J. Clin. Med. 2019, 8, 2217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García de la Torre, N.; Fernández-Durango, R.; Gómez, R.; Fuentes, M.; Roldán-Pallarés, M.; Donate, J.; Barabash, A.; Alonso, B.; Runkle, I.; Durán, A.; et al. Expression of Angiogenic MicroRNAs in Endothelial Progenitor Cells from Type 1 Diabetic Patients With and Without Diabetic Retinopathy. Investig. Ophthalmol. Vis. Sci. 2015, 56, 4090–4098. [Google Scholar] [CrossRef] [Green Version]
- Barutta, F.; Bruno, G.; Matullo, G.; Chaturvedi, N.; Grimaldi, S.; Schalkwijk, C.; Stehouwer, C.D.; Fuller, J.H.; Gruden, G. MicroRNA-126 and micro-/macrovascular complications of type 1 diabetes in the EURODIAB Prospective Complications Study. Acta Diabetol. 2017, 54, 133–139. [Google Scholar] [CrossRef]
- Jiang, Q.; Lyu, X.M.; Yuan, Y.; Wang, L. Plasma. Biosci. Rep. 2017, 37, BSR20160589. [Google Scholar] [CrossRef] [Green Version]
- Kamalden, T.A.; Macgregor-Das, A.M.; Kannan, S.M.; Dunkerly-Eyring, B.; Khaliddin, N.; Xu, Z.; Fusco, A.P.; Yazib, S.A.; Chow, R.C.; Duh, E.J.; et al. Exosomal MicroRNA-15a Transfer from the Pancreas Augments Diabetic Complications by Inducing Oxidative Stress. Antioxid. Redox Signal. 2017, 27, 913–930. [Google Scholar] [CrossRef]
- Blum, A.; Meerson, A.; Rohana, H.; Jabaly, H.; Nahul, N.; Celesh, D.; Romanenko, O.; Tamir, S. MicroRNA-423 may regulate diabetic vasculopathy. Clin. Exp. Med. 2019, 19, 469–477. [Google Scholar] [CrossRef]
- Dantas da Costa e Silva, M.E.; Polina, E.R.; Crispim, D.; Sbruzzi, R.C.; Lavinsky, D.; Mallmann, F.; Martinelli, N.C.; Canani, L.H.; Dos Santos, K.G. Plasma levels of miR-29b and miR-200b in type 2 diabetic retinopathy. J. Cell. Mol. Med. 2019, 23, 1280–1287. [Google Scholar] [CrossRef]
- Liang, Z.; Gao, K.P.; Wang, Y.X.; Liu, Z.C.; Tian, L.; Yang, X.Z.; Ding, J.Y.; Wu, W.T.; Yang, W.H.; Li, Y.L.; et al. RNA sequencing identified specific circulating miRNA biomarkers for early detection of diabetes retinopathy. Am. J. Physiol. Endocrinol. Metab. 2018, 315, E374–E385. [Google Scholar] [CrossRef]
- Yang, T.T.; Song, S.J.; Xue, H.B.; Shi, D.F.; Liu, C.M.; Liu, H. Regulatory T cells in the pathogenesis of type 2 diabetes mellitus retinopathy by miR-155. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 2010–2015. [Google Scholar]
- Pastukh, N.; Meerson, A.; Kalish, D.; Jabaly, H.; Blum, A. Serum miR-122 levels correlate with diabetic retinopathy. Clin. Exp. Med. 2019, 19, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Shaker, O.G.; Abdelaleem, O.O.; Mahmoud, R.H.; Abdelghaffar, N.K.; Ahmed, T.I.; Said, O.M.; Zaki, O.M. Diagnostic and prognostic role of serum miR-20b, miR-17-3p, HOTAIR, and MALAT1 in diabetic retinopathy. IUBMB Life 2019, 71, 310–320. [Google Scholar] [CrossRef]
- Ma, J.; Wang, J.; Liu, Y.; Wang, C.; Duan, D.; Lu, N.; Wang, K.; Zhang, L.; Gu, K.; Chen, S.; et al. Comparisons of serum miRNA expression profiles in patients with diabetic retinopathy and type 2 diabetes mellitus. Clinics 2017, 72, 111–115. [Google Scholar] [CrossRef]
- Rovira-Llopis, S.; Escribano-Lopez, I.; Diaz-Morales, N.; Iannantuoni, F.; Lopez-Domenech, S.; Andújar, I.; Jover, A.; Pantoja, J.; Pallardo, L.M.; Bañuls, C.; et al. Downregulation of miR-31 in Diabetic Nephropathy and its Relationship with Inflammation. Cell. Physiol. Biochem. 2018, 50, 1005–1014. [Google Scholar] [CrossRef]
- Chen, S.; Yuan, M.; Liu, Y.; Zhao, X.; Lian, P.; Chen, Y.; Liu, B.; Lu, L. Landscape of microRNA in the aqueous humour of proliferative diabetic retinopathy as assessed by next-generation sequencing. Clin. Exp. Ophthalmol. 2019, 47, 925–936. [Google Scholar] [CrossRef] [PubMed]
- Pinazo-Durán, M.D.; Zanón-Moreno, V.; Lleó-Perez, A.; García-Medina, J.J.; Galbis-Estrada, C.; Roig-Revert, M.J.; Marco-Ramírez, C.; López-Gálvez, M.; Dolz-Marco, R.; Duarte, L.; et al. Genetic systems for a new approach to risk of progression of diabetic retinopathy. Arch. Soc. Española Oftalmol. 2016, 91, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Hirota, K.; Keino, H.; Inoue, M.; Ishida, H.; Hirakata, A. Comparisons of microRNA expression profiles in vitreous humor between eyes with macular hole and eyes with proliferative diabetic retinopathy. Graefe’s Arch. Clin. Exp. Ophthalmol. 2015, 253, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Salam, A.; Mathew, R.; Sivaprasad, S. Treatment of proliferative diabetic retinopathy with anti-VEGF agents. Acta Ophthalmol. 2011, 89, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Song, Y.H.; Li, F.; Yang, T.; Lu, Y.W.; Geng, Y.J. MicroRNA-221 regulates high glucose-induced endothelial dysfunction. Biochem. Biophys. Res. Commun. 2009, 381, 81–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 2008, 15, 261–271. [Google Scholar] [CrossRef] [Green Version]
- Usui-Ouchi, A.; Ouchi, Y.; Kiyokawa, M.; Sakuma, T.; Ito, R.; Ebihara, N. Upregulation of Mir-21 Levels in the Vitreous Humor Is Associated with Development of Proliferative Vitreoretinal Disease. PLoS ONE 2016, 11, e0158043. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.Z.; Li, C.; Chen, Q.; Jing, Y.; Carpenter, R.; Jiang, Y.; Kung, H.F.; Lai, L.; Jiang, B.H. MiR-21 induced angiogenesis through AKT and ERK activation and HIF-1α expression. PLoS ONE 2011, 6, e19139. [Google Scholar] [CrossRef] [Green Version]
- Qing, S.; Yuan, S.; Yun, C.; Hui, H.; Mao, P.; Wen, F.; Ding, Y.; Liu, Q. Serum miRNA biomarkers serve as a fingerprint for proliferative diabetic retinopathy. Cell. Physiol. Biochem. 2014, 34, 1733–1740. [Google Scholar] [CrossRef]
- Sun, L.L.; Jiang, B.G.; Li, W.T.; Zou, J.J.; Shi, Y.Q.; Liu, Z.M. MicroRNA-15a positively regulates insulin synthesis by inhibiting uncoupling protein-2 expression. Diabetes Res. Clin. Pract. 2011, 91, 94–100. [Google Scholar] [CrossRef]
- Xiao, Q.; Zhao, Y.; Xu, J.; Li, W.J.; Chen, Y.; Sun, H.J. NFE2/miR-423-5p/TFF1 axis regulates high glucose-induced apoptosis in retinal pigment epithelial cells. BMC Mol. Cell Biol. 2019, 20, 39. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Y.; Cui, Z.; Liu, J.; Chen, J.; Tang, S. MicroRNA-29b-3p Promotes Human Retinal Microvascular Endothelial Cell Apoptosis via Blocking SIRT1 in Diabetic Retinopathy. Front. Physiol. 2019, 10, 1621. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.F.; Li, L.; Wang, L.M. miR-155 and miR-146b negatively regulates IL6 in Helicobacter pylori (cagA+) infected gastroduodenal ulcer. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 607–613. [Google Scholar] [PubMed]
- Zha, T.; Su, F.; Liu, X.; Yang, C.; Liu, L. Role of Long Non-Coding RNA (LncRNA) LINC-PINT Downregulation in Cardiomyopathy and Retinopathy Progression Among Patients with Type 2 Diabetes. Med. Sci. Monit. 2019, 25, 8509–8514. [Google Scholar] [CrossRef]
- Toraih, E.A.; Abdelghany, A.A.; Abd El Fadeal, N.M.; Al Ageeli, E.; Fawzy, M.S. Deciphering the role of circulating lncRNAs: RNCR2, NEAT2, CDKN2B-AS1, and PVT1 and the possible prediction of anti-VEGF treatment outcomes in diabetic retinopathy patients. Graefe’s Arch. Clin. Exp. Ophthalmol. 2019, 257, 1897–1913. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Su, N.; Zhang, Y.; Wang, G. Clinical Significance of Serum lncRNA Cancer Susceptibility Candidate 2 (CASC2) for Chronic Renal Failure in Patients with Type 2 Diabetes. Med. Sci. Monit. 2018, 24, 6079–6084. [Google Scholar] [CrossRef] [PubMed]
- Fawzy, M.S.; Abdelghany, A.A.; Toraih, E.A.; Mohamed, A.M. Circulating long noncoding RNAs H19 and GAS5 are associated with type 2 diabetes but not with diabetic retinopathy: A preliminary study. Bosn. J. Basic Med. Sci. 2020, 20, 365–371. [Google Scholar] [CrossRef] [Green Version]
- Yan, B.; Tao, Z.F.; Li, X.M.; Zhang, H.; Yao, J.; Jiang, Q. Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Investig. Ophthalmol. Vis. Sci. 2014, 55, 941–951. [Google Scholar] [CrossRef]
- Biswas, S.; Thomas, A.A.; Chen, S.; Aref-Eshghi, E.; Feng, B.; Gonder, J.; Sadikovic, B.; Chakrabarti, S. MALAT1: An Epigenetic Regulator of Inflammation in Diabetic Retinopathy. Sci. Rep. 2018, 8, 6526. [Google Scholar] [CrossRef]
- Liu, J.Y.; Yao, J.; Li, X.M.; Song, Y.C.; Wang, X.Q.; Li, Y.J.; Yan, B.; Jiang, Q. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014, 5, e1506. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhao, L.; Hambly, B.; Bao, S.; Wang, K. Diabetic retinopathy: Reversibility of epigenetic modifications and new therapeutic targets. Cell Biosci. 2017, 7, 42. [Google Scholar] [CrossRef] [Green Version]
- Portela, A.; Esteller, M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010, 28, 1057–1068. [Google Scholar] [CrossRef]
- Maghbooli, Z.; Hossein-nezhad, A.; Larijani, B.; Amini, M.; Keshtkar, A. Global DNA methylation as a possible biomarker for diabetic retinopathy. Diabetes Metab. Res. Rev. 2015, 31, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos Nunes, M.K.; Silva, A.S.; de Queiroga Evangelista, I.W.; Filho, J.M.; Gomes, C.N.A.P.; do Nascimento, R.A.F.; Luna, R.C.P.; de Carvalho Costa, M.J.; de Oliveira, N.F.P.; Persuhn, D.C. Hypermethylation in the promoter of the MTHFR gene is associated with diabetic complications and biochemical indicators. Diabetol. Metab. Syndr. 2017, 9, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santana Bezerra, H.; Severo de Assis, C.; Dos Santos Nunes, M.K.; Wanderley de Queiroga Evangelista, I.; Modesto Filho, J.; Alves Pegado Gomes, C.N.; Ferreira do Nascimento, R.A.; Pordeus Luna, R.C.; de Carvalho Costa, M.J.; de Oliveira, N.F.P.; et al. The MTHFR promoter hypermethylation pattern associated with the A1298C polymorphism influences lipid parameters and glycemic control in diabetic patients. Diabetol. Metab. Syndr. 2019, 11, 4. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos Nunes, M.K.; Silva, A.S.; Wanderley de Queiroga Evangelista, I.; Modesto Filho, J.; Alves Pegado Gomes, C.N.; Ferreira do Nascimento, R.A.; Pordeus Luna, R.C.; de Carvalho Costa, M.J.; Paulo de Oliveira, N.F.; Camati Persuhn, D. Analysis of the DNA methylation profiles of miR-9-3, miR-34a, and miR-137 promoters in patients with diabetic retinopathy and nephropathy. J. Diabetes Complicat. 2018, 32, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, G.; Sutherland, H.G.; Camilleri, E.T.; Lea, R.A.; Haupt, L.M.; Griffiths, L.R. Genetic and epigenetic variants in the MTHFR gene are not associated with non-Hodgkin lymphoma. Meta Gene 2015, 6, 91–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liew, S.C.; Gupta, E.D. Methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism: Epidemiology, metabolism and the associated diseases. Eur. J. Med. Genet. 2015, 58, 1–10. [Google Scholar] [CrossRef]
- Larroumet, A.; Rigo, M.; Lecocq, M.; Delyfer, M.N.; Korobelnik, J.F.; Monlun, M.; Foussard, N.; Poupon, P.; Blanco, L.; Mohammedi, K.; et al. Previous dramatic reduction of HbA1c and retinopathy in Type 2 Diabetes. J. Diabetes Complicat. 2020, 34, 107604. [Google Scholar] [CrossRef]
- Takao, T.; Inoue, K.; Suka, M.; Yanagisawa, H.; Iwamoto, Y. Optimal cutoff values of fasting plasma glucose (FPG) variability for detecting retinopathy and the threshold of FPG levels for predicting the risk of retinopathy in type 2 diabetes: A longitudinal study over 27 years. Diabetes Res. Clin. Pract. 2018, 140, 228–235. [Google Scholar] [CrossRef]
- Antonetti, D.A.; Klein, R.; Gardner, T.W. Diabetic retinopathy. N. Engl. J. Med. 2012, 366, 1227–1239. [Google Scholar] [CrossRef] [Green Version]
- Gabriel, R.; Boukichou Abdelkader, N.; Acosta, T.; Gilis-Januszewska, A.; Gómez-Huelgas, R.; Makrilakis, K.; Kamenov, Z.; Paulweber, B.; Satman, I.; Djordjevic, P.; et al. Early prevention of diabetes microvascular complications in people with hyperglycaemia in Europe. ePREDICE randomized trial. Study protocol, recruitment and selected baseline data. PLoS ONE 2020, 15, e0231196. [Google Scholar] [CrossRef] [Green Version]
- Tyrberg, M.; Nyström, L.; Arnqvist, H.J.; Bolinder, J.; Gudbjörnsdottir, S.; Landin-Olsson, M.; Eriksson, J.W.; Svensson, M.K. Overweight, hyperglycemia and tobacco use are modifiable risk factors for onset of retinopathy 9 and 17years after the diagnosis of diabetes—A retrospective observational nation-wide cohort study. Diabetes Res. Clin. Pract. 2017, 133, 21–29. [Google Scholar] [CrossRef]
- Das, A.; McGuire, P.G.; Rangasamy, S. Diabetic Macular Edema: Pathophysiology and Novel Therapeutic Targets. Ophthalmology 2015, 122, 1375–1394. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.K.; Keenan, H.A.; Cavallerano, J.D.; Asztalos, B.F.; Schaefer, E.J.; Sell, D.R.; Strauch, C.M.; Monnier, V.M.; Doria, A.; Aiello, L.P.; et al. Protection from retinopathy and other complications in patients with type 1 diabetes of extreme duration: The joslin 50-year medalist study. Diabetes Care 2011, 34, 968–974. [Google Scholar] [CrossRef] [Green Version]
- Sebastiani, G.; Nigi, L.; Grieco, G.E.; Mancarella, F.; Ventriglia, G.; Dotta, F. Circulating microRNAs and diabetes mellitus: A novel tool for disease prediction, diagnosis, and staging? J. Endocrinol. Investig. 2017, 40, 591–610. [Google Scholar] [CrossRef] [PubMed]
- Di Mauro, S.; Scamporrino, A.; Petta, S.; Urbano, F.; Filippello, A.; Ragusa, M.; Di Martino, M.T.; Scionti, F.; Grimaudo, S.; Pipitone, R.M.; et al. Serum coding and non-coding RNAs as biomarkers of NAFLD and fibrosis severity. Liver Int. 2019, 39, 1742–1754. [Google Scholar] [CrossRef] [Green Version]
- Sasso, F.C.; Pafundi, P.C.; Gelso, A.; Bono, V.; Costagliola, C.; Marfella, R.; Sardu, C.; Rinaldi, L.; Galiero, R.; Acierno, C.; et al. Telemedicine for screening diabetic retinopathy: The NO BLIND Italian multicenter study. Diabetes Metab. Res. Rev. 2019, 35, e3113. [Google Scholar] [CrossRef] [PubMed]
- Galiero, R.; Pafundi, P.C.; Nevola, R.; Rinaldi, L.; Acierno, C.; Caturano, A.; Salvatore, T.; Adinolfi, L.E.; Costagliola, C.; Sasso, F.C. The Importance of Telemedicine during COVID-19 Pandemic: A Focus on Diabetic Retinopathy. J. Diabetes Res. 2020, 14, 9036847. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).