Current Understanding of Leaf Senescence in Rice
Abstract
:1. Introduction
2. Chlorophyll Degradation Pathways in Rice
3. Stay-Green Traits in Rice
4. Transcriptional Regulation of Leaf Senescence
5. Leaf Senescence and Nitrogen Metabolism
6. Stress-Induced Senescence
7. Breeding Strategies to Improve Rice Yield
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Woo, H.R.; Kim, H.J.; Lim, P.O.; Nam, H.G. Leaf Senescence: Systems and dynamics aspects. Annu. Rev. Plant Biol. 2019, 70, 347–376. [Google Scholar] [CrossRef] [Green Version]
- Avila-Ospina, L.; Moison, M.; Yoshimoto, K.; Masclaux-Daubresse, C. Autophagy, plant senescence, and nutrient recycling. J. Exp. Bot. 2014, 65, 3799–3811. [Google Scholar] [CrossRef] [Green Version]
- Gan, S. Concepts and types of senescence in plants. Methods Mol. Biol. 2018, 1744, 3–8. [Google Scholar] [PubMed]
- Gan, S.; Hörtensteiner, S. Frontiers in plant senescence research: From bench to bank. Plant Mol. Biol. 2013, 82, 503–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregersen, P.L.; Culetic, A.; Boschian, L.; Krupinska, K. Plant senescence and crop productivity. Plant Mol. Biol. 2013, 82, 603–622. [Google Scholar] [CrossRef]
- Yadava, P.; Singh, A.; Kumar, K.; Singh, I. Plant senescence and agriculture. In Senescence Signalling and Control in Plants; Academic Press: Cambridge, MA, USA, 2019; pp. 283–302. [Google Scholar]
- Wing, R.A.; Purugganan, M.D.; Zhang, Q. The rice genome revolution: From an ancient grain to Green Super Rice. Nat. Rev. Genet. 2018, 19, 505–517. [Google Scholar] [CrossRef]
- Evans, J.R. Improving photosynthesis. Plant Physiol. 2013, 162, 1780–1793. [Google Scholar] [CrossRef] [Green Version]
- Mae, T. Physiological nitrogen efficiency in rice: Nitrogen utilization, photosynthesis, and yield potential. Plant Soil 1997, 196, 201–210. [Google Scholar] [CrossRef]
- Masclaux-Daubresse, C.; Reisdorf-Cren, M.; Orsel, M. Leaf nitrogen remobilisation for plant development and grain filling. Plant Biol. 2008, 10, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Havé, M.; Marmagne, A.; Chardon, F.; Masclaux-Daubresse, C. Nitrogen remobilization during leaf senescence: Lessons from Arabidopsis to crops. J. Exp. Bot. 2017, 68, 2513–2529. [Google Scholar]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J.A. A NAC Gene Regulating Senescence Improves Grain Protein, Zinc, and Iron Content in Wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.; Wang, C.; Huang, L.; Chen, S.G. Leaf senescence in rice plants: Cloning and characterization of senescence up-regulated genes. J. Exp. Bot. 2001, 52, 1117–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Zhou, Y.; Zhou, G.; Ye, R.; Zhao, L.; Li, X.; Lin, Y. Identification of early senescence-associated genes in rice flag leaves. Plant Mol. Biol. 2008, 67, 37–55. [Google Scholar] [CrossRef]
- Woo, H.R.; Kim, H.J.; Nam, H.G.; Lim, P.O. Plant leaf senescence and death–regulation by multiple layers of control and implications for aging in general. J. Cell Sci. 2013, 126, 4823–4833. [Google Scholar] [CrossRef] [Green Version]
- Leng, Y.; Ye, G.; Zeng, D. Genetic Dissection of Leaf Senescence in Rice. Int. J. Mol. Sci. 2017, 18, 2686. [Google Scholar] [CrossRef] [Green Version]
- Kuai, B.; Chen, J.; Hörtensteiner, S. The biochemistry and molecular biology of chlorophyll breakdown. J. Exp. Bot. 2018, 69, 751–767. [Google Scholar] [CrossRef] [PubMed]
- Tamary, E.; Nevo, R.; Naveh, L.; Levin-Zaidman, S.; Kiss, V.; Savidor, A.; Levin, Y.; Eyal, Y.; Reich, Z.; Adam, Z. Chlorophyll catabolism precedes changes in chloroplast structure and proteome during leaf senescence. Plant Direct. 2019, 3, e00127. [Google Scholar] [CrossRef]
- Hörtensteiner, S. Update on the biochemistry of chlorophyll breakdown. Plant Mol. Biol. 2013, 82, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Jeong, H.; Lee, S.; Lee, J.; Kim, S.J.; Park, J.W.; Woo, H.R.; Lim, P.O.; An, G.; Nam, H.G.; et al. Molecular bases for differential aging programs between flag and second leaves during grain-filling in rice. Sci. Rep. 2017, 7, 8792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, Y.; Takehisa, H.; Kamatsuki, K.; Minami, H.; Namiki, N.; Ikawa, H.; Ohyanagi, H.; Sugimoto, K.; Antonio, B.A.; Nagamura, Y. RiceXPro version 3.0: Expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 2013, 41, D1206–D1213. [Google Scholar] [CrossRef] [Green Version]
- Kusaba, M.; Ito, H.; Morita, R.; Lida, S.; Sato, Y.; Fujimoto, M.; Kawasaki, S.; Tanaka, R.; Hirochik, H.; Nishimura, M.; et al. Rice Non-Yellow Coloring1 Is Involved in Light-Harvesting Complex II and Grana Degradation during Leaf Senescence. Plant Cell 2007, 19, 1362–1375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, Y.; Morita, R.; Katsuma, S.; Nishimura, M.; Tanaka, A.; Kusaba, M. Two short-chain dehydrogenase/reductases, Non-Yellowcoloring1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 2009, 57, 120–131. [Google Scholar] [CrossRef]
- Piao, W.; Han, S.H.; Sakuraba, Y.; Paek, N.-C. Rice 7-Hydroxymethyl Chlorophyll a Reductase Is Involved in the Promotion of Chlorophyll Degradation and Modulates Cell Death Signaling. Mol. Cells 2017, 40, 773–786. [Google Scholar] [PubMed] [Green Version]
- Shimoda, Y.; Ito, H.; Tanaka, A. Arabidopsis STAY-GREEN, Mendel’s Green Cotyledon Gene, Encodes Magnesium-Dechelatase. Plant Cell 2016, 28, 2147–2160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakuraba, Y.; Schelbert, S.; Park, S.Y.; Han, S.H.; Lee, B.D.; Andrès, C.B.; Kessler, F.; Hörtensteiner, S.; Paek, N.-C. STAY-GREEN and Chlorophyll Catabolic Enzymes Interact at Light-Harvesting Complex II for Chlorophyll Detoxification during Leaf Senescence in Arabidopsis. Plant Cell 2012, 24, 507–518. [Google Scholar] [CrossRef] [Green Version]
- Sakuraba, Y.; Park, S.Y.; Paek, N.-C. The Divergent Roles of STAYGREEN (SGR) Homologs in Chlorophyll Degradation. Mol. Cells 2015, 38, 390–395. [Google Scholar] [CrossRef] [Green Version]
- Park, S.Y.; Yu, J.W.; Park, J.S.; Li, J.; Yoo, S.C.; Lee, N.Y.; Lee, S.K.; Jeong, S.W.; Seo, H.S.; Koh, H.J.; et al. The Senescence-Induced Staygreen Protein Regulates Chlorophyll Degradation. Plant Cell 2007, 19, 1649–1664. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Li, M.; Liang, N.; Yan, H.; Wei, Y.; Xu, X.; Liu, J.; Xu, Z.; Chen, F.; Wu, G. Molecular cloning and function analysis of the stay green gene in rice. Plant J. 2007, 52, 197–209. [Google Scholar] [CrossRef]
- Shin, D.; Lee, S.; Kim, T.H.; Lee, J.H.; Park, J.; Lee, J.; Lee, J.Y.; Cho, L.H.; Choi, J.Y.; Lee, W.; et al. Natural variations at the Stay-Green gene promoter control lifespan and yield in rice cultivars. Nat. Commun. 2020, 11, 2819. [Google Scholar] [CrossRef]
- Rong, H.; Tang, Y.; Zhang, H.; Wu, P.; Chen, Y.; Li, M.; Wu, G.; Jiang, H. The Stay-Green Rice like (SGRL) gene regulates chlorophyll degradation in rice. J. Plant Physiol. 2013, 170, 1367–1373. [Google Scholar] [CrossRef]
- Schelbert, S.; Aubry, S.; Burla, B.; Agne, B.; Kessler, F.; Krupinska, K.; Hörtensteiner, S. Pheophytin Pheophorbide Hydrolase (Pheophytinase) Is Involved in Chlorophyll Breakdown during Leaf Senescence in Arabidopsis. Plant Cell 2009, 21, 767–785. [Google Scholar] [CrossRef] [Green Version]
- Morita, R.; Sato, Y.; Masuda, Y.; Nishimura, M.; Kusaba, M. Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J. 2009, 59, 940–952. [Google Scholar] [CrossRef]
- Das, A.; Christ, B.; Hörtensteiner, S. Characterization of the pheophorbide a oxygenase/phyllobilin pathway of chlorophyll breakdown in grasses. Planta 2018, 248, 875–892. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Li, M.; Chen, Y.; Wu, P.; Wu, G.; Jiang, H. Knockdown of OsPAO and OsRCCR1 cause different plant death phenotypes in rice. J. Plant Physiol. 2011, 168, 1952–1959. [Google Scholar] [CrossRef]
- Zhang, Z.; He, Y.; Li, L.; Zhang, X.; Xu, X.; Shi, Y.; Wu, J.L. Characterization of a novel allele encoding pheophorbide a oxygenase in rice. Plant Signal Behav. 2021, 16, 1864606. [Google Scholar] [CrossRef] [PubMed]
- Balazadeh, S. Stay-Green Not Always Stays Green. Mol. Plant. 2014, 7, 1264–1266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hörtensteiner, S. Chlorophyll degradation during senescence. Annu. Rev. Plant Biol. 2006, 57, 55–77. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H.; Howarth, C.J. Five ways to stay green. J. Exp. Bot. 2000, 51, 329–337. [Google Scholar] [CrossRef] [Green Version]
- Thomas, H.; Ougham, H. The stay-green trait. J. Exp. Bot. 2014, 65, 3889–3900. [Google Scholar] [CrossRef] [PubMed]
- Borrell, A.K.; Hammer, G.L.; Oosterom, E.V. Stay-green: A consequence of the balance between supply and demand for nitrogen during grain filling. Ann. Appl. Biol. 2001, 138, 91–95. [Google Scholar] [CrossRef]
- Morales, F.; Ancín, M.; Fakhet, D.; González-Torralba, J.; Gámez, A.L.; Seminario, A.; Soba, D.; Ben, M.S.; Garriga, M.; Aranjuelo, I. photosynthetic metabolism under stressful growth conditions as a bases for crop breeding and yield improvement. Plants 2020, 9, 88. [Google Scholar] [CrossRef] [Green Version]
- Kusaba, M.; Tanaka, A.; Tanaka, R. Stay-green plants: What do they tell us about the molecular mechanism of leaf senescence. Photosynth. Res. 2013, 117, 221–234. [Google Scholar] [CrossRef]
- Kamal, N.M.; Alnor Gorafi, Y.S.; Abdelrahman, M.; Abdellatef, E.; Tsujimoto, H. Stay-Green trait: A prospective approach for Yield Potential, and Drought and Heat Stress Adaptation in Globally Important Cereals. Int. J. Mol. Sci. 2019, 20, 5837. [Google Scholar] [CrossRef] [Green Version]
- Tian, F.; Gong, J.; Zhang, J.; Zhang, M.; Wang, G.; Li, A.; Wang, W. Enhanced stability of thylakoid membrane proteins and antioxidant competence contribute to drought stress resistance in the tasg1 wheat stay-green mutant. J. Exp. Bot. 2013, 64, 1509–1520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barry, C.S.; McQuinn, R.P.; Chung, M.Y.; Besuden, A.; Giovannoni, J.J. Amino Acid Substitutions in Homologs of the STAY-GREEN Protein Are Responsible for the green-flesh and chlorophyll retainer Mutations of Tomato and Pepper. Plant Physiol. 2008, 147, 179–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cha, K.W.; Lee, Y.J.; Koh, H.J.; Lee, B.M.; Nam, Y.W.; Paek, N.-C. Isolation, characterization, and mapping of the stay green mutant in rice. Theor. Appl. Genet. 2002, 104, 526–532. [Google Scholar] [CrossRef] [PubMed]
- Duvick, D.; Smith, J.; Cooper, M. Long-term selection in a commercial hybrid maize breeding program. Plant Breed. Rev. 2004, 24, 109–152. [Google Scholar]
- Fang, C.; Li, C.; Li, W.; Wang, Z.; Zhou, Z.; Shen, Y.; Wu, M.; Wu, Y.; Li, G.; Kong, L.A.; et al. Concerted evolution of D1 and D2 to regulate chlorophyll degradation in soybean. Plant J. 2014, 77, 700–712. [Google Scholar] [CrossRef]
- Grbić, V.; Bleecker, A.B. Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 1995, 8, 595–602. [Google Scholar] [CrossRef]
- Rampino, P.; Spano, G.; Pataleo, S.; Mita, G.; Napier, J.A.; di Fonzo, N.; Shewry, P.R.; Perrotta, C. Molecular analysis of a durum wheat ‘stay green’ mutant: Expression pattern of photosynthesis-related genes. J. Cereal Sci. 2006, 43, 160–168. [Google Scholar] [CrossRef]
- Gous, P.W.; Warren, F.; Gilbert, R.; Fox, G.P. Drought-proofing barley (Hordeum vulgare): The effects of stay green on starch and amylose structure. Cereal Chem. 2017, 94, 873–880. [Google Scholar] [CrossRef]
- Jiang, G.H.; He, Y.Q.; Xu, C.G.; Li, X.H.; Zhang, Q. The genetic basis of stay-green in rice analyzed in a population of doubled haploid lines derived from an indica by japonica cross. Theor. Appl. Genet. 2004, 108, 688–698. [Google Scholar] [CrossRef]
- Yoo, S.-C.; Cho, S.-H.; Zhang, H.; Paik, H.-C.; Lee, C.-H.; Li, J.; Yoo, J.-H.; Koh, H.-J.; Seo, H.S.; Paek, N.-C. Quantitative trait loci associated with functional stay-green SNU-SG1 in rice. Mol. Cells 2007, 24, 83–94. [Google Scholar] [PubMed]
- Shim, K.-C.; Kim, S.H.; Jeon, Y.-A.; Lee, H.-S.; Adeva, C.; Kang, J.-W.; Kim, H.-J.; Tai, T.H.; Ahn, S.-N. A RING-Type E3 Ubiquitin Ligase, OsGW2, Controls Chlorophyll Content and Dark-Induced Senescence in Rice. Int. J. Mol. Sci. 2020, 21, 1704. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.D.; Yan, Y.F.; Kim, M.Y.; Lee, S.H.; Lee, B.W. Population-specific quantitative trait loci mapping for functional stay-green trait in rice (Oryza sativa L.). Genome 2011, 5, 235. [Google Scholar]
- Lim, J.H.; Yang, H.J.; Jung, K.H.; Yoo, S.C.; Paek, N.-C. Quantitative Trait Locus Mapping and Candidate Gene Analysis for Plant Architecture Traits Using Whole Genome Re-Sequencing in Rice. Mol. Cells 2014, 37, 149–160. [Google Scholar] [PubMed] [Green Version]
- Singh, U.M.; Sinha, P.; Dixit, S.; Abbai, R.; Venkateshwarlu, C.; Chitikineni, A.; Singh, V.K.; Varshney, R.K.; Kumar, A. Unraveling candidate genomic regions responsible for delayed leaf senescence in rice. PLoS ONE 2020, 15, e0240591. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xie, W.; Xing, H.; Yan, J.; Meng, X.; Li, X.; Fu, X.; Xu, J.; Lian, X.; Yu, S.; et al. Genetic Architecture of Natural Variation in Rice Chlorophyll Content Revealed by a Genome-Wide Association Study. Mol. Plant 2015, 8, 946–957. [Google Scholar] [PubMed] [Green Version]
- Zhao, Y.; Qiang, C.; Wang, X.; Chen, Y.; Deng, J.; Jiang, C.; Sun, X.; Chen, H.; Li, J.; Piao, W.; et al. New alleles for chlorophyll content and stay-green traits revealed by a genome wide association study in rice (Oryza sativa). Sci. Rep. 2019, 9, 2541. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.; Shi, J.; Quan, S.; Cui, B.; Kleessen, S.; Nikoloski, Z.; Tohge, T.; Alexander, D.; Guo, L.; Lin, H.; et al. Metabolic variation between japonica and indica rice cultivars as revealed by non-targeted metabolomics. Sci. Rep. 2014, 4, 5067. [Google Scholar] [CrossRef] [Green Version]
- Abdelkhalik, A.F.; Shishido, R.; Nomura, K.; Ikehashi, H. QTL-based analysis of leaf senescence in an indica/japonica hybrid in rice (Oryza sativa L.). Theor. Appl. Genet. 2005, 110, 1226–1235. [Google Scholar] [CrossRef] [PubMed]
- Ramkumar, M.K.; Senthil Kumar, S.; Gaikwad, K.; Pandey, R.; Chinnusamy, V.; Singh, N.K.; Singh, A.K.; Mohapatra, T.; Sevanthi, A.M. A Novel Stay-Green Mutant of Rice with Delayed Leaf Senescence and Better Harvest Index Confers Drought Tolerance. Plants 2019, 8, 375. [Google Scholar] [CrossRef] [Green Version]
- Sakuraba, Y.; Kim, D.; Han, S.H.; Kim, S.H.; Piao, W.; Yanagisawa, S.; An, G.; Paek, N.-C. Multilayered Regulation of Membrane-Bound ONAC054 is Essential for Abscisic Acid-induced Leaf Senescence in Rice. Plant Cell 2020, 32, 630–649. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.; Shim, Y.; Gi, E.; An, G.; Paek, N.-C. Mutation of ONAC096 Enhances Grain Yield by Increasing Panicle Number and Delaying Leaf Senescence during Grain Filling in Rice. Int. J. Mol. Sci. 2019, 20, 5241. [Google Scholar] [CrossRef] [Green Version]
- Sarwat, M.; Naqvi, A.R.; Ahmad, P.; Ashraf, M.; Akram, N.A. Phytohormones and microRNAs as sensors and regulators of leaf senescence: Assigning macro roles to small molecules. Biotechnol. Adv. 2013, 31, 1153–1171. [Google Scholar] [CrossRef]
- Bengoa Luoni, S.; Astigueta, F.H.; Nicosia, S.; Moschen, S.; Fernandez, P.; Heinz, R. Transcription factors associated with leaf senescence in crops. Plants 2019, 8, 411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.; Koyama, H.; Bhati, K.K.; Alok, A. The biotechnological importance of the plant-specific NAC transcription factor family in crop improvement. J. Plant Res. 2021. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Park, J.H.; Kim, J.; Kim, J.J.; Hong, S.; Kim, J.; Kim, J.H.; Woo, H.R.; Hyeon, C.; Lim, P.O.; et al. Time-evolving genetic networks reveal a NAC troika that negatively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E4930–E4939. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J.; et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Huang, W.; Liu, L.; Chen, T.; Zhou, F.; Lin, Y. Identification and functional characterization of a rice NAC gene involved in the regulation of leaf senescence. BMC Plant Biol. 2013, 13, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Wang, Y.; Lv, B.; Li, J.; Luo, L.; Lu, S.; Zhang, X.; Ma, H.; Ming, F. The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol. 2014, 55, 604–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, C.; Lu, S.; Lv, B.; Zhang, B.; Shen, J.; He, J.; Luo, L.; Xi, D.; Chen, X.; Ming, F. A Rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 2017, 174, 1747–1763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; He, Y.; Zhang, Z.; Shi, Y.; Zhang, X.; Xu, X.; Wu, J.L.; Tang, S. OsNAC109 regulates senescence, growth and development by altering the expression of senescence- and phytohormone-associated genes in rice. Plant Mol. Biol. 2021, 105, 637–654. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Piao, W.; Lim, J.H.; Han, S.H.; Kim, Y.S.; An, G.; Paek, N.-C. Rice ONAC106 Inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol. 2015, 56, 2325–2339. [Google Scholar] [CrossRef]
- El Mannai, Y.; Akabane, K.; Hiratsu, K.; Satoh-Nagasawa, N.; Wabiko, H. The NAC Transcription factor gene OsY37 (ONAC011) promotes leaf senescence and accelerates heading time in rice. Int. J. Mol. Sci. 2017, 18, 2165. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Antonio, B.; Namiki, N.; Motoyama, R.; Sugimoto, K.; Takehisa, H.; Minami, H.; Kamatsuki, K.; Kusaba, M.; Hirochika, H.; et al. Field transcriptome revealed critical developmental and physiological transitions involved in the expression of growth potential in japonica rice. BMC Plant Biol. 2011, 11, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katiyar, A.; Smita, S.; Lenka, S.K.; Rajwanshi, R.; Chinnusamy, V.; Bansal, K.C. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom. 2012, 13, 544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, D.-Y.; Shim, Y.; Gi, E.; Lee, B.-D.; An, G.; Kang, K.; Paek, N.-C. The MYB-related transcription factor RADIALIS-LIKE3 (OsRL3) functions in ABA-induced leaf senescence and salt sensitivity in rice. Environ. Exp. Bot. 2018, 156, 86–95. [Google Scholar] [CrossRef]
- Piao, W.; Kim, S.H.; Lee, B.D.; An, G.; Sakuraba, Y.; Paek, N.-C. Rice transcription factor OsMYB102 delays leaf senescence by down-regulating abscisic acid accumulation and signaling. J. Exp. Bot. 2019, 70, 2699–2715. [Google Scholar] [CrossRef]
- Sahebi, M.; Hanafi, M.M.; Rafii, M.Y.; Mahmud, T.M.M.; Azizi, P.; Osman, M.; Abiri, R.; Taheri, S.; Kalhori, N.; Shabanimofrad, M.; et al. Improvement of Drought Tolerance in Rice (Oryza sativa L.): Genetics, Genomic tools, and the WRKY Gene Family. Biomed. Res. Int. 2018, 2018, 3158474. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Xu, W.; Hu, X.; Liu, H.; Lin, Y. W-box and G-box elements play important roles in early senescence of rice flag leaf. Sci. Rep. 2016, 6, 20881. [Google Scholar] [CrossRef] [Green Version]
- Ricachenevsky, F.K.; Sperotto, R.A.; Menguer, P.K.; Fett, J.P. Identification of Fe-excess-induced genes in rice shoots reveals a WRKY transcription factor responsive to Fe, drought and senescence. Mol. Biol. Rep. 2010, 37, 3735–3745. [Google Scholar] [CrossRef] [PubMed]
- Jing, S.; Zhou, X.; Song, Y.; Yu, D. Heterologous expression of OsWRKY23 gene enhances pathogen defense and dark-induced leaf senescence in Arabidopsis. Plant Growth Regul. 2009, 58, 181–190. [Google Scholar] [CrossRef]
- Han, M.; Kim, C.Y.; Lee, J.; Lee, S.K.; Jeon, J.S. OsWRKY42 represses OsMT1d and induces reactive oxygen species and leaf senescence in rice. Mol. Cells 2014, 37, 532–539. [Google Scholar] [CrossRef]
- Kim, T.; Kang, K.; Kim, S.-H.; An, G.; Paek, N.-C. OsWRKY5 Promotes Rice Leaf Senescence via Senescence-Associated NAC and Abscisic Acid Biosynthesis Pathway. Int. J. Mol. Sci. 2019, 20, 4437. [Google Scholar] [CrossRef] [Green Version]
- Huq, E.; Al-Sady, B.; Hudson, M.; Kim, C.; Apel, K.; Quail, P.H. Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 2004, 305, 1937–1941. [Google Scholar] [CrossRef] [Green Version]
- Sakuraba, Y.; Jeong, J.; Kang, M.Y.; Kim, J.; Paek, N.-C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 4636. [Google Scholar] [CrossRef] [Green Version]
- Sakuraba, Y.; Kim, E.Y.; Paek, N.-C. Roles of rice PHYTOCHROME-INTERACTING FACTOR-LIKE1 (OsPIL1) in leaf senescence. Plant Signal Behav. 2017, 12, e1362522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakuraba, Y.; Kim, E.Y.; Han, S.H.; Piao, W.; An, G.; Todaka, D.; Yamaguchi-Shinozaki, K.; Paek, N.-C. Rice Phytochrome-Interacting Factor-Like1 (OsPIL1) is involved in the promotion of chlorophyll biosynthesis through feed-forward regulatory loops. J. Exp. Bot. 2017, 68, 4103–4114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jan, A.; Maruyama, K.; Todaka, D.; Kidokoro, S.; Abo, M.; Yoshimura, E.; Shinozaki, K.; Nakashima, K.; Yamaguchi-Shinozaki, K. OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol. 2013, 161, 1202–1216. [Google Scholar] [CrossRef] [Green Version]
- Kong, Z.; Li, M.; Yang, W.; Xu, W.; Xue, Y. A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 2006, 141, 1376–1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, C.; Kang, K.; Shim, Y.; Sakuraba, Y.; An, G.; Paek, N.-C. Rice ETHYLENE RESPONSE FACTOR 101 Promotes Leaf Senescence Through Jasmonic Acid-Mediated Regulation of OsNAP and OsMYC2. Front. Plant Sci. 2020, 11, 1096. [Google Scholar] [CrossRef] [PubMed]
- Uji, Y.; Akimitsu, K.; Gomi, K. Identification of OsMYC2-regulated senescence-associated genes in rice. Planta 2017, 245, 1241–1246. [Google Scholar] [CrossRef]
- Lee, S.; Marmagne, A.; Park, J.; Fabien, C.; Yim, Y.; Kim, S.J.; Kim, T.H.; Lim, P.O.; Masclaux-Daubresse, C.; Nam, H.G. Concurrent activation of OsAMT1;2 and OsGOGAT1 in rice leads to enhanced nitrogen use efficiency under nitrogen limitation. Plant J. 2020, 103, 7–20. [Google Scholar] [CrossRef]
- Wada, S.; Hayashida, Y.; Izumi, M.; Kurusu, T.; Hanamata, S.; Kanno, K.; Kojima, S.; Yamaya, T.; Kuchitsu, K.; Makino, A.; et al. Autophagy supports biomass production and nitrogen use efficiency at the vegetative stage in rice. Plant Physiol. 2015, 168, 60–73. [Google Scholar] [CrossRef] [Green Version]
- Masclaux-Daubresse, C.; Danielvedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture. Ann. Bot. 2010, 105, 1141–1157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Shinozaki, D.; Luo, J.; Pottier, M.; Havé, M.; Marmagne, A.; Reisdorf-Cren, M.; Chardon, F.; Thomine, S.; Yoshimoto, K.; et al. Autophagy and Nutrients Management in Plants. Cells 2019, 8, 1426. [Google Scholar] [CrossRef] [Green Version]
- Ishida, H.; Yoshimoto, K.; Izumi, M.; Reisen, D.; Yano, Y.; Makino, A.; Ohsumi, Y.; Hanson, M.R.; Mae, T. Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 2008, 148, 142–155. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, S.; Hidema, J.; Sakamoto, W.; Ishida, H.; Izumi, M. Selective Elimination of Membrane-Damaged Chloroplasts via Microautophagy. Plant Physiol. 2018, 177, 1007–1026. [Google Scholar] [CrossRef] [Green Version]
- Guiboileau, A.; Yoshimoto, K.; Soulay, F.; Bataillé, M.P.; Avice, J.C.; Masclaux-Daubresse, C. Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol. 2012, 194, 732–740. [Google Scholar] [CrossRef]
- Pottier, M.; Dumont, J.; Masclaux-Daubresse, C.; Thomine, S. Autophagy is essential for optimal translocation of iron to seeds in Arabidopsis. J. Exp. Bot. 2019, 70, 859–869. [Google Scholar] [CrossRef] [Green Version]
- Lornac, A.; Havé, M.; Chardon, F.; Soulay, F.; Clément, G.; Avice, J.C.; Masclaux-Daubresse, C. Autophagy Controls Sulphur Metabolism in the Rosette Leaves of Arabidopsis and Facilitates S Remobilization to the Seeds. Cells 2020, 9, 332. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Soulay, F.; Saudemont, B.; Elmayan, T.; Marmagne, A.; Masclaux-Daubresse, C. Overexpression of ATG8 in Arabidopsis Stimulates Autophagic Activity and Increases Nitrogen Remobilization Efficiency and Grain Filling. Plant Cell Physiol. 2019, 60, 343–352. [Google Scholar] [CrossRef]
- Xia, K.F.; Liu, T.; Ouyang, J.; Wang, R.; Fan, T.; Zhang, M.Y. Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res. 2011, 18, 363–377. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Zhen, X.; Li, X.; Li, N.; Xu, F. Increased Autophagy of Rice Can Increase Yield and Nitrogen Use Efficiency (NUE). Front. Plant Sci. 2019, 10, 584. [Google Scholar] [CrossRef] [Green Version]
- Fan, T.; Yang, W.; Zeng, X.; Xu, X.; Xu, Y.; Fan, X.; Luo, M.; Tian, C.; Xia, K.; Zhang, M. A Rice Autophagy gene OsATG8b is involved in nitrogen remobilization and control of grain quality. Front. Plant Sci. 2020, 11, 588. [Google Scholar] [CrossRef] [PubMed]
- Zhen, X.; Li, X.; Yu, J.; Xu, F. OsATG8c-mediated increased autophagy regulates the yield and nitrogen use efficiency in rice. Int. J. Mol. Sci. 2019, 20, 4956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhen, X.; Zheng, N.; Yu, J.; Bi, C.; Xu, F. Autophagy mediates grain yield and nitrogen stress resistance by modulating nitrogen remobilization in rice. PLoS ONE 2021, 16, e0244996. [Google Scholar] [CrossRef] [PubMed]
- Havé, M.; Balliau, T.; Cottyn-Boitte, B.; Dérond, E.; Cueff, G.; Soulay, F.; Lornac, A.; Reichman, P.; Dissmeyer, N.; Avice, J.C.; et al. Increases in activity of proteasome and papain-like cysteine protease in Arabidopsis autophagy mutants: Back-up compensatory effect or cell-death promoting effect? J. Exp. Bot. 2018, 69, 1369–1385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, X.; Nie, J.; Bai, R.; Sui, X. Amino Acid Transporters in Plants: Identification and Function. Plants 2020, 9, 972. [Google Scholar] [CrossRef]
- Hayashi, H.; Chino, M. Chemical composition of phloem sap from the upper most internode of the rice plant. Plant Cell Physiol. 1990, 31, 247–251. [Google Scholar]
- Lea, P.J.; Sodek, L.; Parry, M.A.J.; Shewry, P.R.; Halford, N.G. Asparagine in plants. Ann. Appl. Biol. 2007, 150, 1–26. [Google Scholar] [CrossRef]
- Tabuchi, M.; Sugiyama, K.; Ishiyama, K.; Inoue, E.; Sato, T.; Takahashi, H.; Yamaya, T. Severe reduction in growth rate and grain filling of rice mutants lacking OsGS1;1, a cytosolic glutamine synthetase1;1. Plant J. 2005, 42, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Fan, X.; Miller, A.J. Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Nian, J.; Xie, Q.; Feng, J.; Zhang, F.; Jing, H.; Zhang, J.; Dong, G.; Liang, Y.; Peng, J.; et al. Rice ferredoxin-dependent glutamate synthase regulates nitrogen-carbon metabolomes and is genetically differentiated between japonica and indica Subspecies. Mol. Plant. 2016, 9, 1520–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, D.D.; Qin, R.; Li, M.; Alamin, M.; Jin, X.L.; Liu, Y.; Shi, C.H. The ferredoxin-dependent glutamate synthase (OsFd-GOGAT) participates in leaf senescence and the nitrogen remobilization in rice. Mol. Genet. Genom. 2017, 292, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Gaufichon, L.; Reisdorf-Cren, M.; Rothstein, S.J.; Chardon, F.; Suzukia, A. Biological functions of asparagine synthetase in plants. Plant Sci. 2010, 179, 141–153. [Google Scholar] [CrossRef]
- Lee, S.; Park, J.; Lee, J.; Shin, D.; Marmagne, A.; Lim, P.O.; Masclaux-Daubresse, C.; An, G.; Nam, H.G. OsASN1 overexpression in rice increases grain protein content and yield under nitrogen-limiting conditions. Plant Cell Physiol. 2020, 61, 1309–1320. [Google Scholar] [CrossRef]
- Zhao, H.; Ma, H.; Yu, L.; Wang, X.; Zhao, J. Genome-wide survey and expression analysis of amino acid transporter gene family in rice (Oryza sativa L.). PLoS ONE 2012, 7, e49210. [Google Scholar] [CrossRef]
- Santiago, J.P.; Tegeder, M. Connecting Source with Sink: The Role of Arabidopsis AAP8 in Phloem Loading of Amino Acids. Plant Physiol. 2016, 171, 508–521. [Google Scholar] [CrossRef] [Green Version]
- Wei, Q.; Yan, Z.; Xiong, Y.; Fang, Z. Altered Expression of OsAAP3 Influences Rice Lesion Mimic and Leaf Senescence by Regulating Arginine Transport and Nitric Oxide Pathway. Int. J. Mol. Sci. 2021, 22, 2181. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Wu, B.; Wang, J.; Zhu, W.; Nie, H.; Qian, J.; Huang, W.; Fang, Z. Blocking amino acid transporter OsAAP3 improves grain yield by promoting outgrowth buds and increasing tiller number in rice. Plant Biotechnol. J. 2018, 16, 1710–1722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, M.R.; Reinders, A.; Ward, J.M. Transport Function of Rice Amino Acid Permeases (AAPs). Plant Cell Physiol. 2015, 56, 1355–1363. [Google Scholar] [CrossRef] [Green Version]
- Peng, B.; Kong, H.; Li, Y.; Wang, L.; Zhong, M.; Sun, L.; Gao, G.; Zhang, Q.; Luo, L.; Wang, G.; et al. OsAAP6 functions as an important regulator of grain protein content and nutritional quality in rice. Nat. Commun. 2014, 5, 4847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wu, B.; Lu, K.; Wei, Q.; Qian, J.; Chen, Y.; Fang, Z. The Amino Acid Permease 5 (OsAAP5) Regulates Tiller Number and Grain Yield in Rice. Plant Physiol. 2019, 180, 1031–1045. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Huang, W.; Wu, B.; Fang, Z.; Wang, X. The amino acid transporter AAP1 mediates growth and grain yield by regulating neutral amino acid uptake and reallocation in Oryza sativa. J. Exp. Bot. 2020, 71, 4763–4777. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Wu, B.; Ji, Y. The Amino Acid Transporter OsAAP4 Contributes to Rice Tillering and Grain Yield by Regulating Neutral Amino Acid Allocation through Two Splicing Variants. Rice 2021, 14, 2. [Google Scholar] [CrossRef]
- Gull, A.; Lone, A.A.; Wani, N.U.I. Biotic and Abiotic Stresses in Plants. In Abiotic and Biotic Stress in Plants; IntechOpen: London, UK, 2019. [Google Scholar]
- Pradhan, P.; Fischer, G.; van Velthuizen, H.; Reusser, M.E.; Kropp, J.P. Closing yield gaps: How sustainable can we be? PLoS ONE 2015, 10, e0129487. [Google Scholar] [CrossRef] [Green Version]
- Gepstein, S.; Glick, B.R. Strategies to ameliorate abiotic stress-induced plant senescence. Plant Mol. Biol. 2013, 82, 623–633. [Google Scholar] [CrossRef]
- Shen, J.; Lv, B.; Luo, L.; He, J.; Mao, C.; Xi, D.; Ming, F. The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice. Sci. Rep. 2017, 7, 40641. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Wang, H.; Cai, J.; Bi, Y.; Li, D.; Song, F. Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol. 2019, 19, 278. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liao, S.; Mei, P.; Pan, Y.; Zhang, Y.; Zheng, X.; Xie, Y.; Miao, Y. OsWRKY93 Dually Functions Between Leaf Senescence and in Response to Biotic Stress in Rice. Front. Plant Sci. 2021, 12, 643011. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of Climate Change on Crops Adaptation and Strategies to Tackle Its Outcome: A Review. Plants 2019, 8, 34. [Google Scholar] [CrossRef] [Green Version]
- Dhankher, O.P.; Foyer, C.H. Climate resilient crops for improving global food security and safety. Plant Cell Environ. 2018, 5, 877–884. [Google Scholar] [CrossRef]
- Hu, H.; Xiong, L. Genetic engineering and breeding of drought-resistant crops. Annu. Rev. Plant Biol. 2014, 65, 715–741. [Google Scholar] [CrossRef]
- Kilasi, N.L.; Singh, J.; Vallejos, C.E.; Ye, C.; Jagadish, S.V.K.; Kusolwa, P. Heat Stress Tolerance in Rice (Oryza sativa L.): Identification of Quantitative Trait Loci and Candidate Genes for Seedling Growth Under Heat Stress. Front. Plant Sci. 2018, 9, 1578. [Google Scholar] [CrossRef] [Green Version]
- Schramm, F.; Larkindale, J.; Kiehlmann, E.; Ganguli, A.; Englich, E.; Vierling, E.; von Koskull-Döring, P. A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 2008, 53, 264–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, Y.; Huang, Z.; Jiang, H.; Wang, Z.; Wu, F.; Xiong, Y.; Yao, J. A heat stress responsive NAC transcription factor heterodimer plays key roles in rice grain filling. J. Exp. Bot. 2021, 72, 2947–2964. [Google Scholar] [CrossRef]
- Liu, L.; Zhou, Y.; Szczerba, M.W.; Li, X.; Lin, Y. Identification and application of a rice senescence-associated promoter. Plant Physiol. 2010, 153, 1239–1249. [Google Scholar] [CrossRef] [Green Version]
- Zani, D.; Crowther, T.W.; Mo, L.; Renner, S.S.; Zohner, C.M. Increased growing-season productivity drives earlier autumn leaf senescence in temperate trees. Science 2020, 370, 1066–1071. [Google Scholar] [CrossRef] [PubMed]
- Breeze, E.; Harrison, E.; McHattie, S.; Hughes, L.; Hickman, R.; Hill, C. High-Resolution Temporal Profiling of Transcripts during Arabidopsis Leaf Senescence Reveals a Distinct Chronology of Processes and Regulation. Plant Cell 2011, 23, 873–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woo, H.R.; Koo, H.J.; Kim, J.; Jeong, H.; Yang, J.O.; Lee, I.H.; Jun, J.H.; Choi, S.H.; Park, S.J.; Kang, B.; et al. Programming of plant leaf senescence with temporal and inter-organellar coordination of transcriptome in Arabidopsis. Plant Physiol. 2016, 171, 452–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
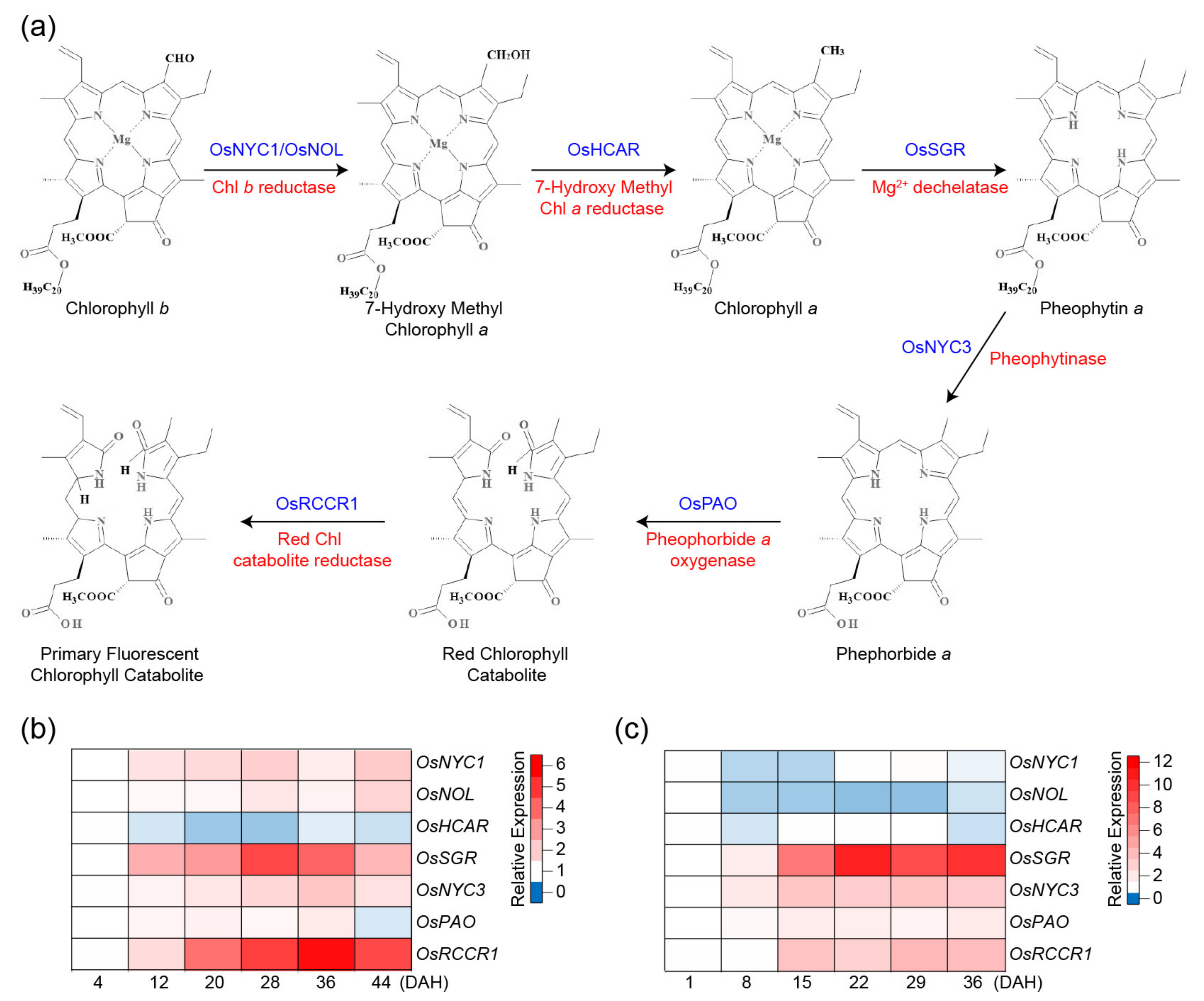
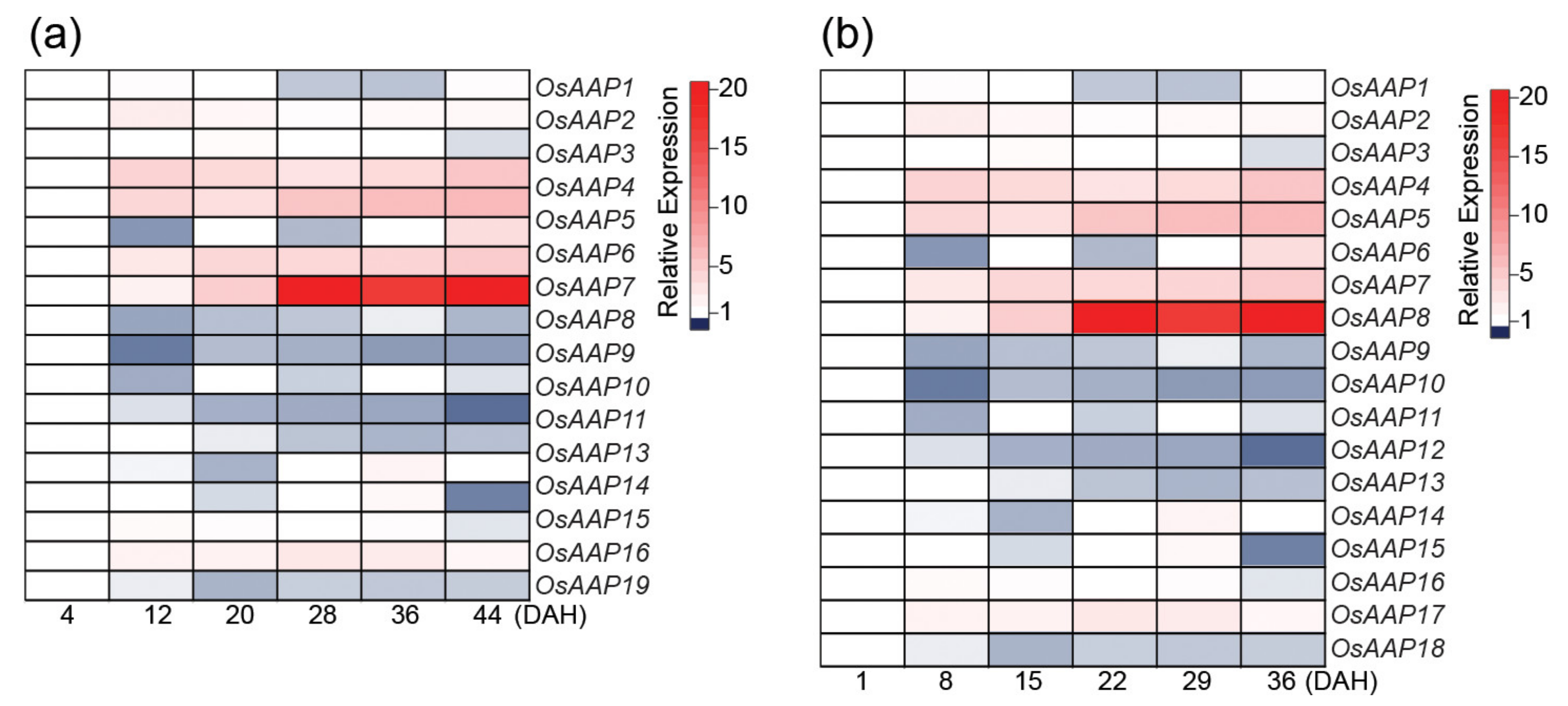
| Gene | Locus Number | Gene Family | Role in Senescence | Pathways Involved | Direct Downstream Targets | References |
|---|---|---|---|---|---|---|
| OsNAP | LOC_Os03g21060 | NAC TF | Positive | Age, ABA, dark, salt, drought | OsSGR, OsNYC1, OsNYC3, OsRCCR1, Osh36, OsI57, Osh69, OsI85 | [71,72,73] |
| OsNAC2 | LOC_Os04g38720 | NAC TF | Positive | Age, ABA, dark | OsSGR, OsNYC3, OsNCED3, OsZEP1 | [74] |
| ONAC106 | LOC_Os08g33670 | NAC TF | Negative | Age, dark, salt | OsSGR, OsNYC1, OsNAC5, OsNAP, OsEIN3, OsS3H, OsDREB2A, OsLEA3, OsbZIP23, LPA1 | [76] |
| ONAC011 (OsY37) | LOC_Os06g46270 | NAC TF | Positive | Age | [77] | |
| ONAC096 | LOC_Os07g04560 | NAC TF | Positive | Age, ABA | OsSGR, OsPAO, OsNYC3, OsRCCR1, OsI85, Osl2, Osl57, OsNAP, ABI5, OsEEL | [65] |
| ONAC054 | LOC_Os03g02800 | NAC TF | positive | Age, ABA, JA | OsNYC1, OsABI5, | [64] |
| OsNAC109 (OsYL3) | LOC_Os09g38000 | NAC TF | Negative | Age, ABA, dark | OsNAP, OsNYC3, OsEATB, OsAMTR1, OsZFP185, OsMPS, OsGA2ox3 | [75] |
| OsRL3 | LOC_Os02g47744 | MYB TF | Positive | ABA, salt, dark | OsSGR, OsNYC1, OsRCCR1, Osl2, Osl43, OsSAG12-2, OsRK1, OsRAB16C, OsRAB16D | [80] |
| OsMYB102 | LOC_Os06g43090 | MYB TF | Negative | Age, ABA, dark | OsSGR, OsNYC1, OsABF4, OsNAP, OsCYP707A6 | [81] |
| OsWRKY80 | LOC_Os09g30400 | WRKY TF | Positive | Dark, ABA, drought, | [84] | |
| OsWRKY23 | LOC_Os01g53260 | WRKY TF | Positive | Age, dark, SA, pathogen | [85] | |
| OsWRKY42 | LOC_Os02g26430 | WRKY TF | Positive | Age, ROS | OsMT1d | [86] |
| OsWRKY5 | LOC_Os05g04640 | WRKY TF | Positive | Age, dark, ABA | CCEs, SAGs, OsNAP, OsNAC2, OsNCED3, OsNCED4, OsNCED5 | [87] |
| OsPIL1 | LOC_Os03g56950 | bHLH TF | Negative | Age, dark | OsPORB, OsCAO1, OsGLK1, OsGLK2 | [90,91] |
| OsMYC2 | LOC_Os10g42430 | MYC TF | Positive | JA, dark | Several SAGs. | [95] |
| OsERF101 | LOC_Os04g32620 | ERF TF | Positive | Age, ABA, drought, JA | OsNAP, OsMYC2, OsJAI1, OsCOI1a, OsSGR, OsNYC1, OsNYC3 | [94] |
| OsTZF1 | LOC_Os01g09620 | CCCH-type zinc finger TF | Negative | Age, ABA, JA, SA, salt, drought, dark | Stress-responsive genes | [92] |
| OsTZF2 (OsDOS) | LOC_Os05g10670 | CCCH-type zinc finger TF | Negative | Age, JA | [93] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Masclaux-Daubresse, C. Current Understanding of Leaf Senescence in Rice. Int. J. Mol. Sci. 2021, 22, 4515. https://doi.org/10.3390/ijms22094515
Lee S, Masclaux-Daubresse C. Current Understanding of Leaf Senescence in Rice. International Journal of Molecular Sciences. 2021; 22(9):4515. https://doi.org/10.3390/ijms22094515
Chicago/Turabian StyleLee, Sichul, and Celine Masclaux-Daubresse. 2021. "Current Understanding of Leaf Senescence in Rice" International Journal of Molecular Sciences 22, no. 9: 4515. https://doi.org/10.3390/ijms22094515






