Characterization of Gut Microbiota Composition in Type 2 Diabetes Patients: A Population-Based Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Population
2.2. Fecal DNA Extraction and 16s rRNA Gene Sequencing
2.3. Sequencing Data Analysis
2.4. Statistical Analysis
3. Results
3.1. Epidemiological Data
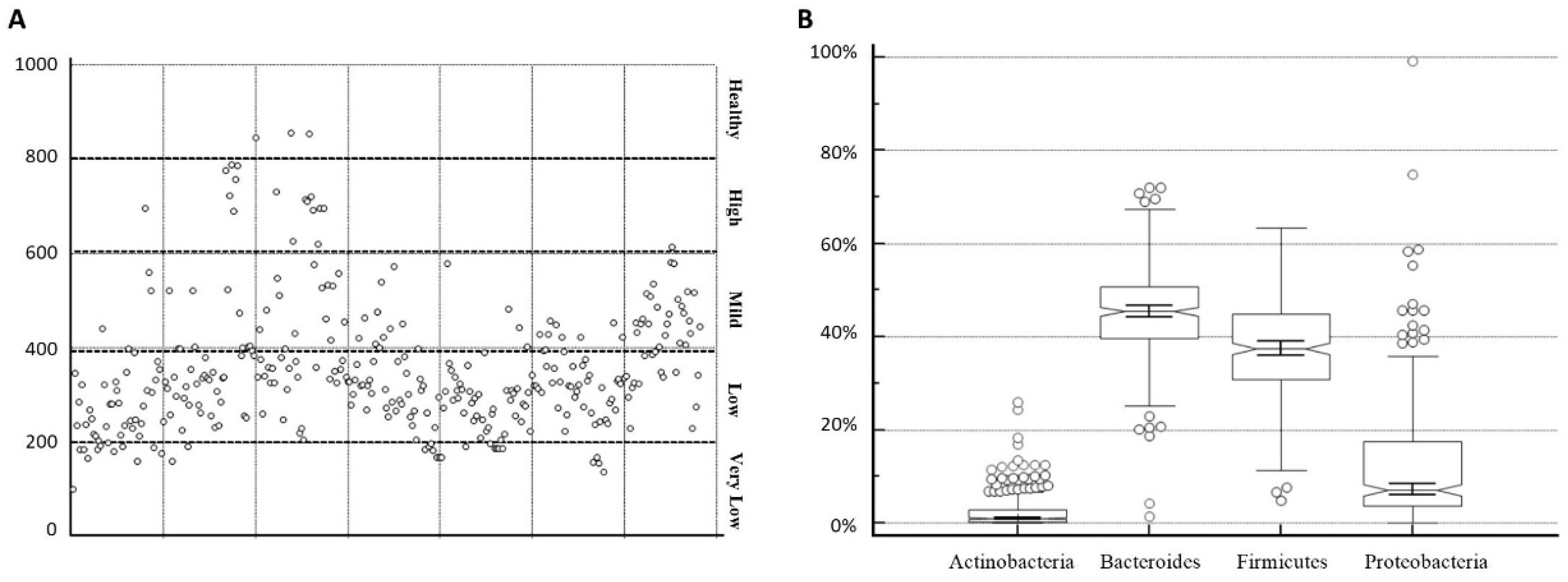
3.2. Microbiota Heterogeneity
- -
- Under 200: very low biodiversity
- -
- 200–400: low biodiversity
- -
- 400–600: mild biodiversity
- -
- 600–800: high biodiversity
- -
- Over 800: healthy biodiversity
3.3. Microbiota Abundance and Richness
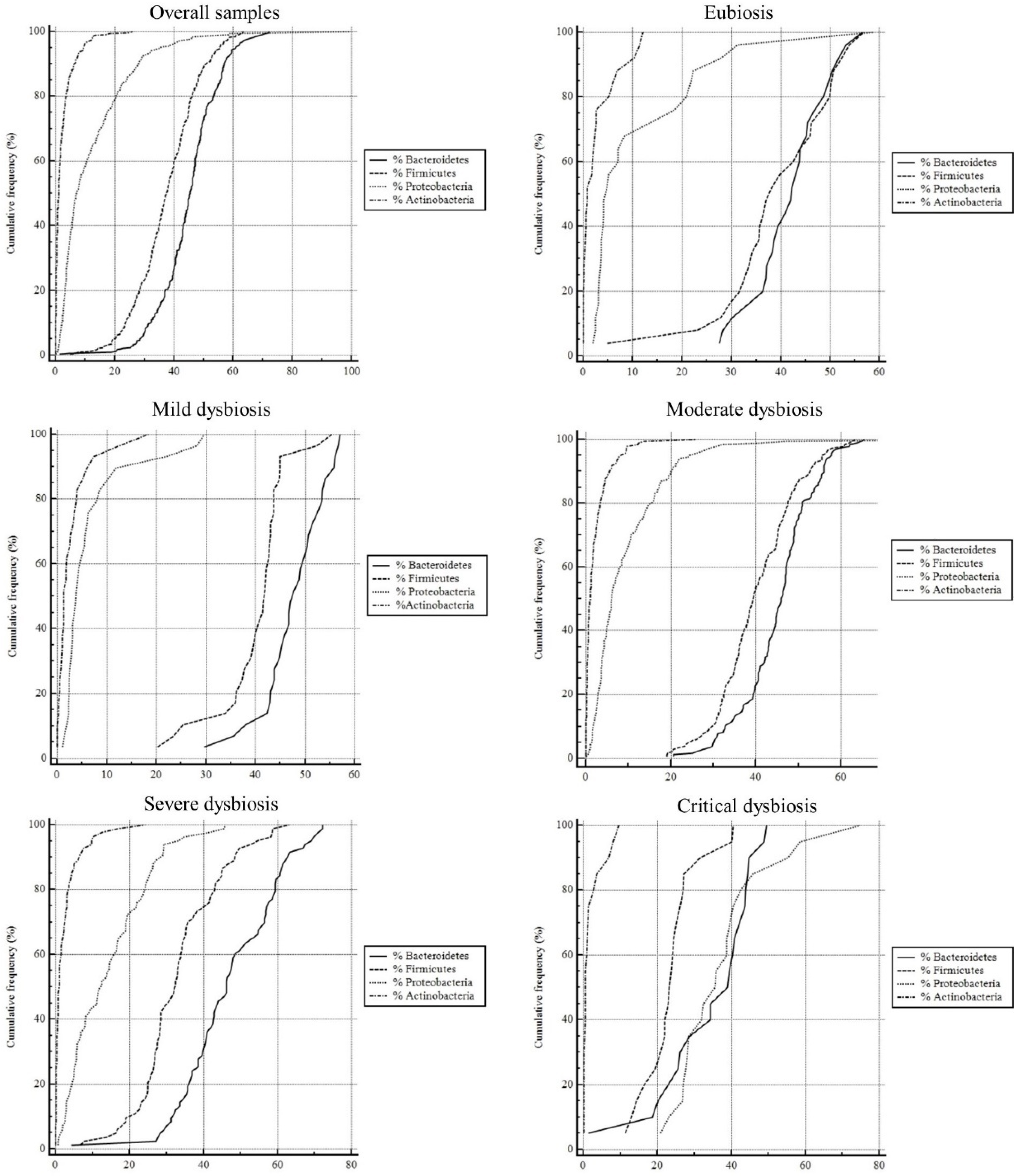
3.4. Dysbiosis and Pathologies
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sircana, A.; Framarin, L.; Leone, N.; Berrutti, M.; Castellino, F.; Parente, R.; de Michieli, F.; Paschetta, E.; Musso, G. Altered Gut Microbiota in Type 2 Diabetes: Just a Coincidence? Curr. Diabetes Rep. 2018, 18, 98. [Google Scholar] [CrossRef] [PubMed]
- Binda, C.; Gibiino, G.; Coluccio, C.; Sbrancia, M.; Dajti, E.; Sinagra, E.; Capurso, G.; Sambri, V.; Cucchetti, A.; Ercolani, G.; et al. Biliary Diseases from the Microbiome Perspective: How Microorganisms Could Change the Approach to Benign and Malignant Diseases. Microorganisms 2022, 10, 312. [Google Scholar] [CrossRef] [PubMed]
- Knezevic, J.; Starchl, C.; Berisha, A.T.; Amrein, K. Thyroid-Gut-Axis: How Does the Microbiota Influence Thyroid Function? Nutrients 2020, 12, 1769. [Google Scholar] [CrossRef] [PubMed]
- Riccio, P.; Rossano, R. The Human Gut Microbiota Is Neither an Organ nor a Commensal. FEBS Lett. 2020, 594, 3262–3271. [Google Scholar] [CrossRef]
- Rodríguez, J.M.; Murphy, K.; Stanton, C.; Ross, R.P.; Kober, O.I.; Juge, N.; Avershina, E.; Rudi, K.; Narbad, A.; Jenmalm, M.C.; et al. The Composition of the Gut Microbiota throughout Life, with an Emphasis on Early Life. Microb. Ecol. Health Dis. 2015, 26, 26050. [Google Scholar] [CrossRef]
- Farooq, R.K.; Alamoudi, W.; Alhibshi, A.; Rehman, S.; Sharma, A.R.; Abdulla, F.A. Varied Composition and Underlying Mechanisms of Gut Microbiome in Neuroinflammation. Microorganisms 2022, 10, 705. [Google Scholar] [CrossRef]
- Shreiner, A.B.; Kao, J.Y.; Young, V.B. The Gut Microbiome in Health and in Disease. Curr. Opin. Gastroenterol. 2015, 31, 69–75. [Google Scholar] [CrossRef]
- Bargiel, P.; Szczuko, M.; Stachowska, L.; Prowans, P.; Czapla, N.; Markowska, M.; Petriczko, J.; Kledzik, J.; Jędrzejczyk-Kledzik, A.; Palma, J.; et al. Microbiome Metabolites and Thyroid Dysfunction. J. Clin. Med. 2021, 10, 3609. [Google Scholar] [CrossRef]
- Schippa, S.; Conte, M.P. Dysbiotic Events in Gut Microbiota: Impact on Human Health. Nutrients 2014, 6, 5786–5805. [Google Scholar] [CrossRef] [Green Version]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut Microbiota Functions: Metabolism of Nutrients and Other Food Components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, X.; Li, T.; Zhang, S.; Wang, L.; Wu, X.; Liu, J. The Genetic Sequence, Origin, and Diagnosis of SARS-CoV-2. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1629–1635. [Google Scholar] [CrossRef] [PubMed]
- González-Regueiro, J.A.; Moreno-Castañeda, L.; Uribe, M.; Chávez-Tapia, N.C. The Role of Bile Acids in Glucose Metabolism and Their Relation with Diabetes. Ann. Hepatol. 2017, 16, s15–s20. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Lei, O.K.; Nie, J.; Shi, Q.; Xu, Y.; Kong, Z. Effects of Low-Carbohydrate Diet and Exercise Training on Gut Microbiota. Front. Nutr. 2022, 9, 884550. [Google Scholar] [CrossRef]
- Naud, S.; Ibrahim, A.; Valles, C.; Maatouk, M.; Bittar, F.; Tidjani Alou, M.; Raoult, D. Candidate Phyla Radiation, an Underappreciated Division of the Human Microbiome, and Its Impact on Health and Disease. Clin. Microbiol. Rev. 2022, 35, e0014021. [Google Scholar] [CrossRef]
- Van Zyl, K.N.; Whitelaw, A.C.; Newton-Foot, M. The Effect of Storage Conditions on Microbial Communities in Stool. PLoS ONE 2020, 15, e0227486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, V.M.; Brito, A.K.P.; Amorim, A.T.; Souza, I.R.; Santos, M.B.; Campos, G.B.; dos Santos, D.C.; Júnior, A.C.R.B.; Santana, J.M.; Santos, D.B.; et al. Evaluation of Fecal Microbiota and Its Correlation with Inflammatory, Hormonal, and Nutritional Profiles in Women. Braz. J. Microbiol. 2022, 53, 1001–1009. [Google Scholar] [CrossRef]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel Disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef]
- Wang, J.; Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; et al. A Metagenome-Wide Association Study of Gut Microbiota in Type 2 Diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Dehghanbanadaki, H.; Aazami, H.; Ejtahed, H.S.; Sohrabi, A.; Raftar, S.K.A.; Tarashi, S.; Tabatabaei-Malazy, O.; Bahramali, G.; Siadat, S.D.; Esfahani, E.N.; et al. The Global Scientific Publications on Gut Microbiota in Type 2 Diabetes; a Bibliometric, Scientometric, and Descriptive Analysis. J. Diabetes. Metab. Disord. 2022, 21, 13–32. [Google Scholar] [CrossRef]
- Wong-Chew, R.M.; de Castro, J.-A.A.; Morelli, L.; Perez, M.; Ozen, M. Gut Immune Homeostasis: The Immunomodulatory Role of Bacillus Clausii, from Basic to Clinical Evidence. Expert. Rev. Clin. Immunol. 2022, 18, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Sardu, C.; Consiglia Trotta, M.; Santella, B.; D’Onofrio, N.; Barbieri, M.; Rizzo, M.R.; Sasso, F.C.; Scisciola, L.; Turriziani, F.; Torella, M.; et al. Microbiota Thrombus Colonization May Influence Athero-Thrombosis in Hyperglycemic Patients with ST Segment Elevation Myocardialinfarction (STEMI). Marianella Study. Diabetes. Res. Clin. Pract. 2021, 173, 108670. [Google Scholar] [CrossRef] [PubMed]
- Petrillo, F.; Petrillo, A.; Marrapodi, M.; Capristo, C.; Gicchino, M.F.; Montaldo, P.; Caredda, E.; Reibaldi, M.; Boatti, L.M.V.; Dell’Annunziata, F.; et al. Characterization and Comparison of Ocular Surface Microbiome in Newborns. Microorganisms 2022, 10, 1390. [Google Scholar] [CrossRef] [PubMed]
- Bielka, W.; Przezak, A.; Pawlik, A. The Role of the Gut Microbiota in the Pathogenesis of Diabetes. International Journal of Molecular Sciences. Int. J. Mol. Sci. 2022, 23, 480. [Google Scholar] [CrossRef]
- Chen, Z.; Radjabzadeh, D.; Chen, L.; Kurilshikov, A.; Kavousi, M.; Ahmadizar, F.; Ikram, M.A.; Uitterlinden, A.G.; Zhernakova, A.; Fu, J.; et al. Association of Insulin Resistance and Type 2 Diabetes with Gut Microbial Diversity: A Microbiome-Wide Analysis from Population Studies. JAMA Netw. Open 2021, 4, e2118811. [Google Scholar] [CrossRef]
- Raybould, H.E. Gut Microbiota, Epithelial Function and Derangements in Obesity. J. Physiol. 2012, 590, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The Role of Short-Chain Fatty Acids from Gut Microbiota in Gut-Brain Communication. Front. Endocrinol. 2020, 11, 25. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.; Si, J.; Qu, Y.; Jie, L.; He, Y.; Wang, C.; Zhang, Y. Vegetarian Diet Duration’s Influence on Women’s Gut Environment. Genes. Nutr. 2021, 16, 16. [Google Scholar] [CrossRef]
- Prokopidis, K.; Cervo, M.M.; Gandham, A.; Scott, D. Impact of Protein Intake in Older Adults with Sarcopenia and Obesity: A Gut Microbiota Perspective. Nutrients 2020, 12, 2285. [Google Scholar] [CrossRef]
- Wong, M.-W.; Yi, C.-H.; Liu, T.-T.; Lei, W.-Y.; Hung, J.-S.; Lin, C.-L.; Lin, S.-Z.; Chen, C.-L. Impact of Vegan Diets on Gut Microbiota: An Update on the Clinical Implications. Tzu-Chi Med. J. 2018, 30, 200–203. [Google Scholar] [CrossRef]
- Losno, E.A.; Sieferle, K.; Perez-Cueto, F.J.A.; Ritz, C. Vegan Diet and the Gut Microbiota Composition in Healthy Adults. Nutrients 2021, 13, 2402. [Google Scholar] [CrossRef] [PubMed]
- Pérez, J.C. Fungi of the Human Gut Microbiota: Roles and Significance. Int. J. Med. Microbiol. 2021, 311, 151490. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, S.C.; Barbara, G.; Buurman, W.; Ockhuizen, T.; Schulzke, J.-D.; Serino, M.; Tilg, H.; Watson, A.; Wells, J.M. Intestinal Permeability—A New Target for Disease Prevention and Therapy. BMC Gastroenterol. 2014, 14, 189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wozniak, H.; Beckmann, T.S.; Fröhlich, L.; Soccorsi, T.; le Terrier, C.; de Watteville, A.; Schrenzel, J.; Heidegger, C.-P. The Central and Biodynamic Role of Gut Microbiota in Critically Ill Patients. Crit. Care. 2022, 26, 250. [Google Scholar] [CrossRef]
- D’Enfert, C.; Kaune, A.-K.; Alaban, L.-R.; Chakraborty, S.; Cole, N.; Delavy, M.; Kosmala, D.; Marsaux, B.; Fróis-Martins, R.; Morelli, M.; et al. The Impact of the Fungus-Host-Microbiota Interplay upon Candida Albicans Infections: Current Knowledge and New Perspectives. FEMS Microbiol. Rev. 2021, 45, fuaa060. [Google Scholar] [CrossRef] [PubMed]
- Fasano, A. Zonulin and Its Regulation of Intestinal Barrier Function: The Biological Door to Inflammation, Autoimmunity, and Cancer. Physiol. Rev. 2011, 91, 151–175. [Google Scholar] [CrossRef] [Green Version]
- Alcock, J.; Maley, C.C.; Aktipis, C.A. Is Eating Behavior Manipulated by the Gastrointestinal Microbiota? Evolutionary Pressures and Potential Mechanisms. Bioessays 2014, 36, 940–949. [Google Scholar] [CrossRef]


| Descriptive Statistics | |||
|---|---|---|---|
| Females | Males | Total | |
| N | 225 | 89 | 314 |
| AGE y.o. mean (SD) min–max | 46.4 (15.0) 16–77 y.o. | 49.0 (16.7) 17–80 y.o. | n/a |
| BMI mean (SD) | 23.1 (4.4) | 24.3 (3.3) | n/a |
| CVD | 71% (n. 64) | 29% (n. 26) | 90 |
| Diabetes, type 2 | 77% (n. 70) | 23% (n. 21) | 91 |
| Autoimmune disorders | 76% (n. 26) | 29% (n. 8) | 34 |
| Celiac disease | 92% (n. 12) | 8% (n. 1) | 13 |
| Thyroid disorders | 76% (n. 19) | 24% (n. 16) | 25 |
| spp | Health Reference Values | Median Values Obtained |
|---|---|---|
| Bacteroidetes | 50–55% | 45.78% (lower) |
| Firmicutes | 40–45% | 37.63% (lower) |
| Proteobacteria | 2–3% | 6.75% (higher) |
| Actinobacteria | 1% | 0.99% (same) |
| Overall Samples | % Bacteroidetes | % Firmicutes | % Proteobacteria | % Actinobacteria |
|---|---|---|---|---|
| (n = 314) | ||||
| Median | 45.42 | 37.45 | 7.05 | 1.01 |
| IQR (25th–75th quartile) | 39.63 to 50.70 | 30.87 to 44.87 | 3.62 to 17.44 | 0.27 to 2.80 |
| Minimum | 1.34 | 4.88 | 0 | 0 |
| Maximum | 72.12 | 63.3 | 99.28 | 25.94 |
| 95% CI | 43.96 to 46.12 | 36.28 to 38.56 | 10.89 to 13.57 | 1.94 to 2.67 |
| Eubiosis | % Bacteroidetes | % Firmicutes | % Proteobacteria | % Actinobacteria |
| (n = 26) | ||||
| Median | 42.18 | 38.11 | 4.74 | 0.78 |
| IQR (25th–75th quartile) | 37.05 to 46.85 | 33.55 to 48.20 | 3.42 to 18.38 | 0.17 to 2.54 |
| Minimum | 27.48 | 4.88 | 1.89 | 0 |
| Maximum | 56.53 | 56.78 | 58.68 | 11.99 |
| 95% CI | 38.82 to 45.09 | 34.51 to 43.92 | 5.97 to 16.89 | 1.17 to 4.24 |
| Mild dysbiosis | % Bacteroidetes | % Firmicutes | % Proteobacteria | % Actinobacteria |
| (n = 30) | ||||
| Median | 47.83 | 41.89 | 3.76 | 1.34 |
| IQR (25th–75th quartile) | 43.88 to 52.50 | 37.72 to 43.63 | 2.53 to 6.29 | 0.79 to 3.43 |
| Minimum | 29.73 | 20.27 | 1.01 | 0 |
| Maximum | 57.1 | 55.37 | 29.71 | 18.41 |
| 95% CI | 45.22 to 50.12 | 37.32 to 42.91 | 3.86 to 9.52 | 1.39 to 4.44 |
| Moderate dysbiosis | % Bacteroidetes | % Firmicutes | % Proteobacteria | % Actinobacteria |
| (n = 184) | ||||
| Median | 45.99 | 39.39 | 6.12 | 1.04 |
| IQR (25th–75th quartile) | 40.60 to 50.02 | 34.24 to 46.29 | 3.56 to 13.43 | 0.28 to 2.76 |
| Minimum | 20.58 | 18.99 | 0 | 0 |
| Maximum | 65.52 | 63.3 | 99.28 | 25.94 |
| 95% CI | 43.99 to 46.44 | 38.82 to 41.51 | 7.97 to 11.01 | 1.68 to 2.61 |
| Severe dysbiosis | % Bacteroidetes | % Firmicutes | % Proteobacteria | % Actinobacteria |
| (n = 84) | ||||
| Median | 46.24 | 32.36 | 12.45 | 0.97 |
| IQR (25th–75th quartile) | 38.45 to 57.24 | 26.56 to 40.68 | 5.33 to 21.87 | 0.25 to 3.02 |
| Minimum | 4.27 | 6.57 | 0.7 | 0 |
| Maximum | 72.12 | 63.07 | 45.76 | 24.25 |
| 95% CI | 44.42 to 50.01 | 30.76 to 35.68 | 11.97 to 16.67 | 1.57 to 3.28 |
| Critical dysbiosis | % Bacteroidetes | % Firmicutes | % Proteobacteria | % Actinobacteria |
| (n = 21) | ||||
| Median | 39.12 | 23.43 | 35.67 | 0.54 |
| IQR (25th–75th quartile) | 26.01 to 43.67 | 20.42 to 26.30 | 28.06 to 40.97 | 0.21 to 1.76 |
| Minimum | 1.34 | 11.31 | 20.81 | 0.15 |
| Maximum | 49.49 | 40.39 | 74.79 | 9.5 |
| 95% CI | 28.68 to 40.13 | 20.18 to 27.25 | 31.41 to 43.74 | 0.59 to 3.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Polidori, I.; Marullo, L.; Ialongo, C.; Tomassetti, F.; Colombo, R.; di Gaudio, F.; Calugi, G.; Marrone, G.; Noce, A.; Bernardini, S.; et al. Characterization of Gut Microbiota Composition in Type 2 Diabetes Patients: A Population-Based Study. Int. J. Environ. Res. Public Health 2022, 19, 15913. https://doi.org/10.3390/ijerph192315913
Polidori I, Marullo L, Ialongo C, Tomassetti F, Colombo R, di Gaudio F, Calugi G, Marrone G, Noce A, Bernardini S, et al. Characterization of Gut Microbiota Composition in Type 2 Diabetes Patients: A Population-Based Study. International Journal of Environmental Research and Public Health. 2022; 19(23):15913. https://doi.org/10.3390/ijerph192315913
Chicago/Turabian StylePolidori, Isabella, Laura Marullo, Cristiano Ialongo, Flaminia Tomassetti, Roberto Colombo, Francesca di Gaudio, Graziella Calugi, Giulia Marrone, Annalisa Noce, Sergio Bernardini, and et al. 2022. "Characterization of Gut Microbiota Composition in Type 2 Diabetes Patients: A Population-Based Study" International Journal of Environmental Research and Public Health 19, no. 23: 15913. https://doi.org/10.3390/ijerph192315913





