Prediction of Number of Cases of 2019 Novel Coronavirus (COVID-19) Using Social Media Search Index
Abstract
1. Introduction
2. Individuals and Methods
2.1. Database
2.1.1. Baidu Search Index in Social Media
2.1.2. Number of New Suspected Infection Cases
2.2. Method
2.2.1. Model Formulation
2.2.2. Parameter Estimation
- (1)
- Subset selection refers to the task of finding a small subset of available predictors that accurately predict the response. If the model has predictors, then the subset selection method will choose the optimal model from the possible models, based on some criteria such as Akaike’s information criteria, Bayesian information criteria, or adjusted .
- (2)
- Forward selection is a stepwise selection method. It starts with no variables in the model, tests the addition of each variable using a chosen model fit criterion, adds the variable (if any), whose inclusion causes the most statistically significant improvement of the fit, and repeats this process until the model can no longer be improved to a statistically significant extent.
- (3)
- Ridge regression is a method to create a parsimonious model when the number of predictor variables exceeds the number of observations, or when the data set has multi-collinearity. Employing the least-squares method is not possible when the number of predictors exceeds the number of observations, which leads to overfitting a model and the failure to find unique solutions. In contrast to the least-squares method, ridge regression shrinks parameters by L2 penalty, to obtain biased but lower variance estimators; thus, the estimates are reasonably reliable approximations of true population values. In this study, ridge regression solves the following problem:
- (4)
- Lasso regression is also a type of linear regression that uses shrinkage. Lasso regression performs both variable selection and parameter shrinkage by using the L1 penalty, which enhances the prediction accuracy and interpretability of the statistical model it produces. The only difference between lasso regression and ridge regression is the penalty function. In this study, lasso regression solves the following problem:
- (5)
- Elastic net is a regularized regression method that linearly combines the L1 and L2 penalties of the lasso and ridge methods. The elastic net method often outperforms the lasso but has a similar sparsity of representation. In this study, elastic net solves the following problem:
2.2.3. Accuracy Metrics
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Variables | Angina Pectoris | Difficulty Urinating | Impotence | Urinary Incontinence | Dizziness |
|---|---|---|---|---|---|
| Lag 1 Day (p Value) | 0.3243 | 0.7197 | 0.7327 | 0.2646 | 0.8089 |
| (0.1515) | (0.2382) | (0.1137) | (0.2464) | (0.4781) | |
| Lag 2 Day (p Value) | 0.1428 | 0.6323 | 0.6309 | 0.0359 | 0.8702 |
| (0.5368) | (0.7821) | (0.4522) | (0.8772) | (0.1603) | |
| Lag 3 Day (p Value) | 0.0086 | 0.5699 | 0.6210 | −0.0479 | 0.9599 |
| (0.9705) | (0.6870) | (0.1927) | (0.8367) | (0.9775) | |
| Lag 4 Day (p Value) | −0.2584 | 0.3913 | 0.5375 | −0.3196 | 0.9445 |
| (0.2581) | (0.0794) | (0.1120) | (0.1578) | (0.8720) | |
| Lag 5 Day (p Value) | −0.4884 | 0.2344 | 0.3950 | −0.4854 | 0.9082 |
| (0.0747) | (0.3065) | (0.5764) | (0.4257) | (0.0861) | |
| Lag 6 Day (p Value) | −0.5826 | 0.1215 | 0.3021 | −0.6054 | 0.8637 |
| (0.1156) | (0.5998) | (0.1833) | (0.1136) | (0.1561) | |
| Lag 7 Day (p Value) | −0.6768 | −0.0797 | 0.2362 | −0.7190 | 0.8054 |
| (0.7438) | (0.7313) | (0.3026) | (0.9922) | (0.4460) | |
| Lag 8 Day (p Value) | −0.7272 | −0.1196 | 0.1444 | −0.7358 | 0.7309 |
| (0.0965) | (0.6055) | (0.5322) | (0.3351) | (0.1172) | |
| Lag 9 Day (p Value) | −0.6612 | −0.3142 | −0.0412 | −0.7723 | 0.6429 |
| (0.9211) | (0.1654) | (0.8594) | (0.9945) | (0.1779) | |
| Lag 10 Day (p Value) | −0.6386 | −0.2417 | −0.0971 | −0.6962 | 0.5584 |
| (0.6418) | (0.2912) | (0.6754) | (0.6625) | (0.2485) |
References
- Novel Coronavirus COVID-19 in China—Statistics & Facts; Health & Pharmaceuticals: Statistics and Facts on Health and Pharmaceuticals: New York, NY, USA, 2020.
- Liem, A.; Wang, C.; Wariyanti, Y.; Latkin, C.A.; Hall, B.J. The neglected health of international migrant workers in the COVID-19 epidemic. Lancet Psychiatry 2020, 7, 20. [Google Scholar] [CrossRef]
- Duan, L.; Zhu, G. Psychological interventions for people affected by the COVID-19 epidemic. Lancet Psychiatry 2020, 7. [Google Scholar] [CrossRef]
- Lai, C.C.; Shih, T.P.; Ko, W.C.; Tang, H.J.; Hsueh, P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents 2020, 105924. [Google Scholar] [CrossRef] [PubMed]
- Velavan, T.P.; Meyer, C.G. The COVID-19 epidemic. Trop. Med. Int. Health 2020, 25, 278–280. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Infection Prevention and Control during Health Care When Novel Coronavirus (nCoV) Infection Is Suspected. Available online: https://www.who.int/publications-detail/infection-prevention-and-control-during-health-care-when-novel-coronavirus-(ncov)-infection-is-suspected-20200125 (accessed on 4 February 2020).
- Robert, R.R.; Interim U.S. Guidance for Risk Assessment and Public Health Management of Healthcare Personnel with Potential Exposure in a Healthcare Setting to Patients with Coronavirus Disease (COVID-19). Available online: https://www.cdc.gov/coronavirus/2019-ncov/hcp/guidance-risk-assesment-hcp.html (accessed on 7 March 2020).
- World Health Organization. Coronavirus Disease 2019 (COVID-19) Situation Report—36. Available online: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200225-sitrep-36-covid-19.pdf?sfvrsn=2791b4e0_2 (accessed on 26 February 2020).
- Heymann, D.L.; Shindo, N. WHO Scientific and Technical Advisory Group for Infectious Hazards COVID-19: What is next for public health? Lancet 2020, 395. [Google Scholar] [CrossRef]
- Gostin, L.O.; Hodge, J.G., Jr. US emergency legal responses to novel coronavirus: Balancing public health and civil liberties. JAMA 2020, 323, 1131–1132. [Google Scholar] [CrossRef]
- Smith, N.; Fraser, M. Straining the system: Novel coronavirus (COVID-19) and preparedness for concomitant disasters. Am. J. Public Health 2020, e1–e2. [Google Scholar] [CrossRef]
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef]
- Shi, H.; Han, X.; Zheng, C. Evolution of CT manifestations in a patient recovered from 2019 novel coronavirus (2019-nCoV) pneumonia in Wuhan, China. Radiology 2020, 200269. [Google Scholar] [CrossRef]
- Xu, X.; Chen, P.; Wang, J.; Feng, J.; Zhou, H.; Li, X.; Zhong, W.; Hao, P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020, 63, 457–460. [Google Scholar] [CrossRef]
- Lau, S.K.; Lee, P.; Tsang, A.K.; Yip, C.C.; Tse, H.; Lee, R.A.; So, L.Y.; Lau, Y.L.; Chan, K.H.; Woo, P.C.; et al. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J. Virol. 2011, 85, 11325–11337. [Google Scholar] [CrossRef] [PubMed]
- Seo, D.W.; Shin, S.Y. Methods using social media and search queries to predict infectious disease outbreaks. Healthc. Inform. Res. 2017, 23, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Meyers, L.A.; Pourbohloul, B.; Newman, M.E.; Skowronski, D.M.; Brunham, R.C. Network theory and SARS: Predicting outbreak diversity. J. Theory Biol. 2005, 232, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Alessa, A.; Faezipour, M. A review of influenza detection and prediction through social networking sites. Theor. Biol. Med. Model. 2018, 15, 2. [Google Scholar] [CrossRef]
- Liu, D.; Guo, S.; Zou, M.; Chen, C.; Deng, F.; Xie, Z.; Hu, S.; Wu, L. A dengue fever predicting model based on Baidu search index data and climate data in South China. PLoS ONE 2019, 14, e0226841. [Google Scholar] [CrossRef]
- Seo, D.W.; Jo, M.W.; Sohn, C.H.; Shin, S.Y.; Lee, J.; Yu, M.; Kim, W.Y.; Lim, K.S.; Lee, S.I. Cumulative query method for influenza surveillance using search engine data. J. Med. Internet Res. 2014, 16, e289. [Google Scholar] [CrossRef]
- Lazer, D.; Kennedy, R.; King, G.; Vespignani, A. Big data. The parable of Google Flu: Traps in big data analysis. Science 2014, 343, 1203–1205. [Google Scholar] [CrossRef]
- Search Engines in China —Statistics & Facts. Available online: https://www.statista.com/topics/1337/search-engines-in-china/ (accessed on 7 November 2019).
- Zhao, S.; Lin, Q.; Ran, J.; Musa, S.S.; Yang, G.; Wang, W.; Lou, Y.; Gao, D.; Yang, L.; He, D.; et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: A data-driven analysis in the early phase of the outbreak. Int. J. Infect. Dis. 2020, 92, 214–217. [Google Scholar] [CrossRef]
- Gao, J.; Tian, Z.; Yang, X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends 2020, 14. [Google Scholar] [CrossRef]
- Calisher, C.; Carroll, D.; Colwell, R.; Corley, R.B.; Daszak, P.; Drosten, C.; Enjuanes, L.; Farrar, J.; Field, H.; Golding, J.; et al. Statement in support of the scientists, public health professionals, and medical professionals of China combatting COVID-19. Lancet 2020, 395. [Google Scholar] [CrossRef]
- Oh, H.J.; Lee, H. When do people verify and share health rumors on social media? The effects of message importance, health anxiety, and health literacy. J. Health Commun. 2019, 24, 837–847. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.W. Trending now: Using social media to predict and track disease outbreaks. Environ. Health Perspect. 2012, 120, A30–A33. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, J. Consumer health information seeking in social media: A literature review. Health Inf. Libr. J. 2017, 34, 268–283. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Y.; Li, C.M.; Liang, J.C.; Tsai, C.C. Health information obtained from the internet and changes in medical decision making: Questionnaire development and cross-sectional survey. J. Med. Internet Res. 2018, 20, e47. [Google Scholar] [CrossRef] [PubMed]
- Eysenbach, G. Infodemiology: Tracking flu-related searches on the web for syndromic surveillance. AMIA Annu. Symp. Proc. 2006, 244, 244–248. [Google Scholar]
- Polgreen, P.M.; Chen, Y.; Pennock, D.M.; Nelson, F.D. Using internet searches for influenza surveillance. Clin. Infect. Dis. 2008, 47, 1443–1448. [Google Scholar] [CrossRef]
- Nsubuga, P.; White, M.E.; Thacker, S.B.; Anderson, M.A.; Blount, S.B.; Broome, C.V.; Chiller, T.M.; Espitia, V.; Imtiaz, R.; Sosin, D.; et al. Public health surveillance: A tool for targeting and monitoring interventions. In Disease Control Priorities in Developing Countries; Jamison, D.T., Breman, J.G., Measham, A.R., Alleyne, G., Claeson, M., Evans, D.B., Jha, P., Mills, A., Musgrove, P., Eds.; Europe PMC: Washington, DC, USA, 2006. [Google Scholar]
- Aiarzaguena, J.M.; Grandes, G.; Salazar, A.; Gaminde, I.; Sanchez, A. The diagnostic challenges presented by patients with medically unexplained symptoms in general practice. Scand. J. Prim. Health Care 2008, 26, 99–105. [Google Scholar] [CrossRef]
- Mack, A.; Choffnes, E.R.; Sparling, P.F.; Hamburg, M.A.; Lemon, S.M. Global Infectious Disease Surveillance and Detection: Assessing the Challenges-Finding Solutions, Workshop Summary; The National Academy of Science Engineering and Medicine: Washington, DC, USA, 2007. [Google Scholar] [CrossRef]
- Welch, V.; Petkovic, J.; Pardo Pardo, J.; Rader, T.; Tugwell, P. Interactive social media interventions to promote health equity: An overview of reviews. Health Promot. Chronic Dis. Prev. Can. 2016, 36, 63–75. [Google Scholar] [CrossRef]
- Feng, Z.; Li, Q.; Zhang, Y. Novel Coronavirus Pneumonia Emergency Response Epidemiology, The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi 2020, 41, 145–151. [Google Scholar] [CrossRef]
- Christaki, E. New technologies in predicting, preventing and controlling emerging infectious diseases. Virulence 2015, 6, 558–565. [Google Scholar] [CrossRef]
- Madhav, N.; Oppenheim, B.; Gallivan, M.; Mulembakani, P.; Rubin, E.; Wolfe, N. Pandemics: Risks, impacts, and mitigation. In Disease Control Priorities: Improving Health and Reducing Poverty; Jamison, D.T., Gelband, H., Horton, S., Jha, P., Laxminarayan, R., Mock, C.N., Nugent, R., Eds.; World Bank: Washington, DC, USA, 2017. [Google Scholar] [CrossRef]
- Coronavirus Disease (COVID-19) Technical Guidance. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance (accessed on 1 February 2020).
- Farkas, O.; Heberger, K. Comparison of ridge regression, partial least-squares, pairwise correlation, forward- and best subset selection methods for prediction of retention indices for aliphatic alcohols. J. Chem. Inf. Model. 2005, 45, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Degroeve, S.; De Baets, B.; Van de Peer, Y.; Rouze, P. Feature subset selection for splice site prediction. Bioinformatics 2002, 18, S75–S83. [Google Scholar] [CrossRef] [PubMed]
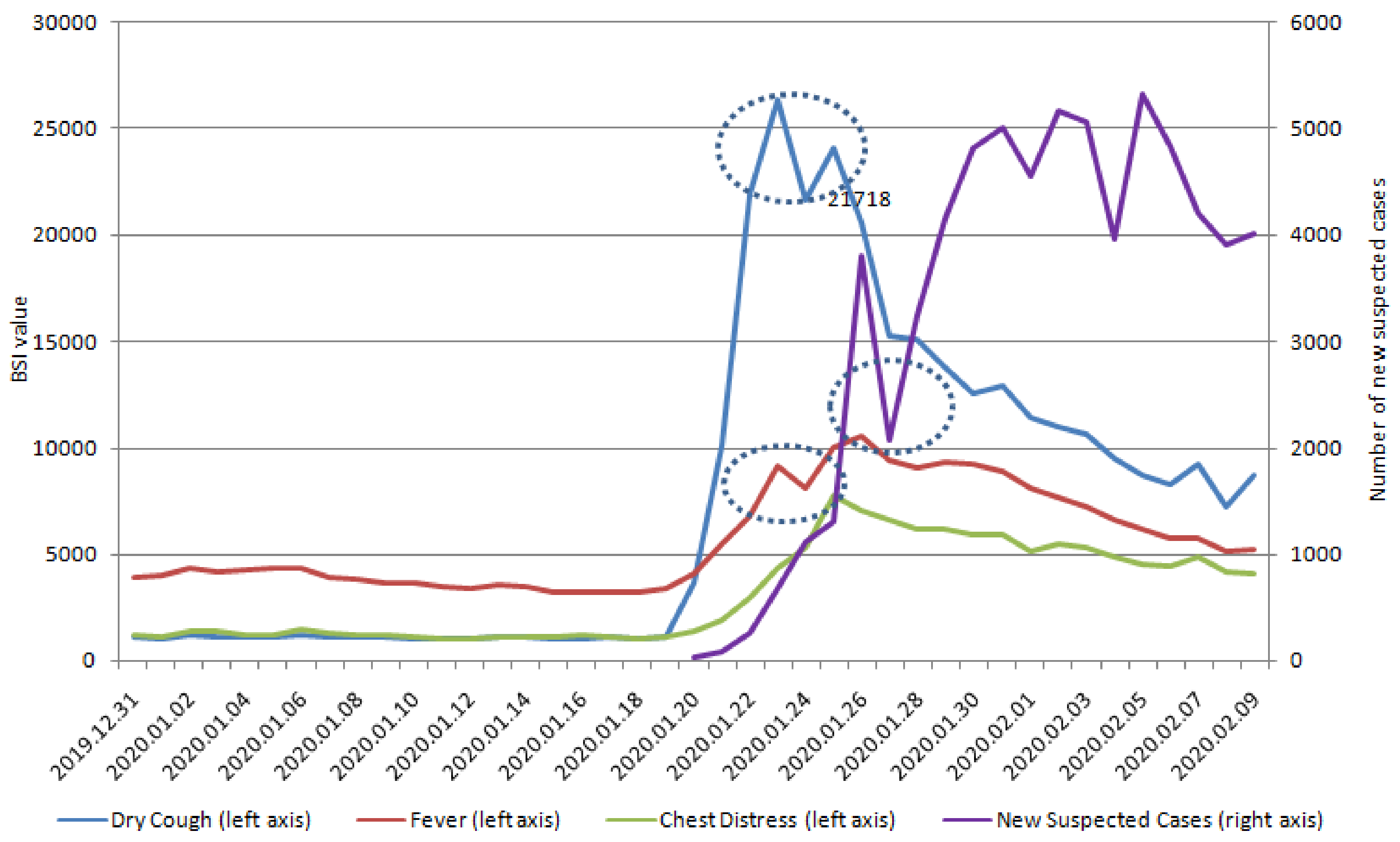
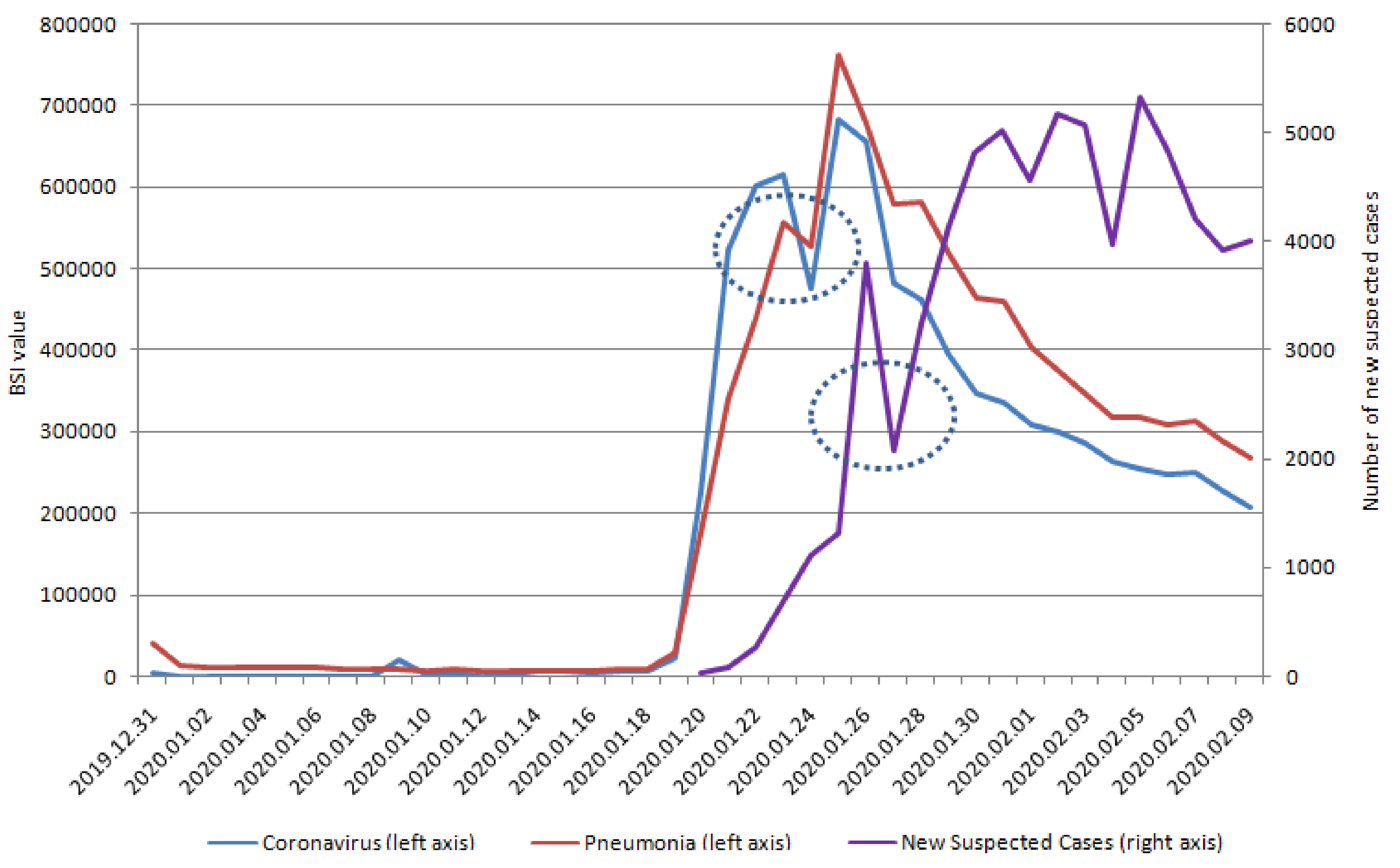
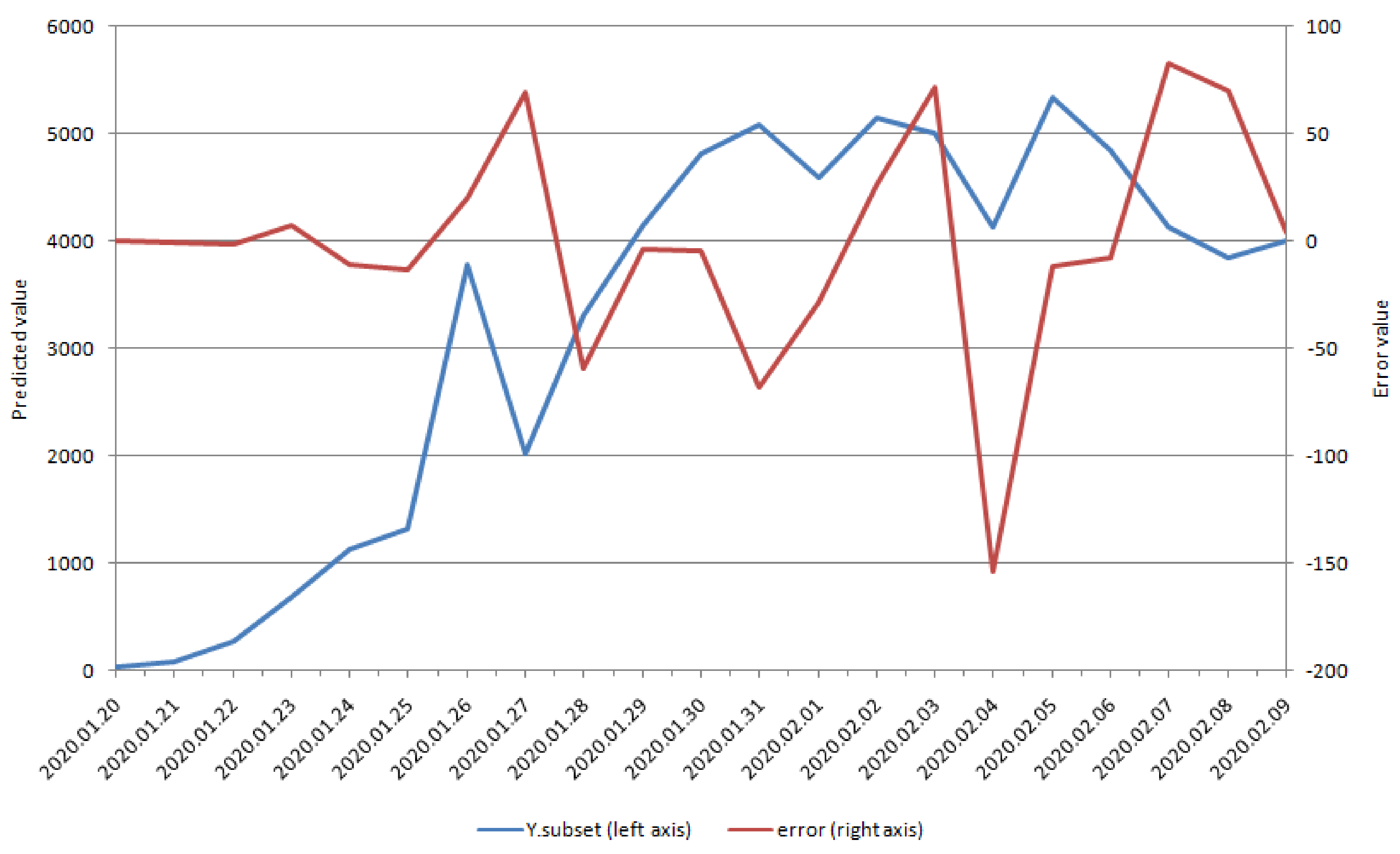
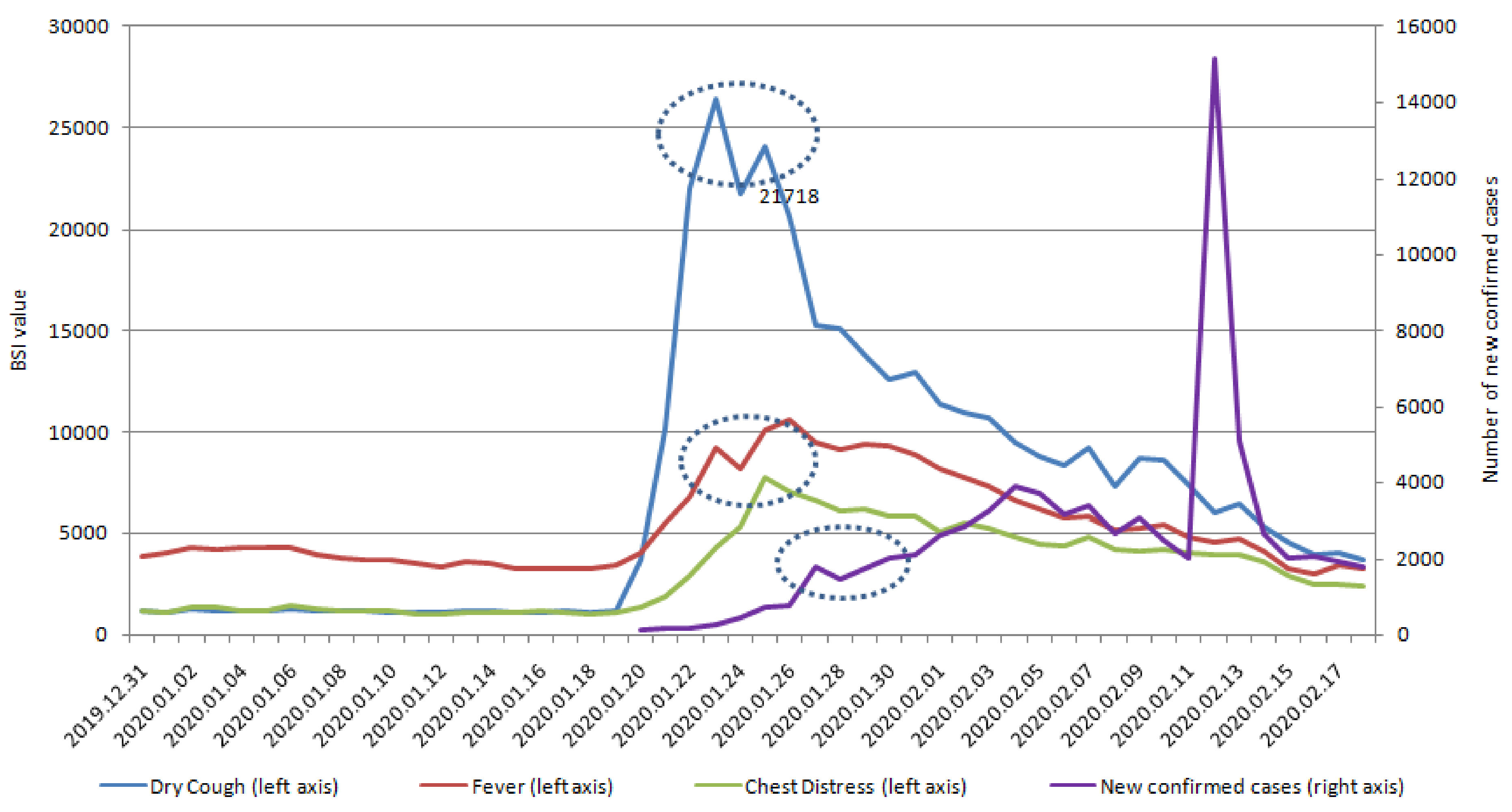
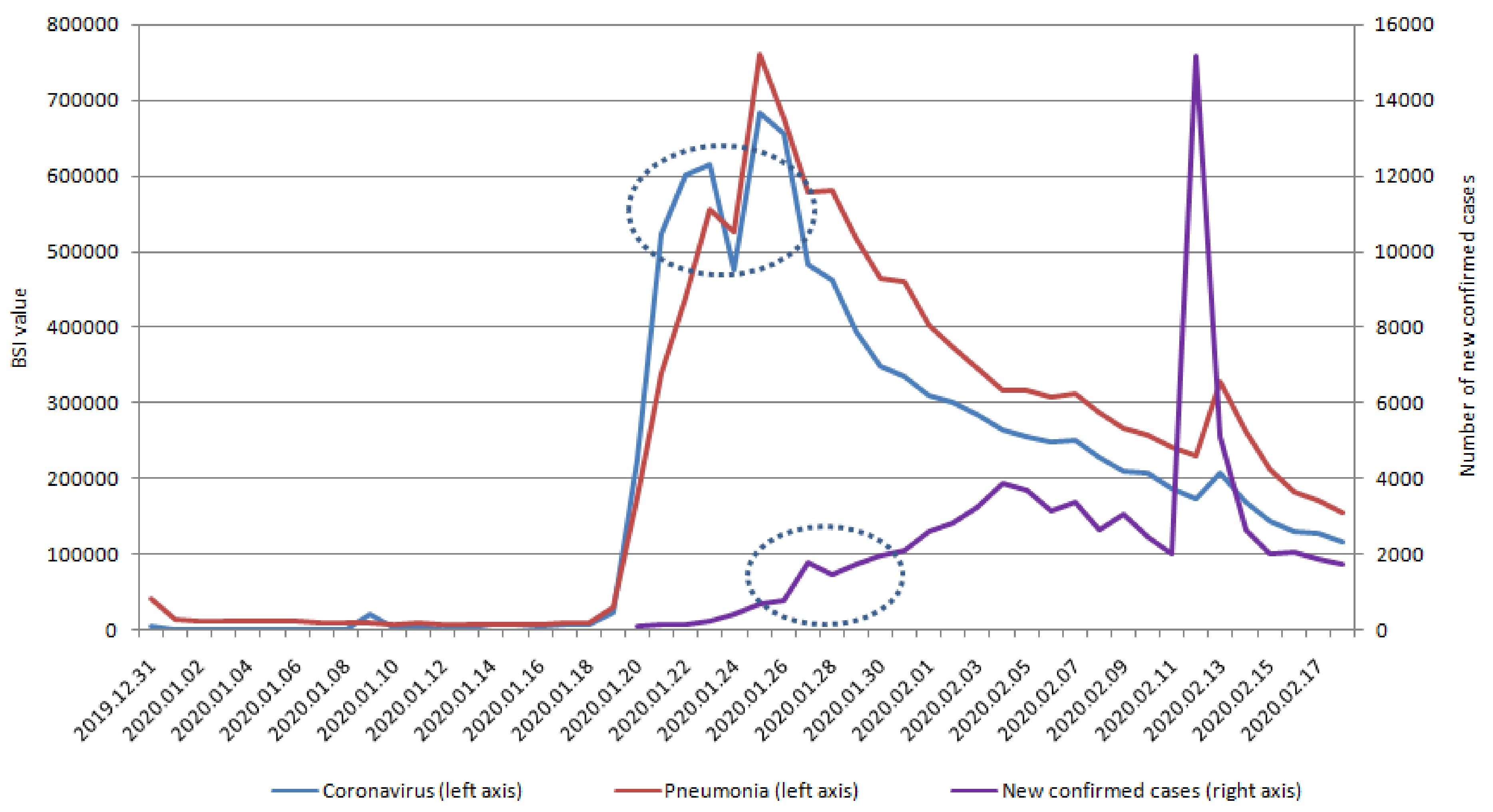
| Variables | Dry Cough | Fever | Chest Distress | Coronavirus | Pneumonia |
|---|---|---|---|---|---|
| Lag 1 Day (p Value) | −0.1070 | 0.3586 | 0.6493 | −0.2094 | 0.1922 |
| (0.6445) | (0.1105) | (0.0014) | (0.3623) | (0.4039) | |
| Lag 2 Day (p Value) | 0.1488 | 0.5650 | 0.7468 | 0.0626 | 0.4111 |
| (0.5198) | (0.0076) | (0.0001) | (0.7876) | (0.0641) | |
| Lag 3 Day (p Value) | 0.4183 | 0.7856 | 0.8590 | 0.3828 | 0.6517 |
| (0.0591) | (<0.0001) | (<0.0001) | (0.0868) | (0.0014) | |
| Lag 4 Day (p Value) | 0.5868 | 0.8596 | 0.9007 | 0.5847 | 0.7824 |
| (0.0052) | (<0.0001) | (<0.0001) | (0.0054) | (<0.0001) | |
| Lag 5 Day (p Value) | 0.6920 | 0.9147 | 0.9175 | 0.7352 | 0.8813 |
| (0.0005) | (<0.0001) | (<0.0001) | (0.0001) | (<0.0001) | |
| Lag 6 Day (p Value) | 0.7779 | 0.9124 | 0.8920 | 0.7831 | 0.9030 |
| (<0.0001) | (<0.0001) | (<0.0001) | (<0.0001) | (<0.0001) | |
| Lag 7 Day (p Value) | 0.8288 | 0.8896 | 0.8396 | 0.8301 | 0.8886 |
| (<0.0001) | (<0.0001) | (<0.0001) | (<0.0001) | (<0.0001) | |
| Lag 8 Day (p Value) | 0.8418 | 0.8361 | 0.7766 | 0.8795 | 0.8832 |
| (<0.0001) | (<0.0001) | (<0.0001) | (<0.0001) | (<0.0001 | |
| Lag 9 Day (p Value) | 0.7758 | 0.7381 | 0.6935 | 0.8325 | 0.8130 |
| (<0.0001) | (0.0001) | (0.0005) | (<0.0001) | (<0.0001) | |
| Lag 10 Day (p Value) | 0.7077 | 0.6647 | 0.6044 | 0.7732 | 0.7306 |
| (0.0003) | (0.0010) | (0.0037) | (<0.0001) | (0.0002) |
| Variables | RMSE | MAE | MAPE | Correlation | Correlation of Increment | Number of Predictor |
|---|---|---|---|---|---|---|
| Subset Selection | 51.6671 | 34.0739 | 0.0107 | 0.9996 | 0.9963 | 10 |
| Forward Selection | 70.0168 | 39.9790 | 0.0113 | 0.9993 | 0.9913 | 15 |
| Ridge Regression | 415.2922 | 279.6788 | 0.0827 | 0.9741 | 0.6937 | 51 |
| Lasso Regression | 519.7440 | 358.0979 | 0.1032 | 0.9597 | 0.4858 | 9 |
| Elastic Net(alpha = 0.2) | 527.4250 | 360.9563 | 0.1085 | 0.9585 | 0.4831 | 24 |
| Elastic Net(alpha = 0.4) | 516.1075 | 347.5939 | 0.1041 | 0.9602 | 0.5037 | 18 |
| Elastic Net(alpha = 0.6) | 514.7714 | 347.7290 | 0.1036 | 0.9604 | 0.4906 | 14 |
| Elastic Net(alpha = 0.8) | 510.1201 | 348.5859 | 0.1033 | 0.9611 | 0.5023 | 11 |
| Variables | Dry Cough | Fever | Chest Distress | Coronavirus | Pneumonia |
|---|---|---|---|---|---|
| Lag 1 Day (p Value) | −0.2444 | −0.1588 | 0.0852 | −0.3125 | −0.2046 |
| (0.1930) | (0.4020) | (0.6544) | (0.0927) | (0.2781) | |
| Lag 2 Day (p Value) | −0.1130 | −0.0186 | 0.1971 | −0.1861 | −0.0720 |
| (0.5523) | (0.9221) | (0.2964) | (0.3248) | (0.7055) | |
| Lag 3 Day (p Value) | −0.0235 | 0.0479 | 0.2392 | −0.0968 | 0.0276 |
| (0.9017) | (0.8014) | (0.2030) | (0.6108) | (0.8849) | |
| Lag 4 Day (p Value) | 0.0257 | 0.1169 | 0.2954 | 0.0144 | 0.1360 |
| (0.8929) | (0.5386) | (0.1130) | (0.9397) | (0.4737) | |
| Lag 5 Day (p Value) | 0.1299 | 0.2169 | 0.3900 | 0.1134 | 0.2269 |
| (0.4938) | (0.2496) | (0.0331) | (0.5506) | (0.2279) | |
| Lag 6 Day (p Value) | 0.1659 | 0.2663 | 0.3895 | 0.1863 | 0.2861 |
| (0.3809) | (0.1549) | (0.0334) | (0.3243) | (0.1253) | |
| Lag 7 Day (p Value) | 0.2190 | 0.3271 | 0.4128 | 0.2442 | 0.3368 |
| (0.2449) | (0.0776) | (0.0234) | (0.1934) | (0.0688) | |
| Lag 8 Day (p Value) | 0.2729 | 0.3757 | 0.4440 | 0.2891 | 0.3621 |
| (0.1446) | (0.0407) | (0.0140) | (0.1213) | (0.0493) | |
| Lag 9 Day (p Value) | 0.3422 | 0.4381 | 0.4879 | 0.3461 | 0.4061 |
| (0.0641) | (0.0155) | (0.0062) | (0.0610) | (0.0260) | |
| Lag 10 Day (p Value) | 0.3823 | 0.4666 | 0.4998 | 0.3843 | 0.4363 |
| (0.0371) | (0.0093) | (0.0049) | (0.0360) | (0.0159) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, L.; Sun, Q.; Wang, Y.; Wu, K.-F.; Chen, M.; Shia, B.-C.; Wu, S.-Y. Prediction of Number of Cases of 2019 Novel Coronavirus (COVID-19) Using Social Media Search Index. Int. J. Environ. Res. Public Health 2020, 17, 2365. https://doi.org/10.3390/ijerph17072365
Qin L, Sun Q, Wang Y, Wu K-F, Chen M, Shia B-C, Wu S-Y. Prediction of Number of Cases of 2019 Novel Coronavirus (COVID-19) Using Social Media Search Index. International Journal of Environmental Research and Public Health. 2020; 17(7):2365. https://doi.org/10.3390/ijerph17072365
Chicago/Turabian StyleQin, Lei, Qiang Sun, Yidan Wang, Ke-Fei Wu, Mingchih Chen, Ben-Chang Shia, and Szu-Yuan Wu. 2020. "Prediction of Number of Cases of 2019 Novel Coronavirus (COVID-19) Using Social Media Search Index" International Journal of Environmental Research and Public Health 17, no. 7: 2365. https://doi.org/10.3390/ijerph17072365
APA StyleQin, L., Sun, Q., Wang, Y., Wu, K.-F., Chen, M., Shia, B.-C., & Wu, S.-Y. (2020). Prediction of Number of Cases of 2019 Novel Coronavirus (COVID-19) Using Social Media Search Index. International Journal of Environmental Research and Public Health, 17(7), 2365. https://doi.org/10.3390/ijerph17072365






