Increased Detection of Viruses in Children with Respiratory Tract Infection Using PCR
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Sample Collection
2.2. Extraction of Viral Nucleic Acids, Reverse Transcription, and Multiplex RT-PCR Analysis
2.3. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lower Respiratory Infections Collaborators. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: A systematic analysis for the global burden of disease study 2016. Lancet Infect. Dis. 2018, 18, 1191–1210. [Google Scholar] [CrossRef]
- van den Hoogen, B.G.; de Jong, J.C.; Groen, J.; Kuiken, T.; de Groot, R.; Fouchier, R.A.; Osterhaus, A.D. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 2001, 7, 719–724. [Google Scholar] [CrossRef] [PubMed]
- van der Hoek, L.; Pyrc, K.; Jebbink, M.F.; Vermeulen-Oost, W.; Berkhout, R.J.; Wolthers, K.C.; Wertheim-van Dillen, P.M.; Kaandorp, J.; Spaargaren, J.; Berkhout, B. Identification of a new human coronavirus. Nat. Med. 2004, 10, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Williams, D.J.; Arnold, S.R.; Ampofo, K.; Bramley, A.M.; Reed, C.; Stockmann, C.; Anderson, E.J.; Grijalva, C.G.; Self, W.H.; et al. Community-acquired pneumonia requiring hospitalization among US Children. N. Engl. J. Med. 2015, 372, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Ruuskanen, O.; Lahti, E.; Jennings, L.C.; Murdoch, D.R. Viral pneumonia. Lancet 2011, 377, 1264–1275. [Google Scholar] [CrossRef]
- Jain, S.; Self, W.H.; Wunderink, R.G.; Fakhran, S.; Balk, R.; Bramley, A.M.; Reed, C.; Grijalva, C.G.; Anderson, E.J.; Courtney, D.M.; et al. Community-acquired pneumonia requiring hospitalization among US. Adults. N. Engl. J. Med. 2015, 373, 415–427. [Google Scholar] [CrossRef]
- Lee, J.H.; Jang, J.H.; Lee, S.H.; Kim, Y.J.; Yoo, K.H.; Sung, K.W.; Lee, N.Y.; Ki, C.S.; Koo, H.H. Respiratory viral infections during the first 28 days after transplantation in pediatric hematopoietic stem cell transplant recipients. Clin. Transplant. 2012, 26, 736–740. [Google Scholar] [CrossRef]
- Bennett, N.J.; Tabarani, C.M.; Bartholoma, N.M.; Wang, D.; Huang, D.; Riddell, S.W.; Kiska, D.L.; Hingre, R.; Rosenberg, H.F.; Domachowske, J.B. Unrecognized viral respiratory tract infections in premature infants during their birth hospitalization: A prospective surveillance study in two neonatal intensive care units. J. Pediatr. 2012, 161, 814–818. [Google Scholar] [CrossRef]
- Minejima, E.; Wong-Beringer, A. Implementation of rapid diagnostics with antimicrobial stewardship. Expert Rev. Anti-Infect. Ther. 2016, 14, 1065–1075. [Google Scholar] [CrossRef]
- Krause, J.C.; Panning, M.; Hengel, H.; Henneke, P. The role of multiplex pcr in respiratory tract infections in children. Dtsch. Arztebl. Int. 2014, 111, 639–645. [Google Scholar] [CrossRef]
- Charles, P.G. Early diagnosis of lower respiratory tract infections (point-of-care tests). Curr. Opin. Pulm. Med. 2008, 14, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Ferronato, A.E.; Gilio, A.E.; Ferraro, A.A.; Paulis, M.; Vieira, S.E. Etiological diagnosis reduces the use of antibiotics in infants with bronchiolitis. Clinics Sao Paulo Brazil 2012, 67, 1001–1006. [Google Scholar] [CrossRef]
- Garcia-Garcia, M.L.; Calvo, C.; Pozo, F.; Villadangos, P.A.; Perez-Brena, P.; Casas, I. Spectrum of respiratory viruses in children with community-acquired pneumonia. Pediatric Infect. Dis. J. 2012, 31, 808–813. [Google Scholar] [CrossRef] [PubMed]
- Tanday, S. Resisting the use of antibiotics for viral infections. Lancet Respir. Med. 2016, 4, 179. [Google Scholar] [CrossRef]
- Huijskens, E.G.; Biesmans, R.C.; Buiting, A.G.; Obihara, C.C.; Rossen, J.W. Diagnostic value of respiratory virus detection in symptomatic children using real-time pcr. Virol. J. 2012, 9, 276. [Google Scholar] [CrossRef]
- Endimiani, A.; Hujer, K.M.; Hujer, A.M.; Kurz, S.; Jacobs, M.R.; Perlin, D.S.; Bonomo, R.A. Are we ready for novel detection methods to treat respiratory pathogens in hospital-acquired pneumonia? Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2011, 52, S373–S383. [Google Scholar] [CrossRef]
- Huang, H.S.; Tsai, C.L.; Chang, J.; Hsu, T.C.; Lin, S.; Lee, C.C. Multiplex pcr system for the rapid diagnosis of respiratory virus infection: Systematic review and meta-analysis. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2018, 24, 1055–1063. [Google Scholar] [CrossRef]
- Chen, H.; Weng, H.; Lin, M.; He, P.; Li, Y.; Xie, Q.; Ke, C.; Jiao, X. The clinical significance of filmarray respiratory panel in diagnosing community-acquired pneumonia. BioMed Res. Int. 2017, 2017, 7320859. [Google Scholar] [CrossRef]
- Kuypers, J.; Wright, N.; Ferrenberg, J.; Huang, M.-L.; Cent, A.; Corey, L.; Morrow, R. Comparison of real-time pcr assays with fluorescent-antibody assays for diagnosis of respiratory virus infections in children. J. Clin. Microbiol. 2006, 44, 2382–2388. [Google Scholar] [CrossRef]
- Templeton, K.E.; Scheltinga, S.A.; Beersma, M.F.; Kroes, A.C.; Claas, E.C. Rapid and sensitive method using multiplex real-time pcr for diagnosis of infections by influenza a and influenza b viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J. Clin. Microbiol. 2004, 42, 1564–1569. [Google Scholar] [CrossRef]
- Huang, A.S.; Chen, W.C.; Huang, W.T.; Huang, S.T.; Lo, Y.C.; Wei, S.H.; Kuo, H.W.; Chan, P.C.; Hung, M.N.; Liu, Y.L.; et al. Public health responses to reemergence of animal rabies, Taiwan, 16 July–28 December, 2013. PLoS ONE 2015, 10, e0132160. [Google Scholar] [CrossRef] [PubMed]
- Bonzel, L.; Tenenbaum, T.; Schroten, H.; Schildgen, O.; Schweitzer-Krantz, S.; Adams, O. Frequent detection of viral coinfection in children hospitalized with acute respiratory tract infection using a real-time polymerase chain reaction. Pediatr. Infect. Dis. J. 2008, 27, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Chidlow, G.R.; Harnett, G.B.; Shellam, G.R.; Smith, D.W. An economical tandem multiplex real-time pcr technique for the detection of a comprehensive range of respiratory pathogens. Viruses 2009, 1, 42–56. [Google Scholar] [CrossRef]
- Maertzdorf, J.; Wang, C.K.; Brown, J.B.; Quinto, J.D.; Chu, M.; de Graaf, M.; van den Hoogen, B.G.; Spaete, R.; Osterhaus, A.D.; Fouchier, R.A. Real-time reverse transcriptase pcr assay for detection of human metapneumoviruses from all known genetic lineages. J. Clin. Microbiol. 2004, 42, 981–986. [Google Scholar] [CrossRef]
- Ward, C.L.; Dempsey, M.H.; Ring, C.J.; Kempson, R.E.; Zhang, L.; Gor, D.; Snowden, B.W.; Tisdale, M. Design and performance testing of quantitative real time pcr assays for influenza a and b viral load measurement. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2004, 29, 179–188. [Google Scholar] [CrossRef]
- Chen, Y.; Cui, D.; Zheng, S.; Yang, S.; Tong, J.; Yang, D.; Fan, J.; Zhang, J.; Lou, B.; Li, X.; et al. Simultaneous detection of influenza a, influenza b, and respiratory syncytial viruses and subtyping of influenza a h3n2 virus and h1n1 (2009) virus by multiplex real-time pcr. J. Clin. Microbiol. 2011, 49, 1653–1656. [Google Scholar] [CrossRef]
- Mills, J.M.; Harper, J.; Broomfield, D.; Templeton, K.E. Rapid testing for respiratory syncytial virus in a paediatric emergency department: Benefits for infection control and bed management. J. Hosp. Infect. 2011, 77, 248–251. [Google Scholar] [CrossRef]
- Sim, S.A.; Leung, V.K.Y.; Ritchie, D.; Slavin, M.A.; Sullivan, S.G.; Teh, B.W. Viral respiratory tract infections in allogeneic hematopoietic stem cell transplantation recipients in the era of molecular testing. Biol. Blood Marrow Transplant. J. Am. Soc. Blood Marrow Transplant. 2018, 24, 1490–1496. [Google Scholar] [CrossRef]
- Shiley, K.T.; Lautenbach, E.; Lee, I. The use of antimicrobial agents after diagnosis of viral respiratory tract infections in hospitalized adults: Antibiotics or anxiolytics? Infect. Control. Hosp. Epidemiol. Off. J. Soc. Hosp. Epidemiol. Am. 2010, 31, 1177–1183. [Google Scholar] [CrossRef]
- McCaig, L.F.; Besser, R.E.; Hughes, J.M. Antimicrobial drug prescription in ambulatory care settings, united states, 1992–2000. Emerg. Infect. Dis. 2003, 9, 432–437. [Google Scholar] [CrossRef]
- Spurling, G.K.; Fonseka, K.; Doust, J.; Del Mar, C. Antibiotics for bronchiolitis in children. Cochrane Database Syst. Rev. 2007, Cd005189. [Google Scholar] [CrossRef]
- Breakell, R.; Thorndyke, B.; Clennett, J.; Harkensee, C. Reducing unnecessary chest x-rays, antibiotics and bronchodilators through implementation of the nice bronchiolitis guideline. Eur. J. Pediatr. 2018, 177, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Sung, C.C.; Chi, H.; Chiu, N.C.; Huang, D.T.; Weng, L.C.; Wang, N.Y.; Huang, F.Y. Viral etiology of acute lower respiratory tract infections in hospitalized young children in northern Taiwan. J. Microbiol. Immunol. Infect. (Wei Mian Yu Gan Ran Za Zhi) 2011, 44, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Bharaj, P.; Sullender, W.M.; Kabra, S.K.; Mani, K.; Cherian, J.; Tyagi, V.; Chahar, H.S.; Kaushik, S.; Dar, L.; Broor, S. Respiratory viral infections detected by multiplex pcr among pediatric patients with lower respiratory tract infections seen at an urban hospital in delhi from 2005 to 2007. Virol. J. 2009, 6, 89. [Google Scholar] [CrossRef]
- Jiang, W.; Wu, M.; Zhou, J.; Wang, Y.; Hao, C.; Ji, W.; Zhang, X.; Gu, W.; Shao, X. Etiologic spectrum and occurrence of coinfections in children hospitalized with community-acquired pneumonia. BMC Infect. Dis. 2017, 17, 787. [Google Scholar] [CrossRef]
- Kim, H.J.; Choi, S.M.; Lee, J.; Park, Y.S.; Lee, C.H.; Yim, J.J.; Yoo, C.G.; Kim, Y.W.; Han, S.K.; Lee, S.M. Respiratory virus of severe pneumonia in south korea: Prevalence and clinical implications. PLoS ONE 2018, 13, e0198902. [Google Scholar] [CrossRef]
- Ortiz-Hernandez, A.A.; Nishimura, K.K.; Noyola, D.E.; Moreno-Espinosa, S.; Gamino, A.; Galindo-Fraga, A.; Valdez Vazquez, R.; Magana Aquino, M.; Ramirez-Venegas, A.; Valdes Salgado, R.; et al. Differential risk of hospitalization among single virus infections causing influenza like illnesses. Influenza Respir. Viruses 2018, 13, 36–43. [Google Scholar] [CrossRef]
- Chi, H.; Chang, I.S.; Tsai, F.Y.; Huang, L.M.; Shao, P.L.; Chiu, N.C.; Chang, L.Y.; Huang, F.Y. Epidemiological study of hospitalization associated with respiratory syncytial virus infection in Taiwanese children between 2004 and 2007. J. Formos. Med Assoc. (Taiwan Yi Zhi) 2011, 110, 388–396. [Google Scholar] [CrossRef]
- Hsu, C.H.; Lin, C.Y.; Chi, H.; Chang, J.H.; Hung, H.Y.; Kao, H.A.; Peng, C.C.; Jim, W.T. Prolonged seasonality of respiratory syncytial virus infection among preterm infants in a subtropical climate. PLoS ONE 2014, 9, e110166. [Google Scholar] [CrossRef]
- Scheltema, N.M.; Gentile, A.; Lucion, F.; Nokes, D.J.; Munywoki, P.K.; Madhi, S.A.; Groome, M.J.; Cohen, C.; Moyes, J.; Thorburn, K.; et al. Global respiratory syncytial virus-associated mortality in young children (rsv gold): A retrospective case series. Lancet Global Health 2017, 5, e984–e991. [Google Scholar] [CrossRef]
- Michelow, I.C.; Olsen, K.; Lozano, J.; Rollins, N.K.; Duffy, L.B.; Ziegler, T.; Kauppila, J.; Leinonen, M.; McCracken, G.H., Jr. Epidemiology and clinical characteristics of community-acquired pneumonia in hospitalized children. Pediatrics 2004, 113, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Nolan, V.G.; Arnold, S.R.; Bramley, A.M.; Ampofo, K.; Williams, D.J.; Grijalva, C.G.; Self, W.H.; Anderson, E.J.; Wunderink, R.G.; Edwards, K.M.; et al. Etiology and impact of coinfections in children hospitalized with community-acquired pneumonia. J. Infect. Dis. 2018, 218, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Debiaggi, M.; Canducci, F.; Ceresola, E.R.; Clementi, M. The role of infections and coinfections with newly identified and emerging respiratory viruses in children. Virol. J. 2012, 9, 247. [Google Scholar] [CrossRef]
- Stefanska, I.; Romanowska, M.; Donevski, S.; Gawryluk, D.; Brydak, L.B. Co-infections with influenza and other respiratory viruses. Adv. Exp. Med. Biol. 2013, 756, 291–301. [Google Scholar]
- Yoshida, L.M.; Suzuki, M.; Nguyen, H.A.; Le, M.N.; Dinh Vu, T.; Yoshino, H.; Schmidt, W.P.; Nguyen, T.T.; Le, H.T.; Morimoto, K.; et al. Respiratory syncytial virus: Co-infection and paediatric lower respiratory tract infections. Eur. Respir. J. 2013, 42, 461–469. [Google Scholar] [CrossRef]
- Semple, M.G.; Cowell, A.; Dove, W.; Greensill, J.; McNamara, P.S.; Halfhide, C.; Shears, P.; Smyth, R.L.; Hart, C.A. Dual infection of infants by human metapneumovirus and human respiratory syncytial virus is strongly associated with severe bronchiolitis. J. Infect. Dis. 2005, 191, 382–386. [Google Scholar] [CrossRef]
- Xepapadaki, P.; Psarras, S.; Bossios, A.; Tsolia, M.; Gourgiotis, D.; Liapi-Adamidou, G.; Constantopoulos, A.G.; Kafetzis, D.; Papadopoulos, N.G. Human metapneumovirus as a causative agent of acute bronchiolitis in infants. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2004, 30, 267–270. [Google Scholar] [CrossRef]
- Lau, S.K.; Yip, C.C.; Tsoi, H.W.; Lee, R.A.; So, L.Y.; Lau, Y.L.; Chan, K.H.; Woo, P.C.; Yuen, K.Y. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected hrv species, hrv-c, associated with acute respiratory illness in children. J. Clin. Microbiol. 2007, 45, 3655–3664. [Google Scholar] [CrossRef]
- Ma, Y.J.; Wang, S.M.; Cho, Y.H.; Shen, C.F.; Liu, C.C.; Chi, H.; Huang, Y.C.; Huang, L.M.; Huang, Y.C.; Lin, H.C.; et al. Clinical and epidemiological characteristics in children with community-acquired mycoplasma pneumonia in Taiwan: A nationwide surveillance. J. Microbiol. Immunol. Infect. (Wei Mian Yu Gan Ran Za Zhi) 2015, 48, 632–638. [Google Scholar] [CrossRef]
- Babakir-Mina, M.; Ciccozzi, M.; Perno, C.F.; Ciotti, M. The human polyomaviruses ki and wu: Virological background and clinical implications. APMIS Acta Pathol. Microbiol. et Immunol. Scand. 2013, 121, 746–754. [Google Scholar] [CrossRef]
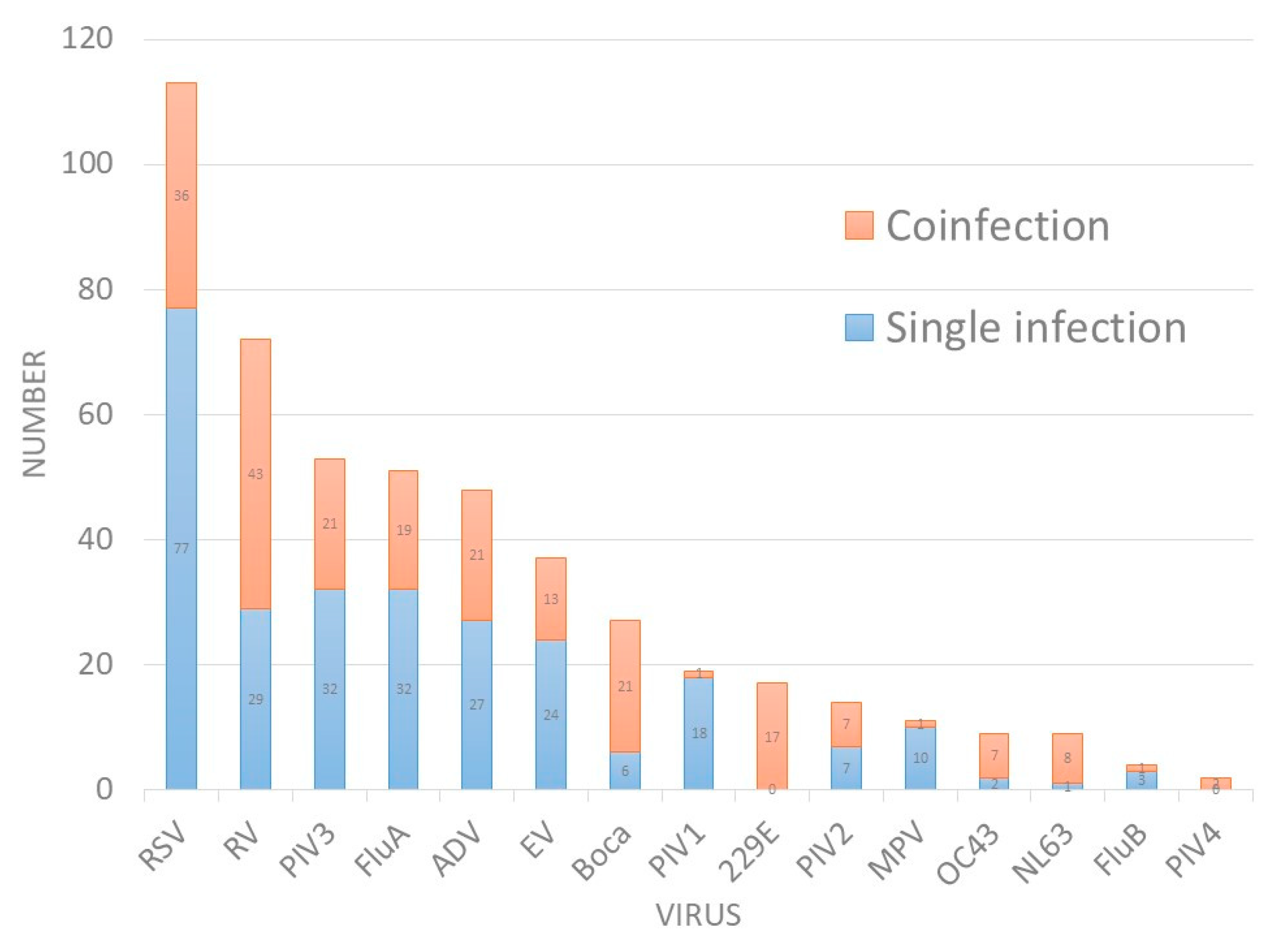
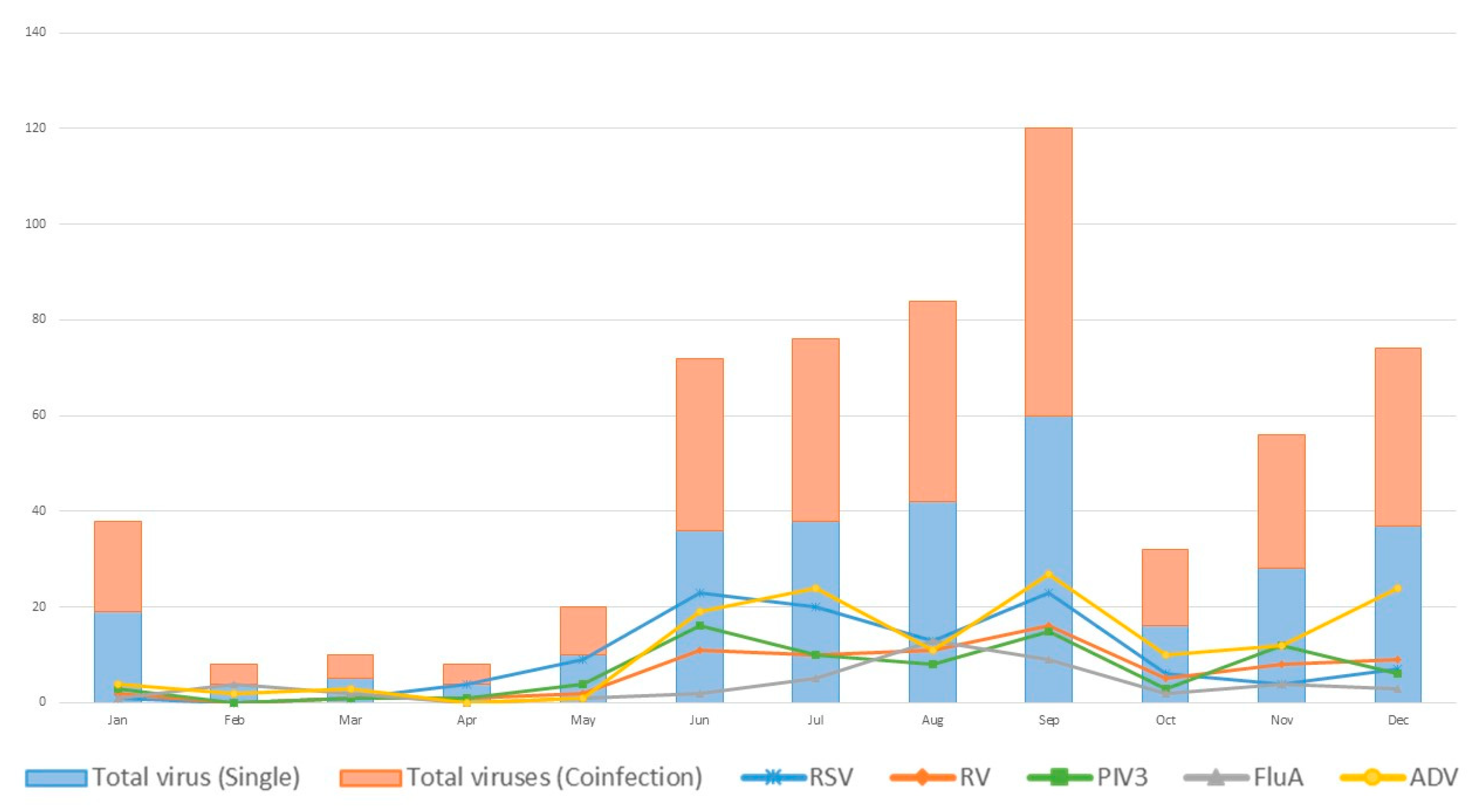
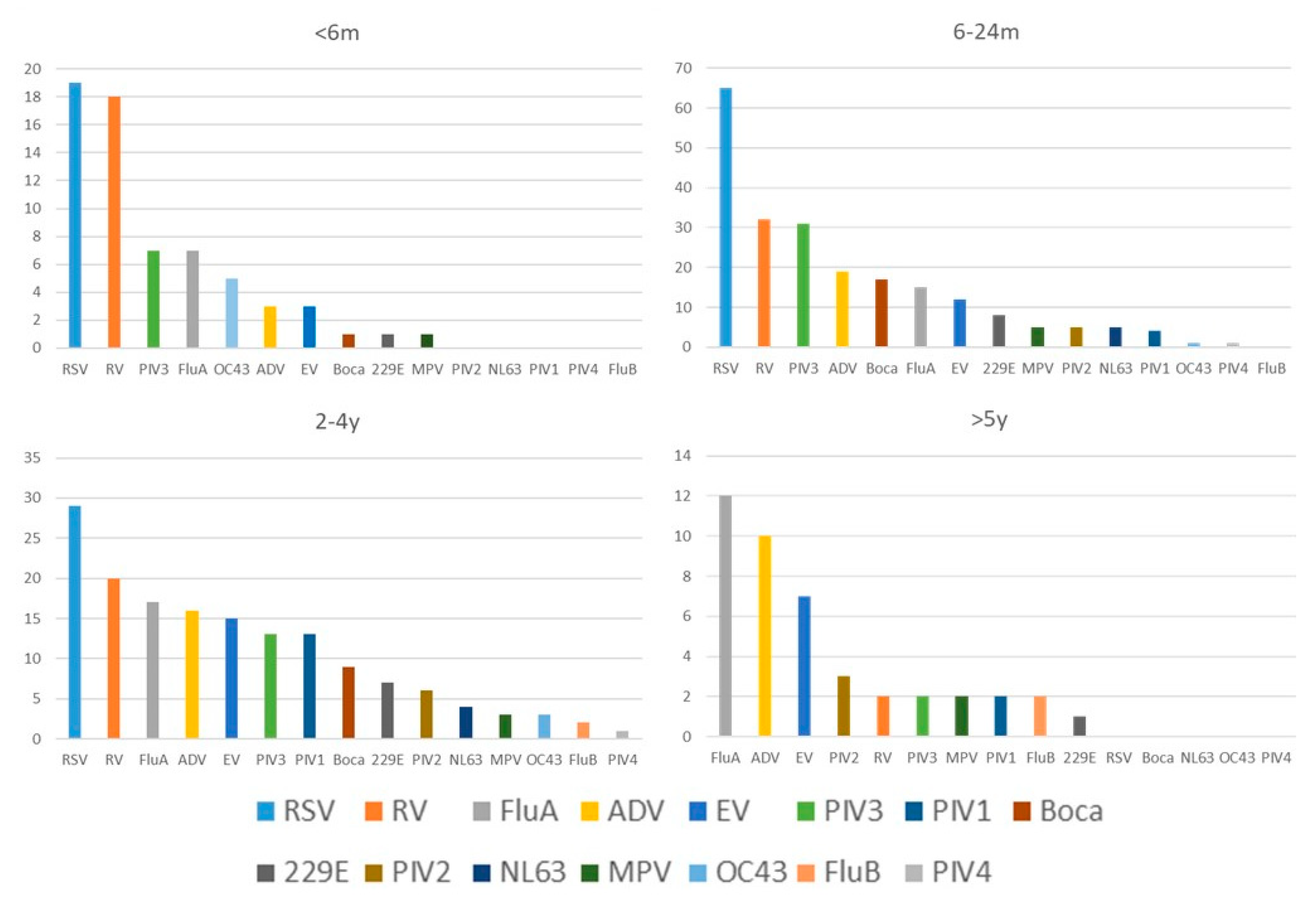
| Viruses | PCR | Traditional Tests | Coefficient k | p for k | |||
|---|---|---|---|---|---|---|---|
| Positive No (%) | Coinfection | Coinfection Rates | Positive No (%) | Coinfection | |||
| Total | 357 (75.3) | 92 | 25.8% | 229 (48.3) | 30 | ||
| RSV | 113 (23.8) | 36 | 31.9% | 70 (9.6) | 4 | 0.72 | <0.01 |
| RV | 72 (15.2) | 43 | 59.7% | - | - | ||
| PIV3 | 53 (11.2) | 21 | 39.6% | 21 (6.9) | 3 | 0.77 | <0.01 |
| FluA | 51 (10.8) | 19 | 37.3% | 47 (18.1) | 5 | 0.92 | <0.01 |
| ADV | 48 (10.1) | 21 | 43.8% | 30 (11.5) | 9 | 0.8 | <0.01 |
| EV | 37 (7.8) | 13 | 35.1% | 29 (11.2) | 7 | 0.87 | <0.01 |
| Boca | 27 (5.7) | 21 | 77.8% | - | - | ||
| PIV1 | 19 (4) | 1 | 5.3% | 13 (5.0) | 3 | 0.925 | <0.01 |
| 229E | 17 (3.6) | 17 | 100.0% | - | - | ||
| PIV2 | 14 (3.0) | 7 | 50.0% | 13 (5.0) | 3 | 0.961 | <0.01 |
| MPV | 11 (2.3) | 1 | 9.1% | - | - | ||
| OC43 | 9 (1.9) | 7 | 77.8% | - | - | ||
| NL63 | 9 (1.9) | 8 | 88.9% | - | - | ||
| FluB | 4 (0.8) | 1 | 25.0% | 12 (4.6) | 5 | 0.49 | <0.01 |
| PIV4 | 2 (0.4) | 2 | 100.0% | - | - | ||
| Viruses | Age (m/o) | BW (kg) | Hospital Days (day) | ICU | Hb | Hct | Plt | WBC | Neut (%) | CRP (mg/dL) |
|---|---|---|---|---|---|---|---|---|---|---|
| RSV | 16.63 | 9.72 | 5.85 | 5 | 11.85 | 35.57 | 336,439 | 10,812 | 43 | 1.47 |
| RV | 19.25 | 10.21 | 5.93 | 4 | 11.96 | 35.88 | 350,282 | 12,949 | 50 | 1.66 |
| PIV3 | 21.4 | 10.75 | 6.96 | 0 | 11.83 | 35.5 | 311,000 | 10,492 | 46.8 | 2.1 |
| FluA | 38.98 | 14.27 | 4.88 | 0 | 11.84 | 35.25 | 261,645 | 9156 | 54.78 | 2.59 |
| ADV | 39 | 15.02 | 6.46 | 2 | 11.68 | 35.1 | 298,575 | 13,415 | 57.84 | 4.65 |
| EV | 35.16 | 14.33 | 4.22 | 0 | 11.93 | 34.63 | 260,028 | 11,689 | 61.6 | 3.19 |
| Boca | 19.41 | 10 | 5.22 | 1 | 12.27 | 36.91 | 309,154 | 10,198 | 47.2 | 2.39 |
| PIV1 | 34.42 | 13.94 | 6.74 | 0 | 12.17 | 36.44 | 233,737 | 7900 | 54.17 | 1.37 |
| 229E | 27.53 | 12.1 | 4.41 | 1 | 12.2 | 36.03 | 323,765 | 13,912 | 54.6 | 2.29 |
| PIV2 | 48.7 | 17.08 | 5.14 | 0 | 11.8 | 35.39 | 243,500 | 9350 | 57 | 2.07 |
| MPV | 26.55 | 12.05 | 6.55 | 0 | 12.03 | 35.75 | 255,455 | 7427 | 46.1 | 2.03 |
| OC43 | 13.78 | 9.96 | 9.56 | 1 | 11.56 | 34.18 | 321,889 | 10,533 | 41.9 | 1.19 |
| NL63 | 21.56 | 11.72 | 3.89 | 0 | 12 | 36 | 319,333 | 14,800 | 65.78 | 2.81 |
| FluB | 86 | 27.2 | 4.5 | 0 | 12.83 | 37.98 | 233,250 | 7800 | 74 | 1.985 |
| PIV4 | 21.5 | 9.7 | 8 | 0 | 12.35 | 38.05 | 21900 | 8000 | 49.5 | 1.13 |
| Variables | Coinfection (N = 92) | Single Pathogen (N = 265) | Negative (N = 117) | p1 | p2 |
|---|---|---|---|---|---|
| Age (m/o) | 29.03 ± 34.2 | 27.06 ± 27.8 | 29.22 ± 35.19 | 0.582 | 0.617 |
| BW (kg) | 13.0 ± 10.3 | 12.2 ± 6.7 | 12.4 ± 8.8 | 0.419 | 0.971 |
| Hospital days (day) | 5.6 ± 4.06 | 5.7 ± 4.99 | 7.7 ± 11.56 | 0.852 | 0.063 |
| ICU | 2 | 10 | 5 | 0.924 | - |
| ICU days | 8.7 ± 14.2 | 3.5 ± 5 | 0 | 0.498 | - |
| Hb | 11.73 ± 1.82 | 11.78 ± 1.82 | 11.53 ± 3.27 | 0.841 | 0.454 |
| Hct | 34.75 ± 6.4 | 34.63 ± 7.18 | 34.44 ± 9.89 | 0.886 | 0.825 |
| Plt | 308,141 ± 133,332 | 286,935 ± 118,557 | 251,730 ± 117,792 | 0.154 | 0.002 |
| WBC | 10,693 ± 5499 | 10,746 ± 6039 | 10,436 ± 6137 | 0.942 | 0.643 |
| ANC | 5676 ± 4612 | 5802 ± 4803 | 5490 ± 4594 | 0.828 | 0.579 |
| Band (%) | 0.55 | 0.74 | 0.97 | 0.356 | 0.297 |
| Neut (%) | 49.5 | 50.7 | 47.7 | 0.648 | 0.27 |
| Eos (%) | 1.13 | 0.98 | 1.36 | 0.474 | 0.132 |
| Baso (%) | 0.27 | 0.17 | 0.18 | 0.08 | 0.693 |
| Mono (%) | 10.04 | 9.91 | 10.2 | 0.834 | 0.692 |
| Lym (%) | 36.78 | 34.7 | 34 | 0.394 | 0.56 |
| Atyp Lym (%) | 0.63 | 0.81 | 0.97 | 0.304 | 0.237 |
| CRP (mg/dL) | 2.03 ± 3.3 | 2.02 ± 3.25 | 2.59 ± 4.66 | 0.981 | 0.227 |
| Study | Country | Study Period | Study Population | Patient No. | Diagnosis | Positive Rate for Virus | 1st Detected Virus | 2nd Detected Virus | 3rd Detected Virus | Coinfection Rate |
|---|---|---|---|---|---|---|---|---|---|---|
| Preeti 2009 [34] | India | 2005–2007 | Children | 301 | Lower RTI | 35.2% | RSV 20.3% | PIV3 7.3% | PIV2 5.6% | 18.8% |
| Childlow 2009 [23] | Australia | Jul 2006–Sep 2006 | Children | 121 | RTI | 71% | RSV 33.9% | RV 30.6% | ADV 8.3% | 34% |
| Huijskens 2012 [15] | Netherland | Jan 2010–Dec 2010 | Children | 177 | RTI | 73% | RSV 36.6% | RV 24% | EV 8.5% | 36.2% |
| Garcia-Garcia 2012 [13] | Spain | Sep 2004–Jul 2010 | Children | 884 | CAP | 73.4% | RSV 41.6% | RV 26.2% | Boca/ ADV 17.8% | 30% |
| Jain 2015 [4] | USA | Jan 2010–Jun 2012 | Children | 2012 | CAP | 66% | RSV 28% | RV 27% | MPV 13% | 26% |
| Jain 2015 [6] | USA | Jan 2010–Jun 2012 | Adults | 2320 | CAP | 23% | RV 9% | Flu A or B 6% | MPV 4% | 8.3% |
| Jiang 2017 [35] | China | Jan 2015–Dec 2015 | Children | 846 | CAP | 70.1% | RSV 22.9% | RV 22.1% | Boca 6% | 34.6% |
| Present study | Taiwan | Aug 2012–Jul 2014 | Children | 474 | RTI | 75.3% | RSV23.8% | RV15.2% | PIV3 11.2% | 25.8% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.-Y.; Hwang, D.; Chiu, N.-C.; Weng, L.-C.; Liu, H.-F.; Mu, J.-J.; Liu, C.-P.; Chi, H. Increased Detection of Viruses in Children with Respiratory Tract Infection Using PCR. Int. J. Environ. Res. Public Health 2020, 17, 564. https://doi.org/10.3390/ijerph17020564
Lin C-Y, Hwang D, Chiu N-C, Weng L-C, Liu H-F, Mu J-J, Liu C-P, Chi H. Increased Detection of Viruses in Children with Respiratory Tract Infection Using PCR. International Journal of Environmental Research and Public Health. 2020; 17(2):564. https://doi.org/10.3390/ijerph17020564
Chicago/Turabian StyleLin, Chien-Yu, David Hwang, Nan-Chang Chiu, Li-Chuan Weng, Hsin-Fu Liu, Jung-Jung Mu, Chang-Pan Liu, and Hsin Chi. 2020. "Increased Detection of Viruses in Children with Respiratory Tract Infection Using PCR" International Journal of Environmental Research and Public Health 17, no. 2: 564. https://doi.org/10.3390/ijerph17020564
APA StyleLin, C.-Y., Hwang, D., Chiu, N.-C., Weng, L.-C., Liu, H.-F., Mu, J.-J., Liu, C.-P., & Chi, H. (2020). Increased Detection of Viruses in Children with Respiratory Tract Infection Using PCR. International Journal of Environmental Research and Public Health, 17(2), 564. https://doi.org/10.3390/ijerph17020564






