A Long Contiguous Stretch of Homozygosity Disclosed a Novel STAG3 Biallelic Pathogenic Variant Causing Primary Ovarian Insufficiency: A Case Report and Review of the Literature
Abstract
1. Introduction
2. Materials and Methods
2.1. Case Presentation
2.2. CGH-SNP Microarray Analysis
2.3. Clinical Exome Sequencing(CES)
2.4. Sanger Sequencing
3. Results
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huhtaniemi, I.; Hovatta, O.; La Marca, A.; Livera, G.; Monniaux, D.; Persani, L.; Heddar, A.; Jarzabek, K.; Laisk-Podar, T.; Salumets, A.; et al. Advances in the Molecular Pathophysiology, Genetics, and Treatment of Primary Ovarian Insufficiency. Trends Endocrinol. Metab. 2018, 29, 400–419. [Google Scholar] [CrossRef] [PubMed]
- Nelson, L.M. Clinical practice. Primary Ovarian Insufficiency. N. Engl. J. Med. 2009, 360, 606–614. [Google Scholar] [CrossRef]
- Shelling, A.N. Premature ovarian failure. Reproduction 2010, 140, 633–641. [Google Scholar] [CrossRef]
- Tucker, E.J.; Grover, S.R.; Bachelot, A.; Touraine, P.; Sinclair, A.H. Premature Ovarian Insufficiency: New Perspectives on Genetic Cause and Phenotypic Spectrum. Endocr. Rev. 2016, 37, 609–635. [Google Scholar] [CrossRef]
- Golezar, S.; Tehrani, F.R.; Khazaei, S.; Ebadi, A.; Keshavarz, Z. The global prevalence of primary ovarian insufficiency and early menopause: A meta-analysis. Climacteric 2019, 22, 403–411. [Google Scholar] [CrossRef]
- Caburet, S.; Arboleda, V.; Llano, E.; Overbeek, P.; Barbero, J.L.; Oka, K.; Harrison, W.; Vaiman, D.; Ben-Neriah, Z.; García-Tuñón, I.; et al. Mutant Cohesin in Premature Ovarian Failure. N. Engl. J. Med. 2014, 370, 943–949. [Google Scholar] [CrossRef]
- Rossetti, R.; Ferrari, I.; Bonomi, M.; Persani, L. Genetics of primary ovarian insufficiency. Clin. Genet. 2016, 91, 183–198. [Google Scholar] [CrossRef]
- França, M.M.; Nishi, M.Y.; Funari, M.F.; Lerario, A.M.; Baracat, E.C.; Hayashida, S.A.; Maciel, G.A.; Jorge, A.A.; Mendonca, B.B. Two rare loss-of-function variants in the STAG3 gene leading to primary ovarian insufficiency. Eur. J. Med. Genet. 2018, 62, 186–189. [Google Scholar] [CrossRef] [PubMed]
- Webber, L.; Davies, M.; Anderson, R.; Bartlett, J.; Braat, D.; Cartwright, B.; Cífková, R.; Keizer-Schrama, S.D.M.; Hogervorst, E.; Janse, F.; et al. ESHRE Guideline: Management of women with premature ovarian insufficiency. Hum. Reprod. 2016, 31, 926–937. [Google Scholar] [CrossRef]
- Qin, Y.; Jiao, X.; Simpson, J.L.; Chen, Z.-J. Genetics of primary ovarian insufficiency: New developments and opportunities. Hum. Reprod. Updat. 2015, 21, 787–808. [Google Scholar] [CrossRef]
- Jaillard, S.; Akloul, L.; Beaumont, M.; Hamdi-Roze, H.; Dubourg, C.; Odent, S.; Duros, S.; Dejucq-Rainsford, N.; Belaud-Rotureau, M.A.; Ravel, C. Array-CGH diagnosis in ovarian failure: Identification of new molecular actors for ovarian physiology. J. Ovarian Res. 2016, 9, 63. [Google Scholar] [CrossRef] [PubMed]
- AlAsiri, S.; Basit, S.; Wood-Trageser, M.; Yatsenko, S.A.; Jeffries, E.P.; Surti, U.; Ketterer, D.M.; Afzal, S.; Ramzan, K.; Haque, M.F.-U.; et al. Exome sequencing reveals MCM8 mutation underlies ovarian failure and chromosomal instability. J. Clin. Investig. 2014, 125, 258–262. [Google Scholar] [CrossRef]
- Heddar, A.; Dessen, P.; Flatters, D.; Misrahi, M. Novel STAG3 mutations in a Caucasian family with primary ovarian insufficiency. Mol. Genet. Genom. 2019, 294, 1527–1534. [Google Scholar] [CrossRef]
- Wood-Trageser, M.; Gürbüz, F.; Yatsenko, S.A.; Jeffries, E.P.; Kotan, L.D.; Surti, U.; Ketterer, D.M.; Matic, J.; Chipkin, J.; Jiang, H.; et al. MCM9 Mutations Are Associated with Ovarian Failure, Short Stature, and Chromosomal Instability. Am. J. Hum. Genet. 2014, 95, 754–762. [Google Scholar] [CrossRef]
- Colombo, R.; Pontoglio, A.; Bini, M. A STAG3 missense mutation in two sisters with primary ovarian insufficiency. Eur. J. Obstet. Gynecol. Reprod. Biol. 2017, 216, 269–271. [Google Scholar] [CrossRef]
- He, W.-B.; Banerjee, S.; Meng, L.-L.; Du, J.; Gong, F.; Huang, H.; Zhang, X.-X.; Wang, Y.-Y.; Lu, G.-X.; Lin, G.; et al. Whole-exome sequencing identifies a homozygous donor splice-site mutation inSTAG3that causes primary ovarian insufficiency. Clin. Genet. 2017, 93, 340–344. [Google Scholar] [CrossRef]
- Stabej, P.L.Q.; Williams, H.; James, C.; Tekman, M.; Stanescu, H.C.; Kleta, R.; Ocaka, L.; Lescai, F.; Storr, H.; Bitner-Glindzicz, M.; et al. STAG3 truncating variant as the cause of primary ovarian insufficiency. Eur. J. Hum. Genet. 2015, 24, 135–138. [Google Scholar] [CrossRef]
- Xiao, W.-J.; He, W.-B.; Zhang, Y.-X.; Meng, L.-L.; Lu, G.-X.; Lin, G.; Tan, Y.-Q.; Du, J. In-Frame Variants in STAG3 Gene Cause Premature Ovarian Insufficiency. Front. Genet. 2019, 10, 1016. [Google Scholar] [CrossRef]
- Demain, L.A.; Boetje, E.; Edgerley, J.J.; Miles, E.; Fitzgerald, C.T.; Busby, G.; Beaman, G.M.; O’Sullivan, J.; O’Keefe, R.T.; Newman, W.G. Biallelic loss of function variants in STAG3 result in primary ovarian insufficiency. Reprod. Biomed. Online 2021, 16, S1472–S6483. [Google Scholar] [CrossRef]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Aguilera, M.A.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2018, 35, 1978–1980. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef]
- França, M.M.; Mendonca, B.B. Genetics of Primary Ovarian Insufficiency in the Next-Generation Sequencing Era. J. Endocr. Soc. 2019, 4, bvz037. [Google Scholar] [CrossRef]
- Ledig, S.; Röpke, A.; Wieacker, P. Copy Number Variants in Premature Ovarian Failure and Ovarian Dysgenesis. Sex. Dev. 2010, 4, 225–232. [Google Scholar] [CrossRef]
- D’Amours, G.; Langlois, M.; Mathonnet, G.; Fetni, R.; Nizard, S.; Srour, M.; Tihy, F.; Phillips, M.S.; Michaud, J.L.; Lemyre, E. SNP arrays: Comparing diagnostic yields for four platforms in children with developmental delay. BMC Med. Genom. 2014, 7, 70. [Google Scholar] [CrossRef]
- Hopkins, J.; Hwang, G.; Jacob, J.; Sapp, N.; Bedigian, R.; Oka, K.; Overbeek, P.; Murray, S.; Jordan, P.W. Meiosis-Specific Cohesin Component, Stag3 Is Essential for Maintaining Centromere Chromatid Cohesion, and Required for DNA Repair and Synapsis between Homologous Chromosomes. PLoS Genet. 2014, 10, e1004413. [Google Scholar] [CrossRef]
- Zhang, M.; Dai, X.; Sun, Y.; Lu, Y.; Zhou, C.; Miao, Y.; Wang, Y.; Xiong, B. Stag3 regulates microtubule stability to maintain euploidy during mouse oocyte meiotic maturation. Oncotarget 2016, 8, 1593–1602. [Google Scholar] [CrossRef]
- Winters, T.; McNicoll, F.; Jessberger, R. Meiotic cohesin STAG 3 is required for chromosome axis formation and sister chromatid cohesion. EMBO J. 2014, 33, 1256–1270. [Google Scholar] [CrossRef] [PubMed]
- Riera-Escamilla, A.; Enguita-Marruedo, A.; Moreno-Mendoza, D.; Chianese, C.; Sleddens-Linkels, E.; Contini, E.; Benelli, M.; Natali, A.; Colpi, G.M.; Ruiz-Castañé, E.; et al. Sequencing of a ‘mouse azoospermia’ gene panel in azoospermic men: Identification of RNF212 and STAG3 mutations as novel genetic causes of meiotic arrest. Hum. Reprod. 2019, 34, 978–988. [Google Scholar] [CrossRef]
- Tüttelmann, F.; Ruckert, C.; Röpke, A. Disorders of spermatogenesis: Perspectives for novel genetic diagnostics after 20 years of unchanged routine. J. Med. Genet. 2018, 30, 12–20. [Google Scholar] [CrossRef]
- van der Bijl, N.; Röpke, A.; Biswas, U.; Wöste, M.; Jessberger, R.; Kliesch, S.; Friedrich, C.; Tüttelmann, F. Mutations in the stromal antigen 3 (STAG3) gene cause male infertility due to meiotic arrest. Hum. Reprod. 2019, 34, 2112–2119. [Google Scholar] [CrossRef]
- Vivenza, D.; Godi, M.; Faienza, M.F.; Mellone, S.; Moia, S.; Rapa, A.; Petri, A.; Bellone, S.; Riccomagno, S.; Cavallo, L.; et al. A novel HESX1 splice mutation causes isolated GH deficiency by interfering with mRNA processing. Eur. J. Endocrinol. 2011, 164, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Jiang, Y.; Mao, Q.; Demeler, B.; Tao, Y.J.; Pati, D. Characterization of the Interaction between the Cohesin Subunits Rad21 and SA1/2. PLoS ONE 2013, 8, e69458. [Google Scholar] [CrossRef] [PubMed]
- Gelbaya, T.; Vitthala, S.; Nardo, L.; Seif, M. Optimizing hormone therapy for future reproductive performance in women with premature ovarian failure. Gynecol. Endocrinol. 2010, 27, 1–7. [Google Scholar] [CrossRef]

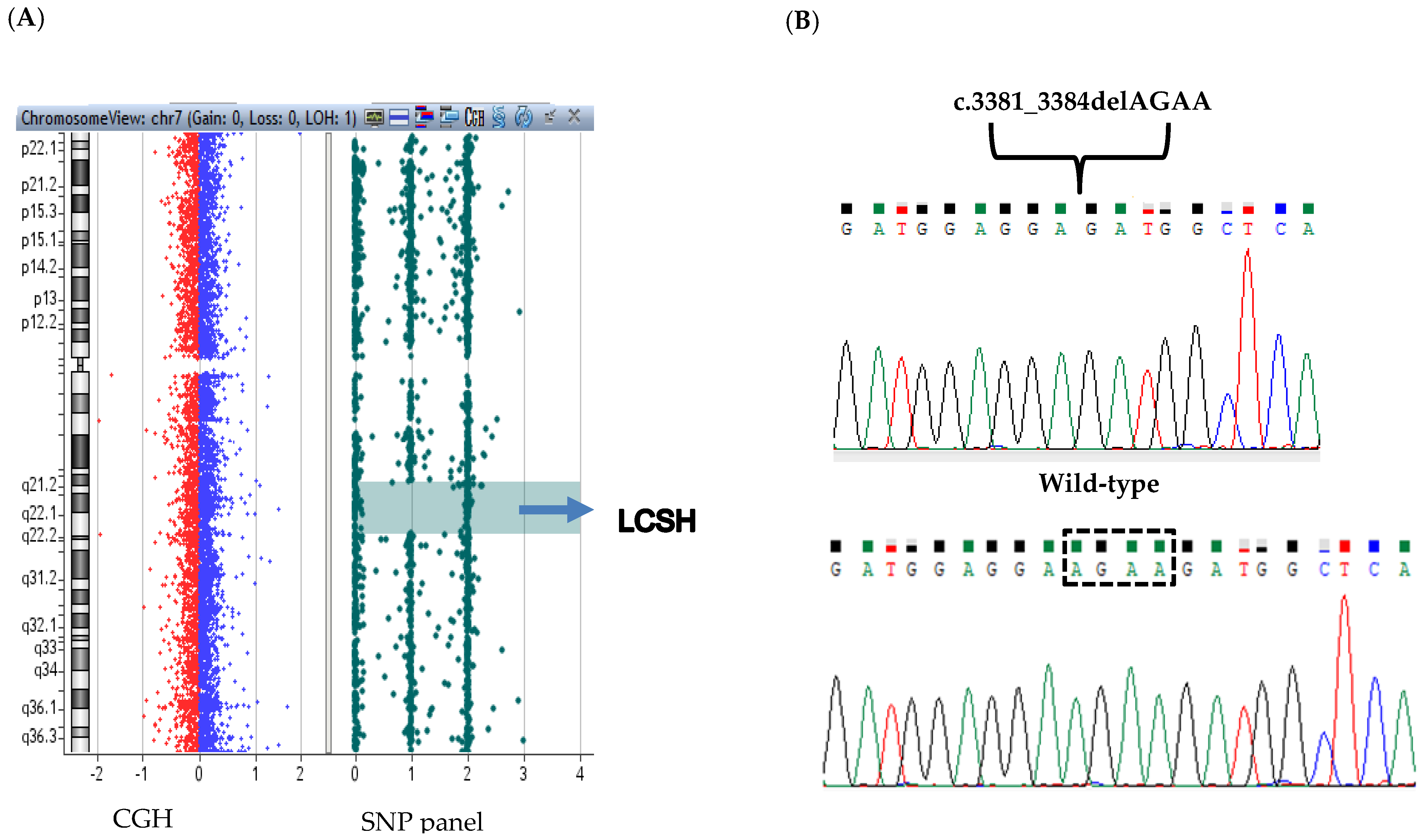
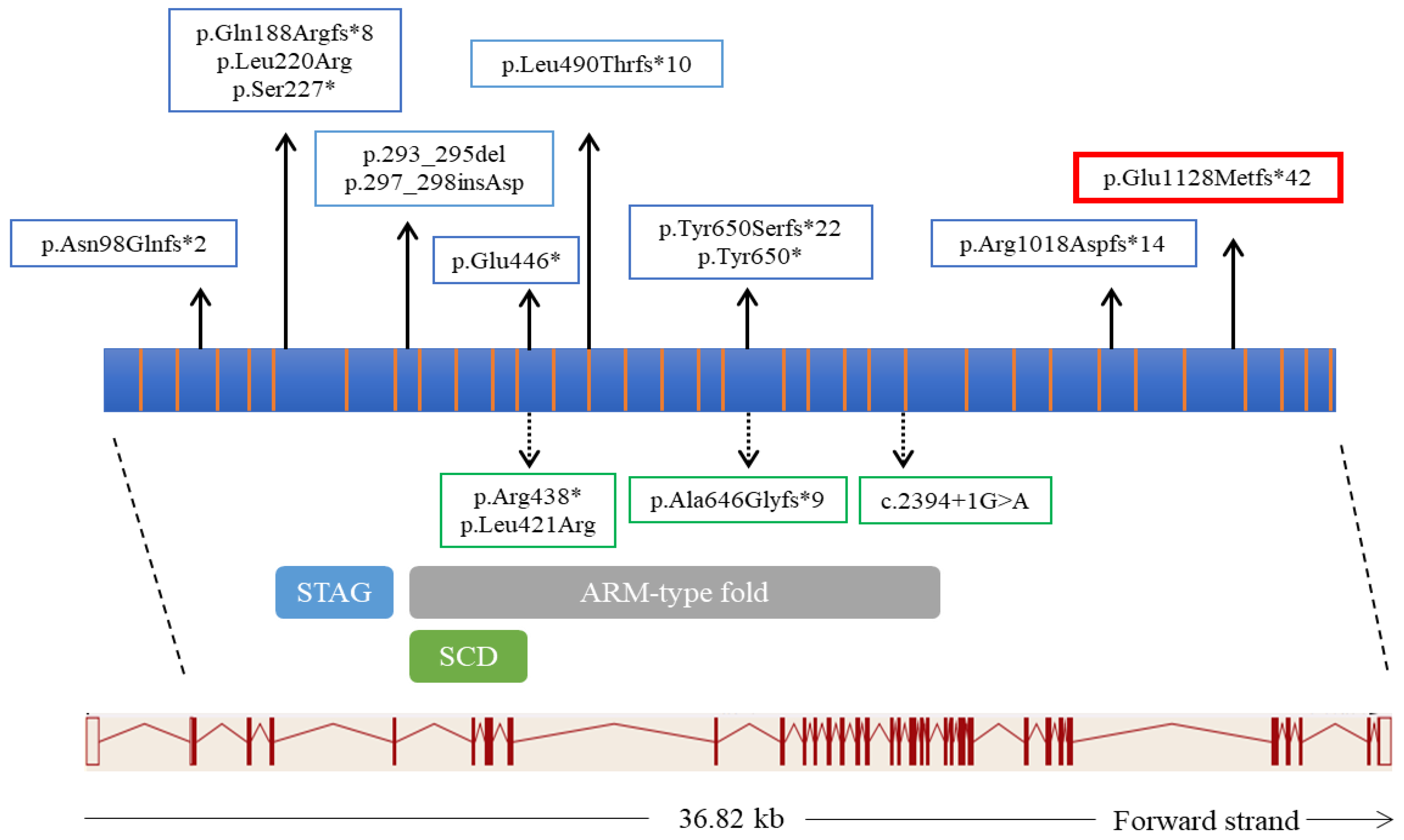
| November 2018 (Staring Therapy) | February 2019 (+3 Months) | May 2019 (+9 Months) | September 2019 (+10 Months) | June 2020 (+18 Months) | November 2020 (+24 Months) | |
|---|---|---|---|---|---|---|
| Tanner stage (BP) | Stage 1 (B1,P1) | Stage 2 (B2,P2) | Stage 3 (B3,P2-3) | Stage 4 (B4,P3) | Stage 4 (B4,P4) | Stage 5 (B5,P4) |
| 17 β-oestradial(pg/mL) | 20 | - | 75 | - | 27 | 40 |
| FSH(mU/mL) | 88.1 | - | 4.7 | - | 55.6 | 51.1 |
| LH(mU/mL) | 28.7 | - | 0.4 | - | 24.2 | 29.9 |
| Menstrual cycle | Amenorrhea | Amenorrhea | Menarche | Oligomenorrhea | Euhenorrhea | Oligomenorrhea |
| Therapy -oestradial hemyhidrate -progesterone | 25 μg/3 days | 50 μg/3 days | 50 μg/3 days | 50 μg/3 days | 50 μg/3 days | 50 μg/3 days 100 mg/14 days |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mellone, S.; Zavattaro, M.; Vurchio, D.; Ronzani, S.; Caputo, M.; Leone, I.; Prodam, F.; Giordano, M. A Long Contiguous Stretch of Homozygosity Disclosed a Novel STAG3 Biallelic Pathogenic Variant Causing Primary Ovarian Insufficiency: A Case Report and Review of the Literature. Genes 2021, 12, 1709. https://doi.org/10.3390/genes12111709
Mellone S, Zavattaro M, Vurchio D, Ronzani S, Caputo M, Leone I, Prodam F, Giordano M. A Long Contiguous Stretch of Homozygosity Disclosed a Novel STAG3 Biallelic Pathogenic Variant Causing Primary Ovarian Insufficiency: A Case Report and Review of the Literature. Genes. 2021; 12(11):1709. https://doi.org/10.3390/genes12111709
Chicago/Turabian StyleMellone, Simona, Marco Zavattaro, Denise Vurchio, Sara Ronzani, Marina Caputo, Ilaria Leone, Flavia Prodam, and Mara Giordano. 2021. "A Long Contiguous Stretch of Homozygosity Disclosed a Novel STAG3 Biallelic Pathogenic Variant Causing Primary Ovarian Insufficiency: A Case Report and Review of the Literature" Genes 12, no. 11: 1709. https://doi.org/10.3390/genes12111709
APA StyleMellone, S., Zavattaro, M., Vurchio, D., Ronzani, S., Caputo, M., Leone, I., Prodam, F., & Giordano, M. (2021). A Long Contiguous Stretch of Homozygosity Disclosed a Novel STAG3 Biallelic Pathogenic Variant Causing Primary Ovarian Insufficiency: A Case Report and Review of the Literature. Genes, 12(11), 1709. https://doi.org/10.3390/genes12111709






