Expression Profile of New Gene Markers and Signaling Pathways Involved in Immunological Processes in Human Cumulus-Oophorus Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cumulus Cell Collection
2.2. Primary Long-Term In Vitro Culture of Cumulus Cells
2.3. Total RNA Extraction
2.4. Microarray Expression Analysis
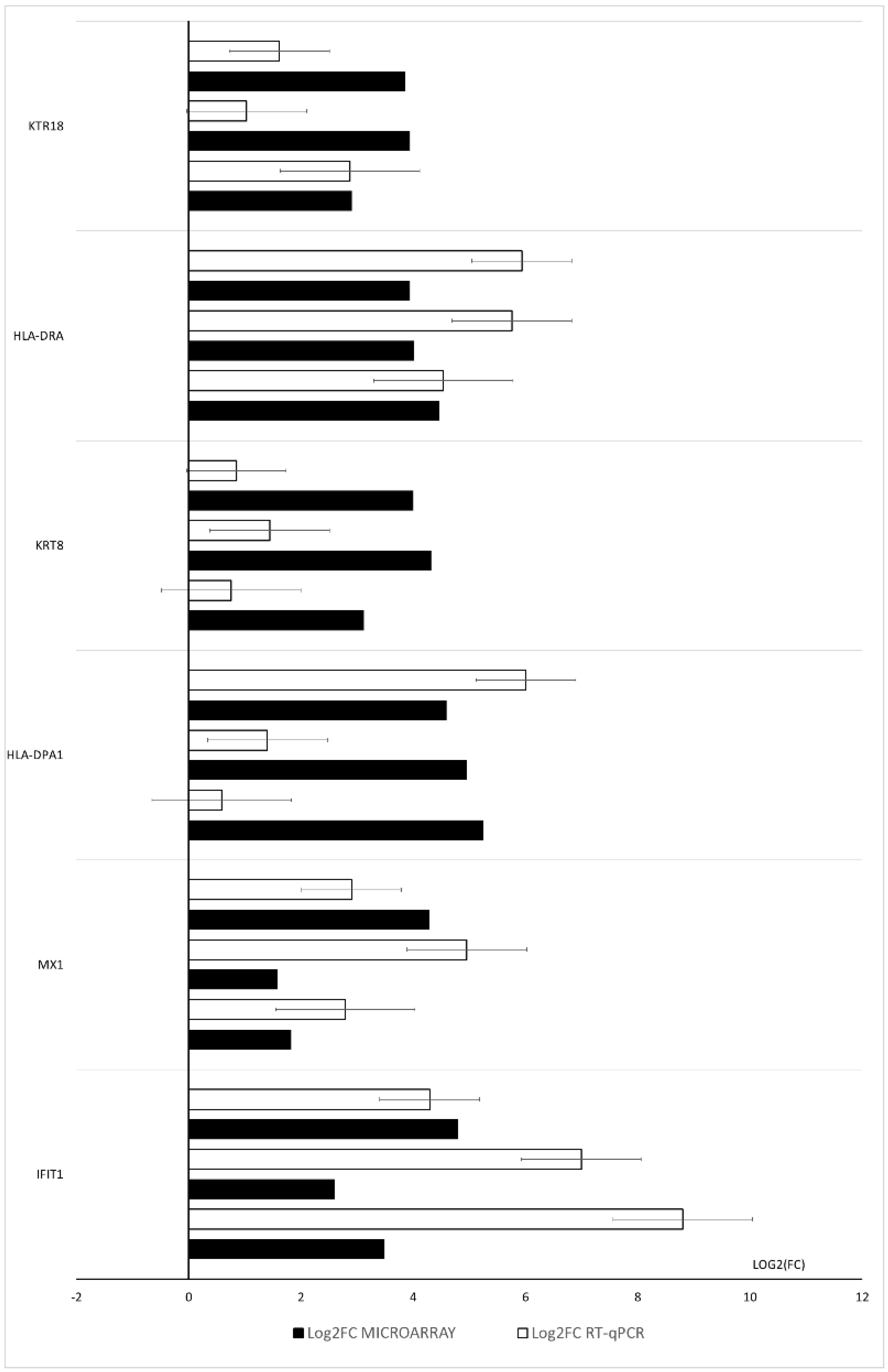
2.5. RT-qPCR Analysis
2.6. Statistical Analysis
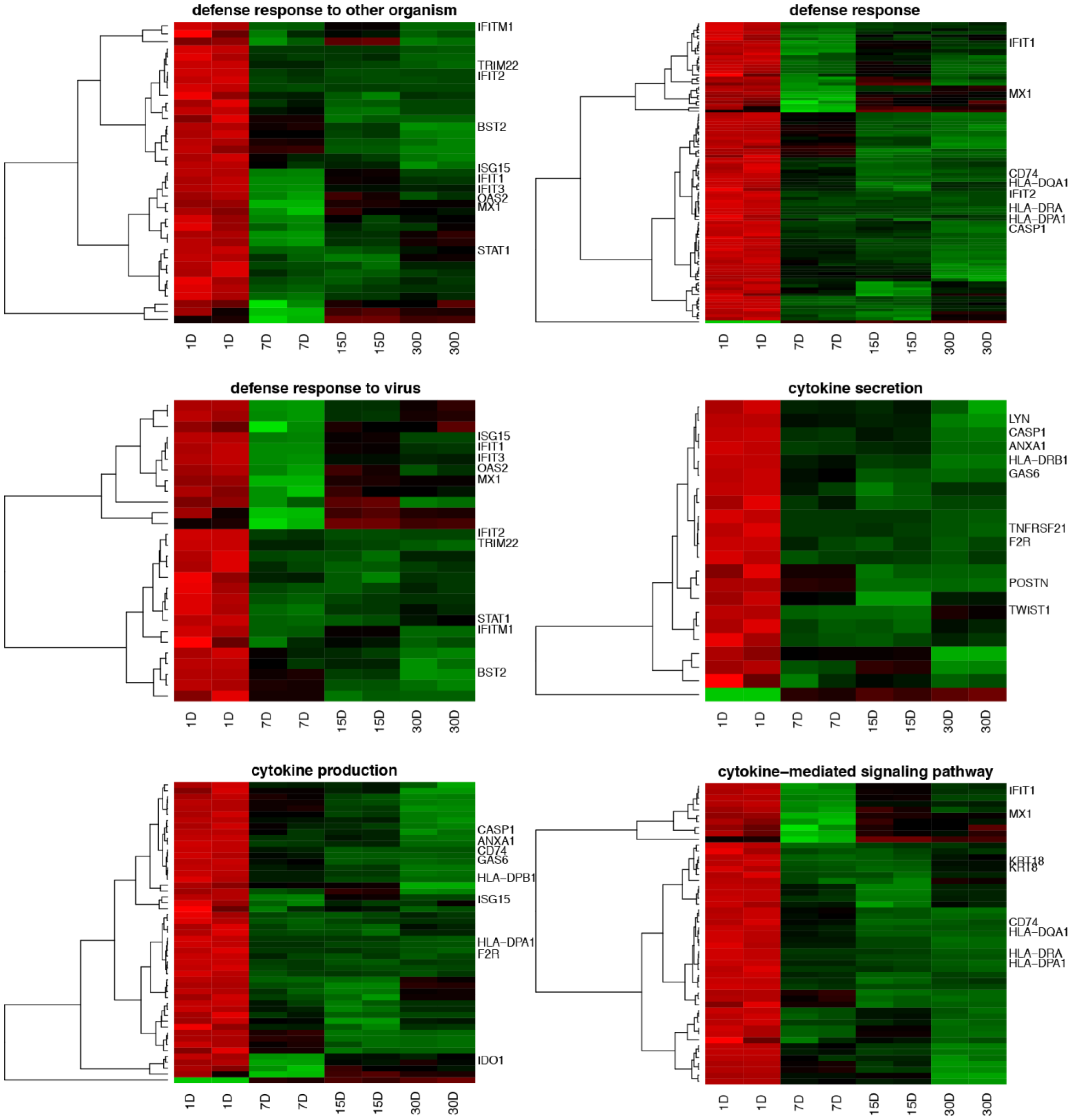
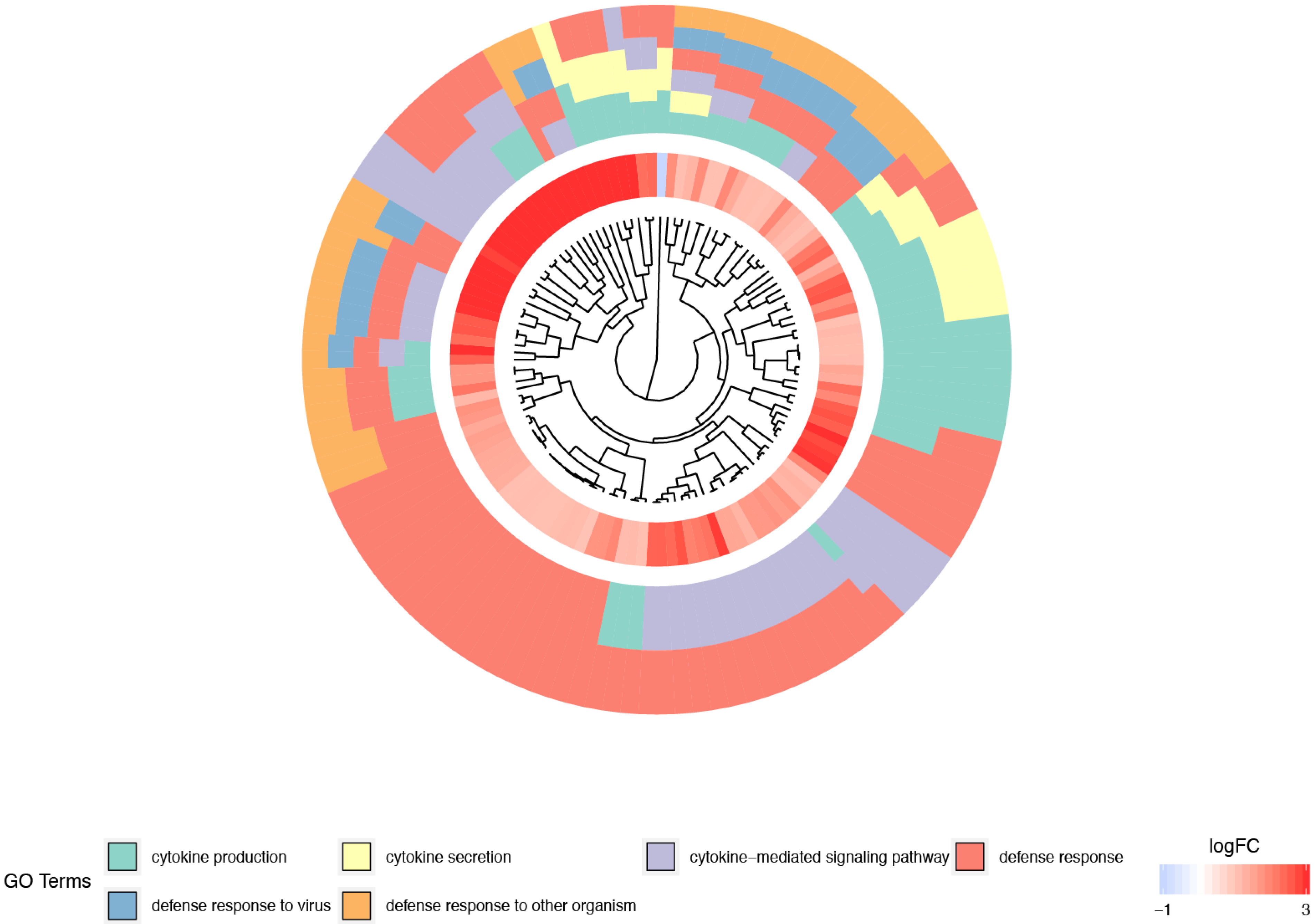
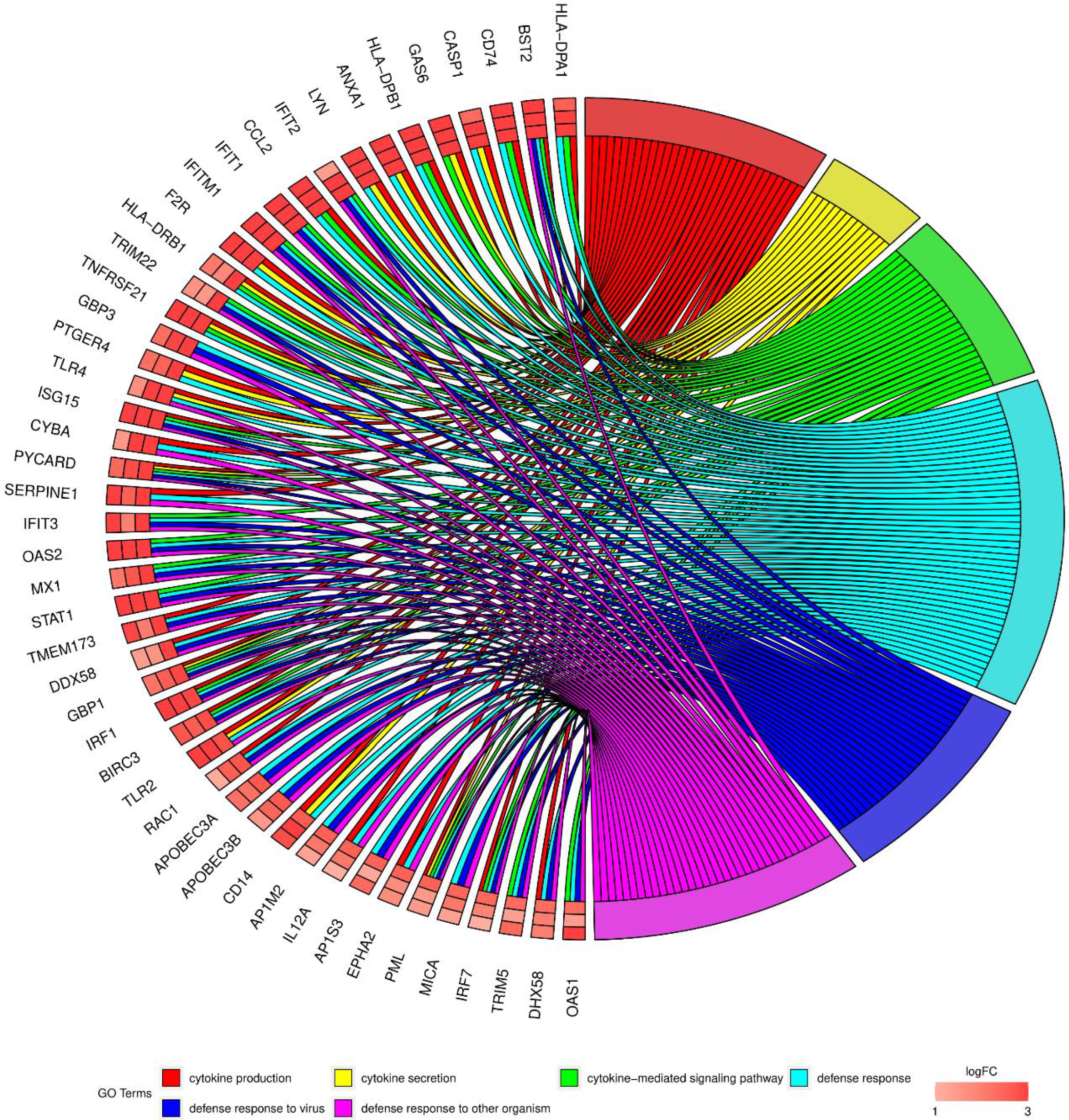
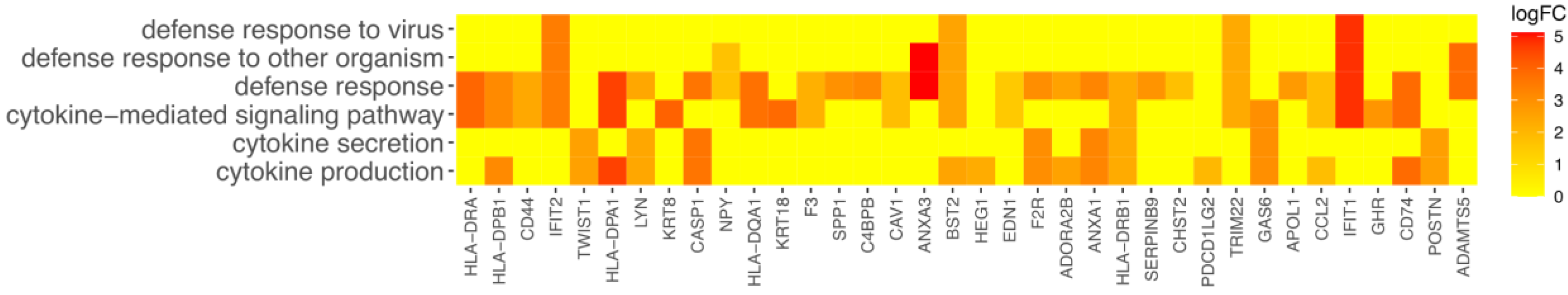
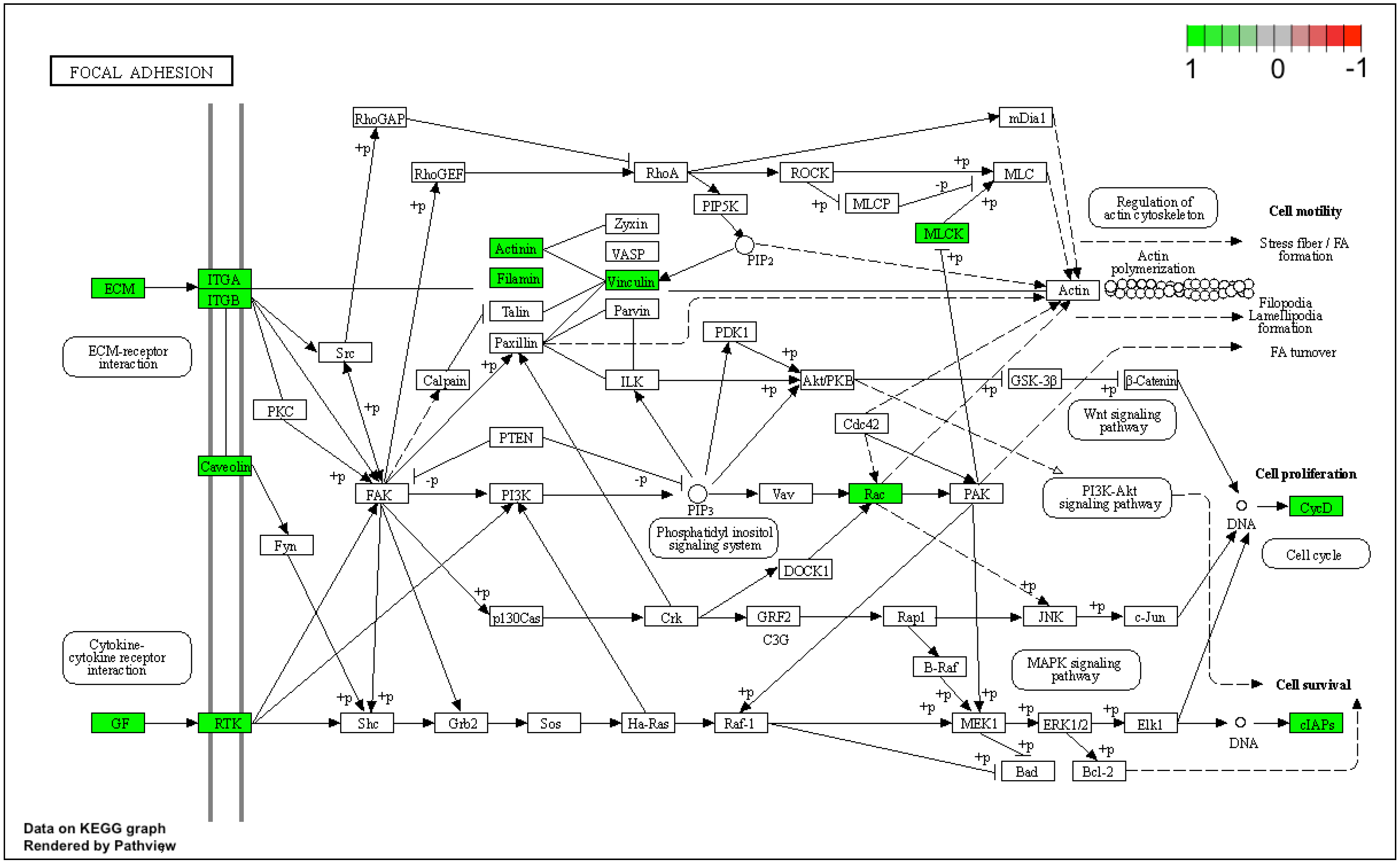
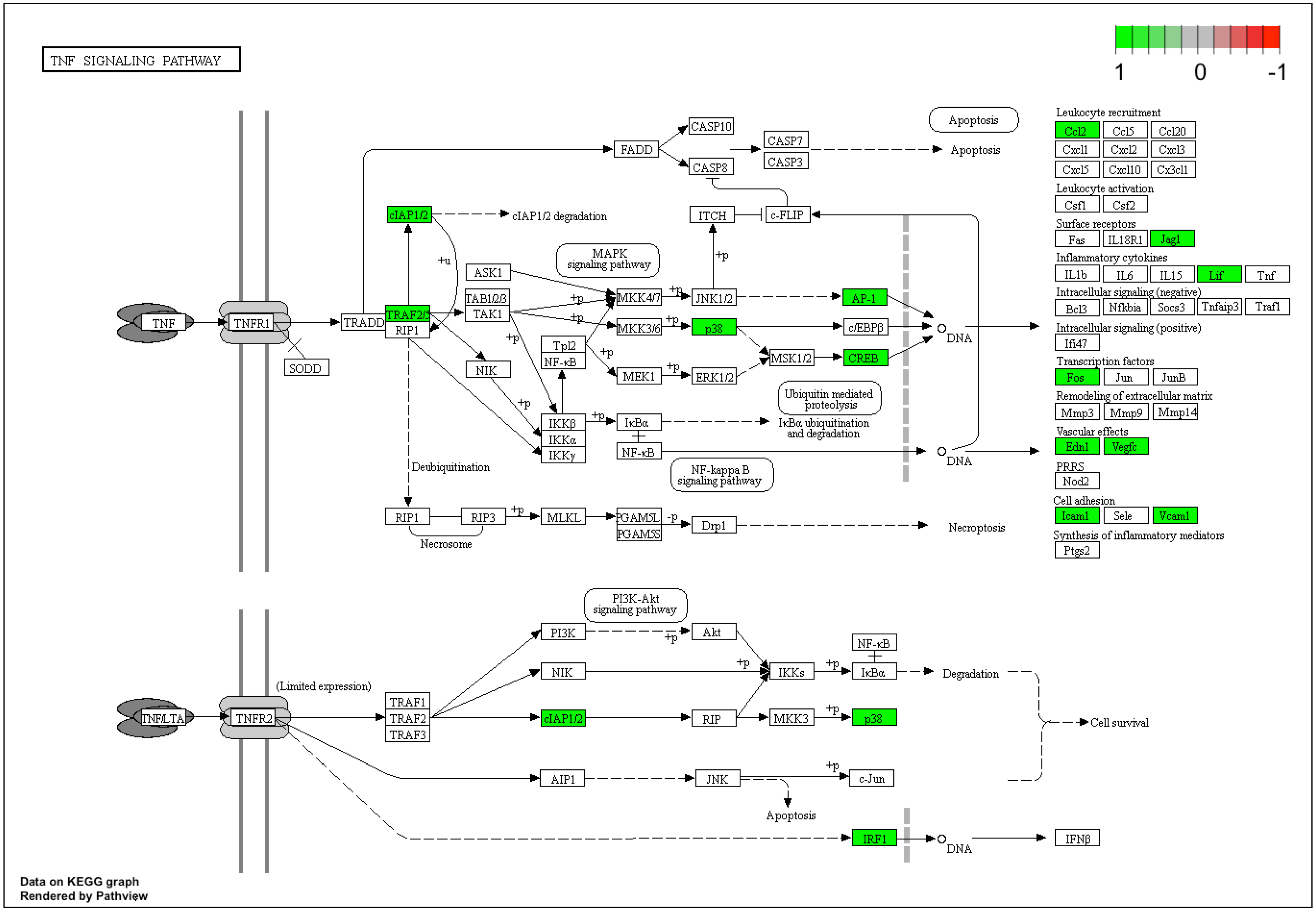
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wigglesworth, K.; Lee, K.-B.; Emori, C.; Sugiura, K.; Eppig, J.J. Transcriptomic diversification of developing cumulus and mural granulosa cells in mouse ovarian follicles. Biol. Reprod. 2015, 92, 23. [Google Scholar] [CrossRef]
- Chermuła, B.; Kranc, W.; Jopek, K.; Budna-Tukan, J.; Hutchings, G.; Dompe, C.; Moncrieff, L.; Janowicz, K.; Józkowiak, M.; Jeseta, M.; et al. Human Cumulus Cells in Long-Term In Vitro Culture Reflect Differential Expression Profile of Genes Responsible for Planned Cell Death and Aging-A Study of New Molecular Markers. Cells 2020, 9, 1265. [Google Scholar] [CrossRef] [PubMed]
- Monniaux, D. Driving folliculogenesis by the oocyte-somatic cell dialog: Lessons from genetic models. Theriogenology 2016, 86, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Bukovsky, A.; Caudle, M.R. Immune physiology of the mammalian ovary—A review. Am. J. Reprod. Immunol. 2008, 59, 12–26. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Li, X.; Zheng, T.; Liang, X.; Li, J.; Huang, J.; Pan, Z.; Zheng, Y. The effect of the immune system on ovarian function and features of ovarian germline stem cells. Springerplus 2016, 5, 1–6. [Google Scholar] [CrossRef] [PubMed]
- La Poulsen, L.C.; Englund, A.L.M.; Wissing, M.L.M.; Yding Andersen, C.; Borup, R.; Grøndahl, M.L. Human granulosa cells function as innate immune cells executing an inflammatory reaction during ovulation: A microarray analysis. Mol. Cell. Endocrinol. 2019, 486, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Plancha, C.E.; Rodrigues, P.; Marques, M.; Almeida, J.M.; Navarro-Costa, P. The time is ripe for oocyte in vitro maturation. J. Assist. Reprod. Genet. 2021, 38, 1281–1283. [Google Scholar] [CrossRef]
- Practice Committees of the American Society for Reproductive Medicine, the Society of Reproductive Biologists and Technologists and the Society for Assisted Reproductive Technology. In vitro maturation: A committee opinion. Fertil. Steril. 2021, 115, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Brazert, M.; Kranc, W.; Jopek, K.; Kempisty, B.; Pawelczyk, L. New markers of human cumulus oophorus cells cultured in vitro-transcriptomic profile. Med. J. Cell Biol. 2020, 8, 60–72. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Bryl, R.; Stefańska, K.; Chermuła, B.; Stelmach, B.; Pieńkowski, W.; Kulus, J.; Perek, J.; Wieczorkiewicz, M.; Wąsiatycz, G.; Ratajczak, K.; et al. Histone demethylases JHDM1D, PHF2 and PHF8 expression pattern in granulosa cells obtained from patients undergoing IVF procedure during short-term IVC. Med. J. Cell Biol. 2021, 9, 1–7. [Google Scholar] [CrossRef]
- Sibiak, R.; Bryl, R.; Stefańska, K.; Chermuła, B.; Pieńkowski, W.; Jeseta, M.; Pawelczyk, L.; Mozdziak, P.; Spaczyński, R.Z.; Kempisty, B. Expression of the apoptosis regulatory gene family in the long-term in vitro cultured human cumulus cells. Med. J. Cell Biol. 2021, 9, 8–13. [Google Scholar] [CrossRef]
- Walter, W.; Sánchez-Cabo, F.; Ricote, M. GOplot: An R package for visually combining expression data with functional analysis: Fig. 1. Bioinformatics 2015, 31, 2912–2914. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinforma. Biomath. 2013, 3, 71–85. [Google Scholar] [PubMed]
- Stefańska, K.; Józkowiak, M.; Antosik, P.; Bukowska, D.; Celichowski, P.; Bruska, M.; Nowicki, M.; Kempisty, B.; Zakova, J.; MacHatkova, M.; et al. Genes regulating programmed cell death are significantly upregulated in porcine immature oocytes. Med. J. Cell Biol. 2019, 7, 1–10. [Google Scholar] [CrossRef]
- Kranc, W.; Brązert, M.; Ożegowska, K.; Budna-Tukan, J.; Celichowski, P.; Jankowski, M.; Bryja, A.; Nawrocki, M.J.; Popis, M.; Jeseta, M.; et al. Response to abiotic and organic substances stimulation belongs to ontologic groups significantly up-regulated in porcine immature oocytes. Med. J. Cell Biol. 2018, 6, 91–100. [Google Scholar] [CrossRef]
- Singh, A.K.; Dutta, M.; Chattopadhyay, R.; Chakravarty, B.; Chaudhury, K. Intrafollicular interleukin-8, interleukin-12, and adrenomedullin are the promising prognostic markers of oocyte and embryo quality in women with endometriosis. J. Assist. Reprod. Genet. 2016, 33, 1363. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.S.; Joo, B.S.; Na, Y.J.; Yoon, M.S.; Choi, O.H.; Kim, W.W. Relationships Between Concentrations of Tumor Necrosis Factor-α and Nitric Oxide in Follicular Fluid and Oocyte Quality. J. Assist. Reprod. Genet. 2000, 17, 222. [Google Scholar] [CrossRef]
- Wunder, D.M.; Muller, M.D.; Birkhäuser, M.H.; Bersinger, N.A. Increased ENA-78 in the follicular fluid of patients with endometriosis. Acta Obstet. Gynecol. Scand. 2006, 85, 336–342. [Google Scholar] [CrossRef]
- Park, J.E.; Do, H.L.; Lee, J.A.; Sung, G.P.; Kim, N.S.; Park, B.C.; Cho, S. Annexin A3 is a potential angiogenic mediator. Biochem. Biophys. Res. Commun. 2005, 337, 1283–1287. [Google Scholar] [CrossRef]
- Du, R.; Liu, B.; Zhou, L.; Wang, D.; He, X.; Xu, X.; Zhang, L.; Niu, C.; Liu, S. Downregulation of annexin A3 inhibits tumor metastasis and decreases drug resistance in breast cancer. Cell Death Dis. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Zhou, Y.; Liu, Z.; Zhang, P. Interaction between ANXA1 and GATA-3 in Immunosuppression of CD4+ T Cells. Mediators Inflamm. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Toufiq, M.; Roelands, J.; Alfaki, M.; Syed Ahamed Kabeer, B.; Saadaoui, M.; Lakshmanan, A.P.; Bangarusamy, D.K.; Murugesan, S.; Bedognetti, D.; Hendrickx, W.; et al. Annexin A3 in sepsis: Novel perspectives from an exploration of public transcriptome data. Immunology 2020, 161, 291–302. [Google Scholar] [CrossRef]
- Meng, H.; Zhang, Y.; An, S.T.; Chen, Y. Annexin A3 gene silencing promotes myocardial cell repair through activation of the PI3K/Akt signaling pathway in rats with acute myocardial infarction. J. Cell. Physiol. 2019, 234, 10535–10546. [Google Scholar] [CrossRef] [PubMed]
- Talevi, R.; Gualtieri, R. Molecules involved in sperm-oviduct adhesion and release. Theriogenology 2010, 73, 796–801. [Google Scholar] [CrossRef]
- Le Cabec, V.; Maridonneau-Parini, I. Annexin 3 is associated with cytoplasmic granules in neutrophils and monocytes and translocates to the plasma membrane in activated cells. Biochem. J. 1994, 303, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Daugherty, M.D.; Schaller, A.M.; Geballe, A.P.; Malik, H.S. Evolution-guided functional analyses reveal diverse antiviral specificities encoded by ifit1 genes in mammals. eLife 2016, 5, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W. Interferon-stimulated genes: Roles in viral pathogenesis. Curr. Opin. Virol. 2014, 6, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Nakaranurack, C.; Puttilerpong, C.; Suwanpimolkul, G. Advance Publication by J-STAGE. J. Reprod. Dev. 2016. [Google Scholar] [CrossRef]
- Wasityastuti, W.; Yano, Y.; Ratnasari, N.; Triyono, T.; Triwikatmani, C.; Indrarti, F.; Heriyanto, D.S.; Yamani, L.N.; Liang, Y.; Utsumi, T.; et al. Protective effects of HLA-DPA1/DPB1 variants against Hepatitis B virus infection in an Indonesian population. Infect. Genet. Evol. 2016, 41, 177–184. [Google Scholar] [CrossRef]
- Mazzoni, G.; Razza, E.; Pedersen, H.S.; Secher, J.; Kadarmideen, H.N.; Callesen, H.; Stroebech, L.; Freude, K.; Hyttel, P. In vitro production of bovine embryos: Cumulus/granulosa cell gene expression patterns point to early atresia as beneficial for oocyte competence. Anim. Reprod. 2017, 14, 482–489. [Google Scholar] [CrossRef]
- Trisdale, S.K.; Schwab, N.M.; Hou, X.; Davis, J.S.; Townson, D.H. Molecular manipulation of keratin 8/18 intermediate filaments: Modulators of FAS-mediated death signaling in human ovarian granulosa tumor cells. J. Ovarian Res. 2016, 9, 1–11. [Google Scholar] [CrossRef]
- Sarapik, A.; Velthut, A.; Haller-Kikkatalo, K.; Faure, G.C.; Béné, M.C.; De Carvalho Bittencourt, M.; Massin, F.; Uibo, R.; Salumets, A. Follicular proinflammatory cytokines and chemokines as markers of IVF success. Clin. Dev. Immunol. 2012, 2012. [Google Scholar] [CrossRef]
- Büscher, U.; Chen, F.C.K.; Kentenich, H.; Schmiady, H. Cytokines in the follicular fluid of stimulated and non-stimulated human ovaries; Is ovulation a suppressed inflammatory reaction? Hum. Reprod. 1999, 14, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Okuda, K.; Sakumoto, R. Multiple roles of TNF super family members in corpus luteum function. Reprod. Biol. Endocrinol. 2003, 1, 1–10. [Google Scholar] [CrossRef]
- Chen, H.L.; Marcinkiewicz, J.L.; Sancho-Tello, M.; Hunt, J.S.; Terranova, P.F. Tumor necrosis factor-α gene expression in mouse oocytes and follicular cells. Biol. Reprod. 1993, 48, 707–714. [Google Scholar] [CrossRef][Green Version]
- Roby, K.F.; Weed, J.; Lyles, R.; Terranova, P.F. Immunological evidence for a human ovarian tumor necrosis factor-α. J. Clin. Endocrinol. Metab. 1990, 71, 1096–1102. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.J.; Brannstrom, M.; Robertson, S.A.; Norman, R.J. Tumor necrosis factor α in the human ovary: Presence in follicular fluid and effects on cell proliferation and prostaglandin production. Fertil. Steril. 1992, 58, 934–940. [Google Scholar] [CrossRef]
- Sakumoto, R.; Okuda, K. Possible actions of tumor necrosis factor-α in ovarian function. J. Reprod. Dev. 2004, 50, 39–46. [Google Scholar] [CrossRef]
- Samir, M.; Glister, C.; Mattar, D.; Laird, M.; Knight, P.G. Follicular expression of pro-inflammatory cytokines tumour necrosis factor-α (TNFα), interleukin 6 (IL6) and their receptors in cattle: TNFα IL6 and macrophages suppress thecal androgen production in vitro. Reproduction 2017, 154, 35–49. [Google Scholar] [CrossRef]
- Glister, C.; Hatzirodos, N.; Hummitzsch, K.; Knight, P.G.; Rodgers, R.J. The global effect of follicle-stimulating hormone and tumour necrosis factor α on gene expression in cultured bovine ovarian granulosa cells. BMC Genom. 2014, 15, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Manabe, N.; Matsuda-Minehata, F.; Goto, Y.; Maeda, A.; Cheng, Y.; Nakagawa, S.; Inoue, N.; Wongpanit, K.; Jin, H.; Gonda, H.; et al. Role of Cell Death Ligand and Receptor System on Regulation of Follicular Atresia in Pig Ovaries. Reprod. Domest. Anim. 2008, 43, 268–272. [Google Scholar] [CrossRef]
- Silva, J.R.V.; Lima, F.E.O.; Souza, A.L.P.; Silva, A.W.B. Interleukin-1β and TNF-α systems in ovarian follicles and their roles during follicular development, oocyte maturation and ovulation. Zygote 2020, 28, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Naz, R.K.; Zhu, X.; Menge, A.C. Expression of Tumor Necrosis Factor-α and Its Receptors Type I and Type II in Human Oocytes; Wiley-Liss, Inc.: Hoboken, NJ, USA, 1997; Volume 47. [Google Scholar]
- Terranova, P.F. Potential roles of tumor necrosis factor-α in follicular development, ovulation, and the life span of the corpus luteum. Domest. Anim. Endocrinol. 1997, 14, 1–15. [Google Scholar] [CrossRef]
- Kong, Q.Q.; Wang, J.; Xiao, B.; Lin, F.H.; Zhu, J.; Sun, G.Y.; Luo, M.J.; Tan, J.H. Cumulus cell-released tumor necrosis factor (TNF)-a promotes post-ovulatory aging of mouse oocytes. Aging 2018, 10, 1745–1757. [Google Scholar] [CrossRef] [PubMed]
- Alijotas-Reig, J.; Esteve-Valverde, E.; Ferrer-Oliveras, R.; Llurba, E.; Gris, J.M. Tumor Necrosis Factor-Alpha and Pregnancy: Focus on Biologics. An Updated and Comprehensive Review. Clin. Rev. Allergy Immunol. 2017, 53, 40–53. [Google Scholar] [CrossRef]
- Zollner, U.; Bischofs, S.; Lalic, I.; Zollner, K.P. LIF and TNF alpha concentrations in embryo culture media are predictive for embryo implantation in IVF. Asian Pac. J. Reprod. 2012, 1, 277–282. [Google Scholar] [CrossRef]



| Gene | Primer Sequences (5′-3′) | Product Size (bp) |
|---|---|---|
| KRT18 | CACAGTCTGCTGAGGTTGGA | 164 |
| GAGCTGCTCCATCTGTAGGG | ||
| HLA-DRA | CATGACAAAGCGCTCCAACT | 231 |
| TGGAACTTGCGGAAAAGGTG | ||
| KRT8 | TGAGGTCAAGGCACAGTACG | 161 |
| TGATGTTCCGGTTCATCTCA | ||
| HLA-DPA1 | CAAGAGCCAATCCAGATGCC | 230 |
| ATGGAGTTTGTAGGGCAGCT | ||
| MX1 | CGGAATCTTGACGAAGCCTG | 224 |
| CCTTTCCTTCCTCCAGCAGA | ||
| IFIT1 | GCGGTTTCCACATGACAACT | 242 |
| ATTCATGAGG. GGCAGTCACA | ||
| HPRT | TGGCGTCGTGATTAGTGATG | 141 |
| ACATCTCGAGCAAGACGTTC | ||
| ACTB | AAAGACCTGTACGCCAACAC | 132 |
| CTCAGGAGGAGCAATGATCTTG | ||
| GAPDH | TCAGCCGCATCTTCTTTTGC | 90 |
| ACGACCAAATCCGTTGACTC |
| Gene Name | Gene Symbol | Fold Change | Adj. p. val. |
|---|---|---|---|
| Annexin A3 | ANXA3 | 34.27 | <0.05 |
| Interferon-induced protein with tetratricopeptide repeats 1 | IFIT1 | 27.75 | <0.05 |
| Major histocompatibility complex, class II, DP alpha 1 | HLA-DPA1 | 24.10 | <0.05 |
| Myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) | MX1 | 19.60 | <0.05 |
| Keratin 8 | KRT8 | 16.04 | <0.05 |
| Major histocompatibility complex, class II, DR alpha | HLA-DRA | 15.36 | <0.05 |
| Keratin 18 | KRT18 | 14.55 | <0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chermuła, B.; Hutchings, G.; Kranc, W.; Józkowiak, M.; Jopek, K.; Stelmach, B.; Mozdziak, P.; Pawelczyk, L.; Piotrowska-Kempisty, H.; Spaczyński, R.Z.; et al. Expression Profile of New Gene Markers and Signaling Pathways Involved in Immunological Processes in Human Cumulus-Oophorus Cells. Genes 2021, 12, 1369. https://doi.org/10.3390/genes12091369
Chermuła B, Hutchings G, Kranc W, Józkowiak M, Jopek K, Stelmach B, Mozdziak P, Pawelczyk L, Piotrowska-Kempisty H, Spaczyński RZ, et al. Expression Profile of New Gene Markers and Signaling Pathways Involved in Immunological Processes in Human Cumulus-Oophorus Cells. Genes. 2021; 12(9):1369. https://doi.org/10.3390/genes12091369
Chicago/Turabian StyleChermuła, Błażej, Greg Hutchings, Wiesława Kranc, Małgorzata Józkowiak, Karol Jopek, Bogusława Stelmach, Paul Mozdziak, Leszek Pawelczyk, Hanna Piotrowska-Kempisty, Robert Z. Spaczyński, and et al. 2021. "Expression Profile of New Gene Markers and Signaling Pathways Involved in Immunological Processes in Human Cumulus-Oophorus Cells" Genes 12, no. 9: 1369. https://doi.org/10.3390/genes12091369
APA StyleChermuła, B., Hutchings, G., Kranc, W., Józkowiak, M., Jopek, K., Stelmach, B., Mozdziak, P., Pawelczyk, L., Piotrowska-Kempisty, H., Spaczyński, R. Z., & Kempisty, B. (2021). Expression Profile of New Gene Markers and Signaling Pathways Involved in Immunological Processes in Human Cumulus-Oophorus Cells. Genes, 12(9), 1369. https://doi.org/10.3390/genes12091369








